Abstract
The dynamic expression of AMPA-type glutamate receptors (AMPA-R) at synapses is a key determinant of synaptic plasticity, including neuroadaptations to drugs of abuse. Recently, microRNAs (miRNAs) have emerged as important posttranscriptional regulators of synaptic plasticity, but whether they target glutamate receptors to mediate this effect is not known. Here we used microarray screening to identify miRNAs that regulate synaptic plasticity within the nucleus accumbens, a brain region critical to forming drug-seeking habits. One of the miRNAs that showed a robust enrichment at medium spiny neuron synapses was miR-181a. Using bioinformatics tools, we detected a highly conserved miR-181a binding site within the mRNA encoding the GluA2 subunit of AMPA-Rs. Overexpression and knockdown of miR-181a in primary neurons demonstrated that this miRNA is a negative posttranscriptional regulator of GluA2 expression. Additionally, miR-181a overexpression reduced GluA2 surface expression, spine formation, and miniature excitatory postsynaptic current (mEPSC) frequency in hippocampal neurons, suggesting that miR-181a could regulate synaptic function. Moreover, miR-181a expression was induced by dopamine signaling in primary neurons, as well as by cocaine and amphetamines, in a mouse model of chronic drug treatment. Taken together, our results identify miR-181a as a key regulator of mammalian AMPA-type glutamate receptors, with potential implications for the regulation of drug-induced synaptic plasticity.
INTRODUCTION
Addiction to drugs of abuse is considered a chronic neurological disease characterized by compulsive drug abuse regardless of the consequences to the individual's well-being. Most drugs of abuse share common modes of action on the brain's pathways related to motivation and reward (mesolimbic dopamine system) through the release of the neurotransmitter dopamine (13, 32). Altered dopamine signaling can lead to neuroadaptations at these sites that can manifest in an individual's decreased sensitivity and/or increased propensity for the abused drug. At the cellular level, neuroadaptations consist of functional changes in synaptic connectivity or strength, such as changes in the number and size of dendritic spines, analogous to the long-lasting modulations of synaptic plasticity during learning and memory (42, 62). A hallmark of synaptic plasticity is the differential trafficking and cell surface expression of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPA-R) subunits (25, 37). Moreover, several studies have now identified alterations in synaptic AMPA-R levels as a possible mechanism by which drugs of abuse may elicit long-lasting neuroadaptations (65, 66). AMPA-R subunits (GluA1/2, formerly referred to as GluR1/2 or GluRA/B) are locally translated near synaptic sites in response to stimulation (23, 25, 26, 57). MicroRNAs (miRNAs), an abundant class of posttranscriptional gene regulatory molecules, are present in the vicinity of synapses, where they regulate the local expression of synaptic proteins (48). However, it is not known whether miRNAs are involved in regulating local protein synthesis of mammalian AMPA-R subunits in brain areas associated with addiction.
Recently, evidence for a role of miRNAs in the brain's reward circuitry has emerged. miRNAs have been implicated in mediating the effects of cocaine (8, 9, 18, 22), nicotine (19, 20), alcohol (41, 44), and several other classes of drugs of abuse (17, 33, 67). In the case of cocaine addiction in rats, the upregulation of miR-181a and the downregulation of two other miRNAs, let-7d and miR-124a, has been revealed in addiction-relevant regions of the brain (8). Extended access to the drug has been shown to increase the expression of striatal miR-212 (and the closely related miR-132), leading to downstream consequences on signaling pathways that ultimately decrease cocaine's motivational properties and provide protection against drug overconsumption (18, 22). Manipulating the alterations of miRNAs in the nucleus accumbens appears to be sufficient to either attenuate or enhance drug-seeking behavior (9, 18, 22). In general, it is possible that by targeting even a single miRNA, drugs of abuse may alter the expression levels of a number of downstream target genes that modulate addiction-related neuronal mechanisms.
In the present study, we pursued the following three main objectives: (i) to define the population of miRNAs enriched in the synaptodendritic compartment of a region critical to the mesolimbic dopamine pathway (nucleus accumbens), (ii) to identify physiological miRNA target mRNAs that are relevant to synaptic plasticity, and (iii) to characterize the functional interaction between a synaptic miRNA and its target mRNA within the context of addiction to drugs of abuse. Our results indicate that the modulation of miR-181a, an miRNA strongly enriched in the synaptodendritic compartment of the nucleus accumbens, can regulate the glutamate receptor 2 subunit (GluA2) of AMPA-Rs at the posttranscriptional level. Since AMPA-R dynamics are vitally involved in synaptic plasticity, this regulation has the potential to impinge on the brain reward circuitry and therefore may play a putative role in addiction-related neuroplastic changes.
MATERIALS AND METHODS
Primary neuronal cell culture.
Cultures of dissociated primary cortical and hippocampal neurons from embryonic day 18 (E18) Sprague-Dawley rats (Charles River Laboratories, Sulzfeld, Germany) were prepared and cultured as described previously by Schratt et al. (50).
Preparation of synaptoneurosomes.
Synaptoneurosomes were prepared from the microdissected nucleus accumbens regions of strain P15 Sprague-Dawley rat pups as described previously by Rao and Steward (45), from three independent litters. Specifically, from a single rat litter consisting of 15 pups, 10 were used for the preparation of synaptoneurosomes whereas the remaining nucleus accumbens regions from the other pups of the litter were saved as whole tissue. Briefly, nucleus accumbens regions were microdissected in 50 ml of homogenization buffer (0.32 M sucrose, 0.1 mM EDTA, 0.25 mM dithiothreitol [DTT], 2 mM HEPES, pH 7.4) and disrupted with a Teflon-coated Dounce-Potter homogenizer (Wheaton) by eight up-and-down strokes. Nuclei and cell debris were pelleted by a 2-min centrifugation at 2,000 × g. The supernatant was centrifuged for an additional 10 min at 14,000 × g to pellet a crude synaptoneurosome-containing fraction. The fraction was subsequently layered over a 5 to 13% discontinuous Ficoll gradient that had been equilibrated at 4°C for 1 h. Synaptoneurosomes were collected from the gradient interface after centrifugation at 45,000 × g for 45 min at 4°C. Samples were obtained from every step of the procedure for Western blot analysis to ensure the quality and purity of the preparations.
Total RNA extraction and qRT-PCR.
Total RNA (including small RNA fraction) was purified using either Qiazol (Qiagen) or the miRVana kit (Ambion) and treated with TURBO DNase (Ambion) to remove DNA contamination. Quantitative real-time PCR (qRT-PCR) was performed with a 7300 real time PCR system (Applied Biosystems) using TaqMan MicroRNA assays (Applied Biosystems) and iTaq SybrGreen Supermix with ROX (Bio-Rad) for the detection of miRNAs and mRNAs, respectively. Primers used for the detection of miRNA and mRNA targets are listed in Table S1 in the supplemental material. RT-PCR data were processed and analyzed using the comparative ΔCT method (34). The ΔCT values were then expressed relative to the control sample (defined for each experiment and set to 1) and the mean ± standard deviation (SD) calculated from the separate experiments performed (n ≥ 3).
Microarray analysis.
Total RNA (including small RNA fraction) was extracted from both the nucleus accumbens whole tissue and from synaptoneurosomes prepared from these tissues from P15 rat pups. The extracted RNA was either Cy3 (RNA from synaptoneurosomes) or Cy5 (RNA from whole tissue) labeled and hybridized to miRNA microarrays. Assays were performed using the service provider LC Sciences, LLC (Houston, TX). The microarrays (μParaflo microfluidic chip technology) contained multiple redundant probes for each Rattus norvegicus miRNA listed in Sanger miRBase release 13.0 (http://www.sanger.ac.uk/Software/Rfam/mirna/). Each detection probe consisted of a chemically modified nucleotide coding segment complementary to target miRNA or another RNA control sequence. The detection probes were made by in situ synthesis using photo-generated reagent chemistry. The hybridization melting temperatures were balanced, and hybridization images were collected using a laser scanner and subsequently digitized. Data were analyzed by first subtracting the background and then normalizing signals using a lowess filter (locally weighted regression) (5). Because we conducted two-color experiments, the ratio of the two sets of detected signals (log2 transformed) and P values of the t test were calculated for all three microarrays; differentially detected signals were those with P < 0.01. Samples with dye bias as indicated by the manufacturer were omitted from the analysis. To identify miRNAs that were the most significantly different in expression between the synaptoneurosomes and the whole tissue, a one-class significance analysis of microarrays (SAM analysis) with a false discovery rate (FDR) of <10% was used (64). To further increase the confidence level of our results, only those miRNAs that were consistently identified in all three arrays were used for SAM analysis.
Northern blot analysis.
Northern blots to detect neuronal mature miRNAs and U6 snRNA were performed as described previously (49) with radiolabeled DNA oligonucleotides serving as detection probes. The same RNA that was extracted by Qiazol (Qiagen) for miRNA microarray analysis was used again for Northern blot analysis. The probe sequences are provided in Table S1 in the supplemental material. For molecular markers, the Decade Marker system (Ambion) was utilized.
Western blot analysis.
Proteins were separated by SDS-PAGE and blotted onto a polyvinylidene difluoride (PVDF) membrane. Nonspecific bindings were blocked with Tris-buffered saline plus 5% milk powder and 0.2% Tween 20. The following primary antibodies were used: mouse anti-PSD95 (1:1,000; MA1-046; Dianova), mouse anti-EEA1 (1:10,000; 610456; Becton Dickinson), mouse anti-β-actin (A5441; Sigma), and rabbit anti-GluA2 (AB1768; Millipore). Primary antibodies were recognized by either a horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibody (1:20,000; 401315; Calbiochem) or an HRP-conjugated rabbit anti-mouse antibody (1:20,000; 402335; Calbiochem). Secondary antibodies were detected by enhanced chemiluminescence with the ECL Plus Western blotting detection system (GE Healthcare). Sizes of resolved protein bands were determined using the precision protein dual color standard (Bio-Rad).
Transfections.
Neuronal transfections were performed with Lipofectamine 2000 (Invitrogen). For each well of a 24-well plate, a total of 1 μg of DNA/RNA was mixed with a 1:50 dilution of Lipofectamine 2000 in neurobasal medium, incubated at room temperature (15 to 25°C) for 20 min, and then further diluted 1:5 in neurobasal medium. Neurons were incubated with the transfection mix for 2 h (4 h for small RNA transfections). Nucleofections were performed with the Rat Neuron Nucleofector kit (Lonza) and program O-003. Primary cortical neurons of rat embryos (E18) were nucleofected with 2 to 3 μg of total DNA per condition and plated on six-well dishes in DMEM-Glutamax containing 10% fetal bovine serum (FBS) (Invitrogen). After 4 h, medium was replaced with standard neuronal culture medium. For dual-luciferase assays, transfections were performed on cells at 5 days in vitro (5DIV cells) (for cortical cells) and on 13DIV cells (for hippocampal cells). Cell lysates were harvested at day 2 posttransfection for cortical cells and day 5 posttransfection for hippocampal cells. In total, 20 μl of cell lysates was then used for the dual-luciferase assay following the manufacturer's protocol for the dual-luciferase reporter assay system (Promega).
Immunostaining.
For GluA2-surface staining, 18DIV neurons were incubated with 0.5 μg of mouse anti-GluA2 antibody (clone 6C4; Millipore) in 240 μl of growth medium for 1 h at 37°C (live staining). After this time, the coverslips were washed four times with ice-cold phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde-4% sucrose in PBS for 10 min at room temperature. Coverslips were subsequently washed five times with PBS for 5 min each time and washed once in 1 × GDB (30 mM phosphate buffer at pH 7.4, 0.5 M NaCl, 0.5% Triton X-100, 0.2% gelatin). Incubation with the secondary antibody was performed at room temperature using goat anti-mouse Alexa Red 546 diluted in 1 × GDB (1:1,000; 400 μl/well). Coverslips were then mounted after three rapid and three slow washes in PBS. For staining of permeabilized neurons, the cells were fixed for 15 min, incubated for 30 min with 1 × GDB, and subjected to immunostaining for GluA2 using primary and secondary antibodies as described above.
DNA constructs.
The GluA2 3′ untranslated region (UTR) (full length) was amplified from a mouse brain cDNA library (C57BL/6) and subsequently cloned into the XbaI site of pGL3promoter vector (luciferase vector) (Promega). A version with a mutated miR-181a-binding site within the GluA2 3′ UTR (mutant GluA2-3′ UTR) was generated with the QuikChange site-directed mutagenesis kit (Stratagene) using overlapping PCR. Approximately 25 ng of this and Renilla luciferase plasmid was used per experiment. See Table S1 in the supplemental material for primer sequences.
Image analysis.
For image analysis, hippocampal neurons were transfected at 13DIV with the indicated miRNAs and miRNA inhibitors in combination with an enhanced green fluorescent protein (eGFP) plasmid and processed for confocal microscopy at 18DIV. For spine analysis, high-resolution z-stack images of GFP-positive neurons were taken with a confocal laser scanning microscope (Zeiss). Random neurons displaying pyramidal morphology were chosen from data sets that had been blinded to the experimental conditions. Spine volumes were subsequently analyzed with the ImageJ software (50). At least nine individual neurons per experimental condition were measured in three or more independent experiments. For each of the experimental conditions, 100 to 150 spines per neuron were analyzed. The size of GluA2 surface clusters was determined with the Analyze Particle function of ImageJ, using threshold images taken of at least 12 neurons per condition. Particles smaller than 0.1 μm2 were excluded from the analysis.
Chronic drug treatment in adult mice.
Chronic drug treatments were performed as described by Ziviani et al. (68). In brief, male C57BL6 adult mice (Charles River) were treated with either saline, cocaine hydrochloride (10 mg/kg), or d-amphetamine-d3 sulfate salt (5 mg/kg) via a single intraperitoneal injection once a day for 5 days. Animals were then sacrificed via rapid decapitation and brain regions dissected on ice for immediate RNA extraction.
miRNA target prediction.
To determine the gene targets of the differentially expressed miRNAs, we used three of the leading miRNA target prediction algorithms: miRanda (http://microrna.sanger.ac.uk/sequences/) (24), PicTar (http://pictar.mdc-berlin.de/) (29), and TargetScan (http://www.targetscan.org/) (16). To determine genes that were similarly identified by two or more of these algorithms, the online program Matchminer (http://discover.nci.nih.gov/matchminer/index.jsp) was used (7).
Functional analysis of miRNA target genes.
Functional analysis of miRNA target genes was performed using Ingenuity Pathways Analysis (IPA-Ingenuity Systems). This software analysis lists genes in the context of known biological response and regulatory networks as well as other higher-order response pathways. miRNA targets that were associated with biological functions in the Ingenuity Pathways Knowledge Base were used in the analysis. For all analyses, Fisher's exact test was used to calculate a P value determining the probability that each biological function assigned to that data set was due to chance alone.
Patch-clamp recordings and data analysis.
Spontaneous miniature excitatory postsynaptic currents (mEPSCs) were recorded in the whole-cell voltage-clamp mode using an EPC-10 patch-clamp amplifier and PULSE software (HEKA Elektronik, Lambrecht, Germany) from cultured neurons at 17 to 19DIV visualized with a charge-coupled-device (CCD) camera (VX55, TILL Photonics GmbH, Gräfelfing, Germany) mounted on an upright microscope (BX51WI, Olympus, Hamburg, Germany). Coverslips with transfected cells were constantly superfused at room temperature with a bath solution containing 156 mM NaCl, 2 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 16.5 mM glucose, and 10 mM HEPES (pH 7.3 with NaOH) to which gabazine (5 μM) and tetrodotoxin (0.5 μM) were added during recording. The pipette solution contained 110 mM CsMeSO3, 25 mM CsCl, 30 mM HEPES, 2 mM MgCl2, 0.362 mM CaCl2, 1 mM EGTA, 4 mM MgATP, and 0.1 mM Na2GTP (pH 7.2 with CsOH). Patch pipettes were pulled from borosilicate glass (Science Products, Hofheim, Germany) and had resistances of 3 to 5.5 MΩ when filled with the pipette solution. Neurons were held at a potential of −70 mV, and mEPSCs were analyzed from 100- to 300-s current recordings made after 10 min of equilibration in the whole-cell configuration. Experiments were performed blindly with respect to the transfection (miR-181a or control duplex RNA [control]). Data were acquired at a sampling rate of 20 kHz and filtered at 3 kHz. Series resistance was controlled every 5 min, and only experiments with uncompensated series resistances of <25 MΩ were accepted. Mean event amplitude and frequency were determined off-line with the Mini Analysis program (Synaptosoft Inc.) using an amplitude threshold of −5 pA. The statistical significance of results was tested by Student's two-tailed t test using GraphPad Prism 4 (GraphPad Software, San Diego, CA) and Microsoft Excel. P values of <0.05 were considered statistically significant.
Statistics.
Experiments are reported as mean ± standard deviation (SD) (or standard error of the mean [SEM] for electrophysiology) and based on three or more independent replications. Significance was determined using Student's t test.
RESULTS
High-throughput screening for the identification of miRNAs enriched at nucleus accumbens synapses.
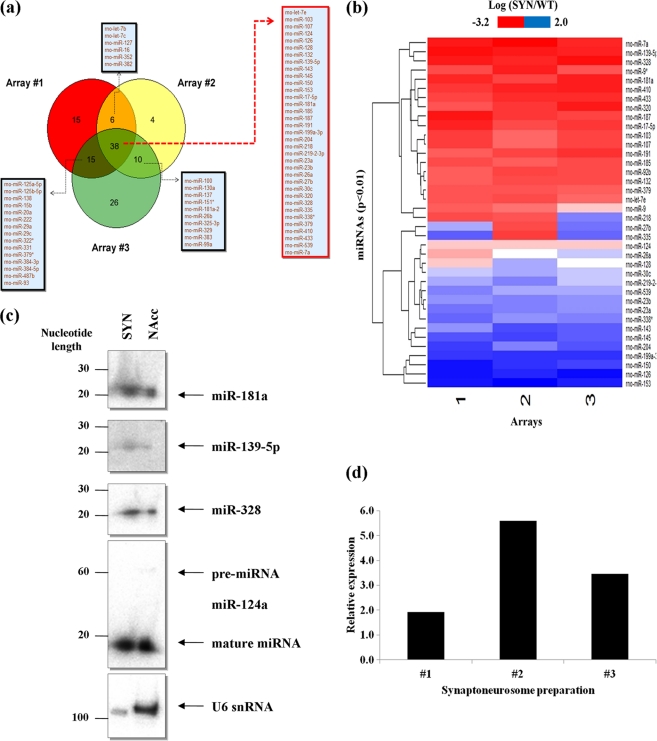
In order to identify miRNAs with functional relevance in drug-induced synaptic plasticity, we chose to profile miRNAs from neurons of the nucleus accumbens, specifically with the emphasis on identifying miRNAs that are enriched within the synaptodendritic compartment. The nucleus accumbens is the primary site of action of most drugs of abuse, and it plays a key role in reward, motivation, and addiction, in response to the release of the neurotransmitter dopamine. To detect and identify miRNAs, the nucleus accumbens from both hemispheres of P15 rat brains were carefully microdissected and synaptoneurosomes were immediately prepared. Synaptoneurosomes are biochemical preparations that are highly enriched in synaptic membranes and also preserve most components of local protein synthesis machinery, including polyribosomes, mRNAs, and regulatory RNAs (such as miRNAs) (14, 45, 49, 50, 53). We confirmed the purity of the synaptoneurosome preparations by the enrichment and depletion of BC1 RNA and U6 snRNA, respectively. Furthermore, the nuclear and soma-resident protein EEA-1 was absent whereas the synaptic proteins PSD-95 and GluA2 were enriched (data not shown). We therefore went on to compare total RNA that was extracted from nucleus accumbens synaptoneurosomes with total RNA prepared from whole tissue by competitive hybridization to microarrays that contained probes for all Rattus norvegicus mature miRNAs listed in the Sanger miRBase release 13.0. In three independent microarray hybridizations, we identified 69 miRNAs that displayed differential expression in at least two hybridizations and 38 miRNAs that were differentially expressed in all three hybridizations (P < 0.01) (Fig. 1a). Of these 38 miRNAs, 20 miRNAs were found to be enriched in the synaptoneurosomes whereas 18 miRNAs were depleted (Fig. 1b) (see Table S2 in the supplemental material). Further stringent filtration of the data to include only those miRNAs with either ≥2-fold enrichment or ≥2-fold depletion from the synaptoneurosomes identified nine enriched miRNAs and seven depleted miRNAs (Table 1). Interestingly, there was almost no overlap between the miRNAs enriched in nucleus accumbens synaptoneurosomes and those previously identified in a screen of rat forebrain synaptoneurosomes (53), suggesting tissue-specific populations of synaptic miRNAs. Of the seven depleted miRNAs, however, four of them (miR-145, miR-126, miR-150, and miR-143) have also been previously documented to be depleted from forebrain synaptoneurosomes, implying a general absence of these miRNAs from the synaptodendritic compartment (see Table S2 in the supplemental material) (53).
Fig 1.
A subset of miRNAs are enriched at nucleus accumbens synapses. (a) miRNAs identified by microarray screening to be consistent between two or more arrays (n = 3; P < 0.01). Total RNA, including small RNA species, was extracted from synaptoneurosomes in three independent preparations and compared to total RNA prepared from whole tissue of the nucleus accumbens via competitive hybridization to microarrays (n = 3). miRNAs that are both enriched and depleted from synaptoneurosomes prepared from the nucleus accumbens region of P15 rats are shown. (b) Differential abundance of miRNAs in synaptoneurosomes. In three separate microarray hybridizations, we identified ∼38 miRNAs that were consistently detected (P < 0.01). Refer to Table 1 for those miRNAs with ≥2-fold enrichment or depletion (P < 0.01) in the synaptoneurosomes. (c) Northern blot validation of miR-181a abundance in synaptoneurosomes (SYN) compared to that in whole tissue (NAcc). Also shown are the expression of miR-124a and U6 snRNA in the SYN and NAcc. Validation of two other miRNAs, miR-139-5p and miR-328, also found to be abundant in the synaptoneurosomes compared to whole tissue. Note the absence of pre-miR-124a from synaptoneurosomes. (d) qRT-PCR validation of miR-181a enrichment in synaptoneurosomes relative to whole tissue in three independent preparations. Data were normalized to the expression of miR-124a, which was equally distributed throughout the cell (Table 1 and Fig. 1c).
Table 1.
miRNAs with ≥2-fold enrichment or depletion from synaptoneurosomes prepared from P15 rat nucleus accumbens region
| miRNA in nucleus accumbens synaptoneurosomesa | Avg fold enrichment or depletion ± SD (n = 3) |
|---|---|
| Enriched miRNAs | |
| rno-miR-139-5p | 3.12 ± 0.59 |
| rno-miR-328 | 3.11 ± 0.76 |
| rno-miR-410 | 2.41 ± 0.47 |
| rno-miR-433 | 2.00 ± 0.32 |
| rno-miR-7a | 3.47 ± 1.36 |
| rno-miR-181a | 2.29 ± 0.55 |
| rno-miR-17-5p | 2.01 ± 0.51 |
| rno-miR-187 | 2.24 ± 0.89 |
| rno-miR-320 | 2.09 ± 0.75 |
| Depleted miRNAs | |
| rno-miR-199a-3p | 0.29 ± 0.02 |
| rno-miR-145 | 0.40 ± 0.04 |
| rno-miR-126 | 0.18 ± 0.05 |
| rno-miR-153 | 0.17 ± 0.05 |
| rno-miR-150 | 0.27 ± 0.10 |
| rno-miR-204 | 0.44 ± 0.13 |
| rno-miR-143 | 0.48 ± 0.13 |
Enrichment or depletion of ≥2-fold; P < 0.01.
Validation of miR-181a enrichment in the synaptodendritic compartment of the nucleus accumbens.
We next proceeded to validate the miRNAs identified in our microarray screen. In accordance with our microarray data, we found higher levels of three selected candidates (miR-139-5p, miR-328, and miR-181a) in total RNA prepared from the synaptoneurosomes compared to that in whole tissue by Northern blotting (Fig. 1c). Moreover, as expected, miR-124 was equally abundant in both the synaptoneurosome preparations and whole tissue, whereas U6 snRNA was strongly depleted from the synaptoneurosomes (Fig. 1c). Of the nine enriched miRNAs, miR-181a showed the highest relative abundance in the microarray screen (see Table S3 in the supplemental material). Additionally, miR-181a was previously found to be associated with Ago2 in the striatum (47), to be expressed in a somatodendritic gradient in hippocampal neurons (31), and to be induced by cocaine (8, 47) (see Table S2 in the supplemental material). Finally, virus-mediated overexpression of this miRNA in the rat nucleus accumbens was recently shown to enhance conditioned place preference (CPP) for cocaine (9). Due to this growing body of evidence in regard to a central role for miR-181a in the addiction process, we focused on this miRNA for further functional analysis. Quantitative RT-PCR (qRT-PCR) analysis of the three independent synaptoneurosome preparations showed a robust abundance of this miRNA, thus further confirming the microarray screen and Northern blot analysis (Fig. 1d). In summation, our results show that miR-181a is strongly enriched in the synaptodendritic compartment of the nucleus accumbens, suggesting that it might be involved in the local regulation of synaptic proteins at this site.
Computational prediction of a putative miR-181a target gene.
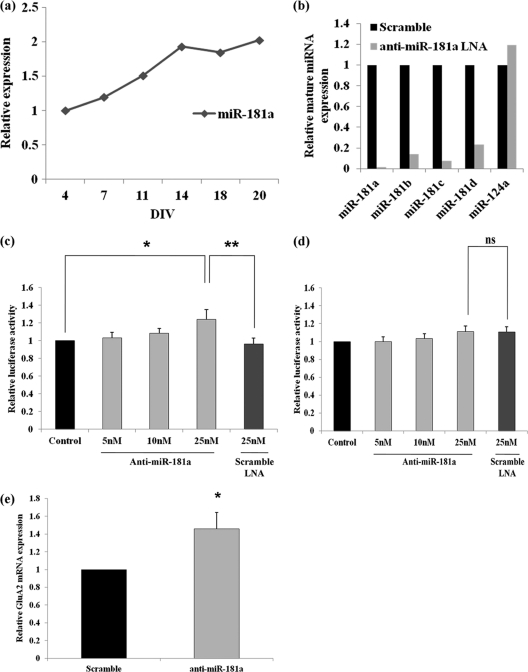
To get insight into the biological function of miR-181a at the synapse, we went on to identify direct target genes of this miRNA. For this purpose, we used a consensus-based approach whereby we compared the target genes predicted by three of the leading target prediction algorithms (TargetScan, PicTar, MiRBase). Based on this integrative strategy, we identified 55 putative target genes of miR-181a that were common to all three algorithms (Fig. 2a) (see Table S4 in the supplemental material). Gene ontology (GO) assignment of these 55 target genes showed a strong enrichment for genes involved in neurological diseases, nervous system development and function, and cell-to-cell signaling and interaction (Fig. 2b) (see Table S5 in the supplemental material). A target gene that was common to all three programs and contained a highly conserved binding site for miR-181a, among several different mammalian species, was the gene encoding GluA2 (Fig. 2c).
Fig 2.
GluA2 is a target of the synaptic miRNA miR-181a. (a) miR-181a target genes similarly predicted by three leading miRNA/target gene prediction algorithms. See Table S3 in the supplemental material for a list and detailed description of these genes. (b) Gene ontology (GO) assignment of the target genes predicted for miR-181a (P = 0.01). See Table S4 in the supplemental material for a list and detailed description of the GO classification. (c) The binding site for the seed sequence (nucleotides 2 to 8 from the 5′ end of the miRNA) of miR-181a in the 3′ UTR of GluA2 is highly conserved among several species. Binding site prediction information and degree of conservation in mammalian species obtained from TargetScan release 5.1. (d) Relative luciferase gene activity of a reporter vector harboring the 3′ UTR of GluA2 mRNA downstream of a luciferase gene in the presence of increasing concentrations of miR-181a or miR-134 duplex (5 to 20 nM). Cotransfection of miR-181a or miR-134 duplex, firefly luciferase reporter, and Renilla luciferase vector was performed on 5DIV cortical cells, and luciferase activity was measured at 7DIV. Bar plots show mean ± SD (n = 4). Significance was determined using Student's t test (*, P < 0.001). (e) Relative luciferase gene activity of a reporter vector harboring the mutant 3′ UTR of GluA2 mRNA downstream of a luciferase gene in the presence of increasing concentrations of miR-181a duplex (5 to 20 nM). Bar plots show mean ± SD (n = 3). (f) miR-181a expression reduces endogenous GluA2 protein levels in neurons. Primary cortical neurons (5DIV) were transfected with 50 nM the indicated duplex RNA and processed for Western blotting with anti-GluA2 and anti-ß-actin antibodies 2 days later. Results from one representative blot are shown. Quantification of the band intensities (normalized to the basal condition) from three independent blots ± SD is shown at the bottom.
The mRNA transcript encoding the GluA2 subunit of AMPA-Rs is a target gene of miR-181a.
The GluA2 protein is an integral subunit of AMPA-R complexes that are highly abundant in the synaptodendritic compartment (data not shown). AMPA-Rs are postsynaptic ionotropic glutamate-gated ion channels that mediate the majority of the fast excitatory neurotransmission in the mammalian central nervous system in the process governing learning, memory and addiction-related behaviors. Interestingly, the AMPA-R subunits GluA1 and GluA2 appear to be locally translated near postsynaptic densities (23, 25, 26, 51, 57, 59, 60), suggesting that they could be subject to regulation by synaptic miRNAs (12, 35, 36, 48, 50, 53). A role for miRNAs in regulating postsynaptic glutamate receptors that show sequence similarities to mammalian AMPA-R genes (56) has recently been identified at the Drosophila neuromuscular junction (27), but whether such regulation occurs in vertebrates is not known. We therefore decided to study a potential functional miR-181a/GluA2 interaction in greater detail.
We used primary cortical and hippocampal cultures for our studies, since GABAergic medium spiny neurons of the nucleus accumbens lose dendritic spines upon prolonged in vitro culture due to the absence of excitatory input (58), thus making them an unsuitable system to study synaptic development in vitro. On the other hand, cortical and hippocampal neurons are widely used to study the mechanisms of plasticity in general and have also proven extremely useful in the context of drug-induced plasticity (15). Most importantly, stimulation of dopamine signaling in hippocampal neurons leads to the activation of D1 family receptors and localized protein synthesis of GluA1 (54). However, the effects of dopamine on GluA2 expression have not been studied.
To verify that the mRNA encoding the GluA2 subunit of AMPA-Rs is a genuine target gene of miR-181a, the full-length 3′ UTR of the transcript was cloned downstream of a luciferase gene in a reporter vector (Fig. 2c). The vector, along with miR-181a duplex, was then cotransfected into primary cortical neurons, and luciferase activity was monitored 48 h later. With increasing concentrations of miR-181a duplex (5→20 nM) there was a robust decrease in luciferase activity (Fig. 2d). Upon cotransfection with miR-134, an miRNA we had previously identified as being enriched in the synaptodendritic compartment of mature neurons (51) but which lacks a well-characterized binding site in the 3′ UTR of GluA2, there was no effect on luciferase activity even with increasing concentrations (5→20 nM) (Fig. 2d). Overall, these results suggest that the repression of luciferase activity in the cortical neurons was a specific effect of miR-181a overexpression.
We next tested whether the predicted miR-181a binding site located within the GluA2-3′ UTR (wild type) is responsible for miR-181a-mediated inhibition of reporter gene expression. Accordingly, point mutations were introduced into the wild-type GluA2-3′ UTR to abolish the predicted seed pairing of miR-181a to GluA2 mRNA (mutant GluA2-3′ UTR). Upon cotransfection of the resulting mutant, GluA2-3′ UTR, with increasing concentrations of miR-181a duplex (5→20 nM), there was no significant change in luciferase activity (Fig. 2e). These results suggest that the repressive effect of miR-181a on wild-type GluA2-3′ UTR is mediated via a single, highly conserved binding site (Fig. 2c). Western blotting using a GluA2-specific antibody confirmed that miR-181a overexpression in primary cortical neurons led to a slight but significant reduction (12% ± 5%) in the expression of total endogenous GluA2 protein levels in primary neurons (Fig. 2f).
The expression of endogenous miR-181a increases during the maturation of hippocampal neurons in cell culture (Fig. 3a). To test whether endogenous miR-181a regulates the translation of GluA2 mRNA in primary neurons, we decided to use antisense molecules that would effectively sequester endogenous miR-181a (so-called power locked nucleic acids [LNAs]; Exiqon). In an initial assessment by qRT-PCR, the anti-miR-181a molecules showed both a strong specificity and an efficacy for sequestering endogenous miR-181a (and other members of the miR-181 family that share a highly conserved seed sequence) without any effect on nonhomologous miRNAs, such as miR-124a (Fig. 3b). Upon cotransfection of the wild-type GluA2-3′ UTR luciferase reporter constructs into cortical neurons alongside anti-miR-181a molecules, there was a significant upregulation in luciferase activity. This activity increased with increasing concentrations of the anti-miR-181a LNA (5→25 nM) (Fig. 3c). Upon cotransfection with an LNA molecule of equal length but a scrambled sequence (scramble power LNA-A; 25 nM), no effect was observed on the activity of the luciferase construct (Fig. 3c). Moreover, the inhibitory function of endogenous miR-181a on the activity of the GluA2 reporter construct was dependent on the presence of an miR-181a binding site, since the mutant GluA2-3′ UTR construct was resistant to increasing amounts of anti-miR181a (5→25 nM) (Fig. 3d).
Fig 3.
Endogenous miR-181a regulates a GluA2-3′ UTR reporter gene in neurons. (a) The expression of miR-181a in dissociated neuronal cell culture as measured by qRT-PCR. Data were normalized to the expression of U6 snRNA and are shown relative to expression level at 4DIV. (b) Effect of antisense-miR-181a LNA on endogenous miR-181a,-181b,-181c, and -181d levels. Data were normalized using U6 snRNA, and data are shown relative to equal amounts of scramble LNA that was nucleofected. The specificity of the miR-181a LNA on the expression of miR-124a is shown for comparison. (c) Luciferase gene activity in a reporter vector harboring the wild-type 3′ UTR of GluA2 mRNA downstream of a luciferase gene in the presence of increasing concentrations of miR-181a LNA (5 to 25 nM). Cotransfection of anti-miR-181a LNA or scramble LNA (25 nM) with firefly luciferase reporter and Renilla luciferase vector was performed on 5DIV cortical cells, and luciferase expression was measured at 7DIV. Data are shown relative to basal conditions for each type of plasmid transfected. Bar plots show mean ± SD (n = 4). Significance was determined using Student's t test (*, P < 0.03; **, P < 0.005). (d) Luciferase gene activity in a reporter vector harboring the mutant 3′ UTR of GluA2 mRNA downstream of a luciferase gene in the presence of increasing concentrations of miR-181a LNA (5 to 25 nM). (e) Change in endogenous GluA2 mRNA levels upon sequestration of endogenous miR-181a through the use of antisense molecules that target miR-181a. Change in mRNA levels was measured by qRT-PCR using GAPDH as the endogenous control, and data are shown relative to equal amounts of scrambled LNA that was nucleofected. Cortical cells were nucleofected with LNA at the time of plating, and mRNA levels were measured at 7DIV. Bar plots show mean ± SD (n = 3); *, P < 0.03.
Next, we used nucleofection of anti-miR-181a (100 nM) into primary cortical neurons in order to measure changes in endogenous GluA2 mRNA and protein levels upon miR-181a inhibition. By nucleofection, it is possible to achieve ∼80% transfection efficiency. In our experiments, we observed a modest but significant increase in GluA2 mRNA levels compared to those under control conditions (Fig. 3e). Additionally, equal concentrations of anti-miR-181a LNA and control LNA that were “spiked” in just prior to RNA extraction from the cortical cells had no effect on the expression level of GluA2 mRNA, eliminating the possibility that the observed increase was an artifact of altered qRT-PCR efficiency due to the presence of anti-miR-181a (data not shown). This suggests that at least part of the miR-181a-dependent repression of GluA2 expression (Fig. 3c) is due to GluA2 mRNA destabilization. We next measured the level of the GluA2 protein in response to the nucleofected anti-miR-181a (100 nM) by Western blotting. In this set of experiments, however, we could not detect significant differences in GluA2 protein levels between anti-miR-181a and control conditions (data not shown).
Taken together, these results identify GluA2 mRNA as a genuine target gene of miR-181a in primary rat neurons. Moreover, they further raise the possibility that miR-181a-mediated modulation of GluA2 expression could be involved in synaptic function.
miR-181a reduces AMPA-R surface clusters, spine volume, and mEPSC frequency in cultured hippocampal neurons.
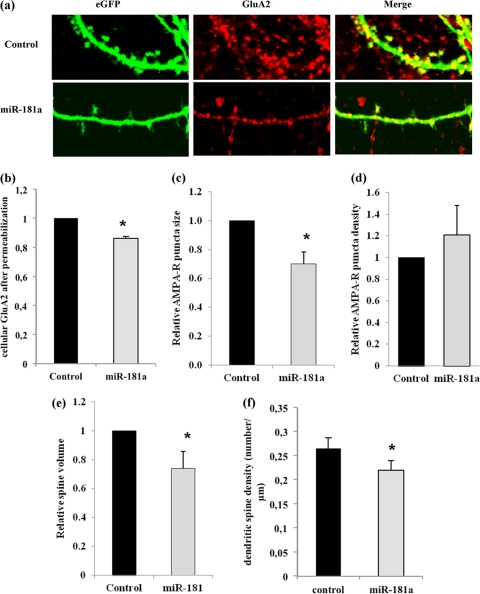
To address whether miR-181a-directed regulation of GluA2 has any consequence on AMPA-R function, we decided to monitor AMPA-R surface expression at synapses in primary hippocampal neurons. It was shown that the number of surface AMPA-R clusters correlates with the number of functional synapses, and the size of individual AMPA-R surface clusters is a good measure for postsynaptic strength (38). For this set of experiments, 13DIV hippocampal neurons were cotransfected with a vector expressing a green fluorescent protein (eGFP) together with either miR-181a duplex or a control duplex RNA of scrambled sequence. The eGFP allowed us to identify those AMPA-R clusters that were present on the dendrites of transfected neurons (Fig. 4a). Using immunocytochemistry (ICC) with an anti-GluA2-antibody on permeabilized hippocampal neurons at 18DIV, we could confirm miR-181a-mediated reduction in total GluA2 levels (Fig. 4b). The number and size of GluA2-containing surface AMPA-R clusters was subsequently measured by ICC on nonpermeabilized neurons (53). Using confocal microscopy, we found a decrease in the average size, but not in the density, of GluA2-containing AMPA-R surface clusters on dendrites of neurons transfected with miR-181a duplex compared to neurons transfected with control duplex RNA (Fig. 4a, c, and d). In accordance with the results obtained from Western blotting (data not shown), we were not able to detect a significant increase in GluA2-containing AMPA-R surface cluster size upon inhibition of miR-181a (data not shown). Additionally, we noticed that miR-181a overexpression significantly reduced dendritic spine volume and density in 18DIV hippocampal neurons in comparison to cells transfected with the control duplex (Fig. 4e and f), suggesting that this miRNA can also specifically interfere with dendritic spine morphogenesis.
Fig 4.
miR-181a reduces GluA2 surface expression and dendritic spine volume in hippocampal neurons. Primary rat hippocampal neurons were transfected with eGFP plasmid and either miR-181a duplex (25 nM) or scramble pre-miR (control, 25 nM) at 11DIV and processed for immunostaining at 18DIV. (a) Representative images of dendrites from transfected neurons stained with anti-GluA2 under nonpermeablizing conditions. (b) Quantification of anti-GluA2 immunostaining performed under permeabilizing conditions. Data are relative to control transfected cells. Bar graphs represent the mean ± SD (n = 4); *, P < 0.05 (Student's t test). (c) Quantification of anti-GluA2 immunostaining performed under nonpermeabilized conditions. Data are relative to control-transfected cells. Bar graphs represent the mean ± SD (n = 4). *, P < 0.01 (Student's t test). (d) Quantification of the average density (total number/cell area [μm2]) of surface GluA2 clusters. Data are relative to control transfected cells. Bar graphs represent the mean ± SD (n = 4). (e) Quantification of the average spine volume (average mean eGFP intensity of individual spines/total intensity of whole cell) (n = 4; *, P < 0.01). (f) Quantification of the average spine density (total number of spines/dendritic length [μm]). Data are relative to control-transfected cells. Bar graphs represent the mean ± SD (n = 4; *, P < 0.05).
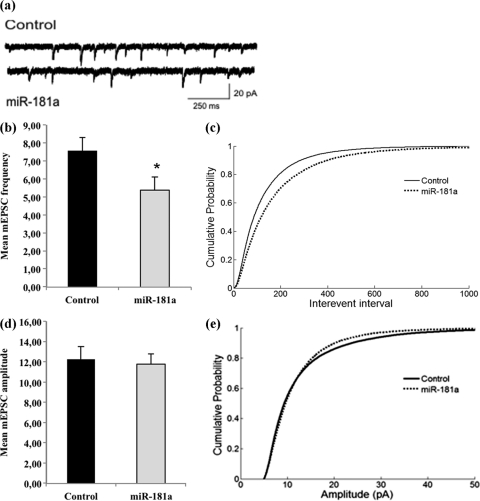
Finally, we recorded mEPSCs from miR-181a overexpression neurons to detect any changes in basal synaptic transmission mediated primarily by AMPA-Rs (Fig. 5). In accordance with decreases in GluA2 surface expression and spine number and volume (Fig. 4), we observed a significant reduction in mEPSC frequency in miR-181a-overexpressing neurons compared to control transfected neurons (Fig. 5a to c). In contrast, miR-181a overexpression led to only a subtle reduction in the average mEPSC amplitude (Fig. 5d and e), which according to the cumulative probability plot appeared to be restricted mostly to events of larger amplitude (Fig. 5e). The potential mechanisms that could underlie these differential effects on frequency versus amplitude are highlighted in the discussion. Together, our results indicate that miR-181a might play a negative regulatory role in the expression of surface GluA2-containing AMPA-R, spine morphogenesis, and basal synaptic transmission, likely due to the inhibition of GluA2 mRNA translation and/or the promotion of GluA2 mRNA degradation at synapses.
Fig 5.
miR-181a reduces the frequency of mEPSCs in hippocampal neurons. Neurons were transfected as described for Fig. 4 and subjected to patch-clamp recordings between 17 and 19DIV. (a) Representative current traces from neurons transfected with either miR-181a or a control RNA. (b) Average mEPSC frequencies of neurons transfected either with miR-181a or a control RNA. n = 13 neurons per condition, originating from three independent transfections. P < 0.05. (c) Cumulative probability of interevent intervals from neurons analyzed in panel b. n = 26224 (control), 18285 (miR-181a). P < 0.001 by Kolmogorov-Smirnov (KS) test. (d) Average mEPSC amplitudes of events from neurons analyzed in panel b. P = 0.78. (e) Cumulative probability plot of amplitudes of events from neurons analyzed in panel b. n = 26309 (control), 18388 (miR-181a). P < 0.001 by KS test.
Stimulation of dopamine signaling induces miR-181a expression in hippocampus neurons.
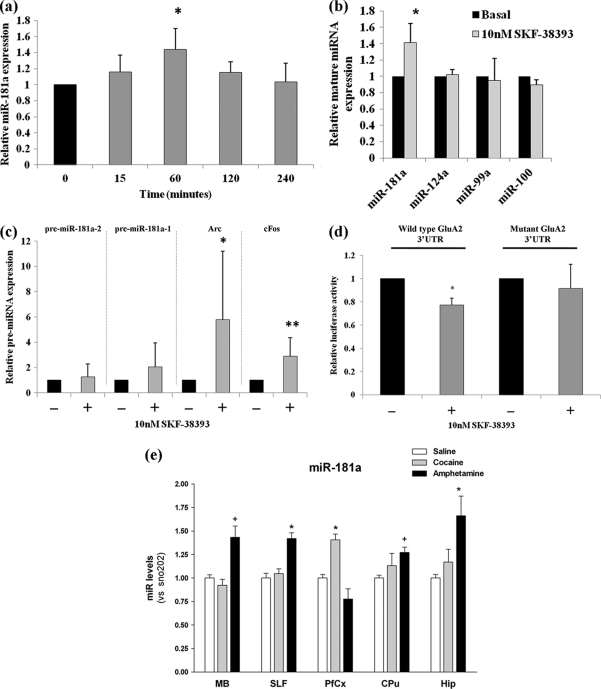
The neurotransmitter dopamine has a central role in motivational control, as most drugs of abuse modulate dopamine signaling in order to elicit persistent structural and functional plasticity in neurons (28, 66). Therefore, we tested whether dopamine signaling would modulate the expression of miR-181a in dissociated hippocampal neurons that express endogenous dopamine receptors (15, 54). For this set of experiments, we simulated dopamine signaling through the bath application of D1/D5 dopamine receptor agonist SKF-38393 (52). When the expression of the mature miR-181a form was measured by qRT-PCR upon SKF-38393 treatment over a period of 4 h, maximum induction was observed after 1 h of treatment (Fig. 6a). Therefore, we used this time point for further studies. Upon stimulation with variable amounts of SKF-38393 (10 nM→100 μM), we observed maximum induction of miR-181a at 10 nM (Fig. 6b and data not shown). At this concentration, other miRNAs that were either enriched (miR-100 and miR-99a) (Fig. 1a) or equally distributed in synaptoneurosomes (miR-124a) (Fig. 1c) were not affected by agonist treatment (Fig. 6b), suggesting that dopamine signaling selectively activates miR-181a. Dopamine signaling was effectively induced as judged by the robust activation of the immediate-early genes Arc and c-Fos (Fig. 6c). Moreover, at 10 nM the agonist did not significantly activate the two potential miR-181a precursors, pre-miR-181a and -b, implying that the activation might involve a posttranscriptional regulation step (Fig. 6c). This is further supported by the presence of pre-miR-181a-2, and not the pre-miR-181a-1, in nucleus accumbens synaptoneurosomes as judged by microarray (Fig. 1a) (see Table S6 in the supplemental material). Taken together, these results suggest that the stimulation of dopamine signaling in hippocampus neurons leads to an increase in mature miR-181a, possibly due to increased pre-miR-181a processing.
Fig 6.
Dopamine agonist induces miR-181a expression in neurons. (a) Expression of mature miR-181a in hippocampal neuronal cells during dopamine treatment. 18DIV hippocampal neurons were treated with 10 nM SKF-38393 for 15 min to 4 h. At each time point, cells were washed and lysed and the expression of miR-181a was measured by qRT-PCR. Data are relative to mock-treated cells and are normalized to U6 snRNA. Bar plots show mean ± SD (n = 3); *, P < 0.05. (b) Expression of mature miR-181a in hippocampal neuronal cells in the presence of 10 nM SKF-38393. Also shown are the expressions of other miRNAs (miR-99a, miR-100, and miR-124a) at this concentration of the agonist. 18DIV hippocampal neurons were treated with 10 nM SKF-38393 for 1 h at 37°C. After 1 h of treatment, cells were washed and lysed and the expression of miR-181a was measured by qRT-PCR. Data are relative to mock-treated cells and are normalized to the expression of U6 snRNA. Bar plots show mean ± SD (n = 3). Significance was determined using Student's t test (*, P < 0.009). (c) Expression of pre-miR-181a-1, pre-miR-181a-2, Arc, and c-Fos transcripts in hippocampal neuronal cells in the presence of 10 nM SKF-38393. 18DIV hippocampal neurons were treated with 10 nM SKF-38393 for 1 h at 37°C. After 1 h of treatment, cells were washed and lysed and the expression of the transcripts was measured by qRT-PCR. Data are relative to mock-treated cells and are normalized to the expression of GAPDH mRNA. Bar plots show mean ± SD (n = 5). *, P = 0.073; **, P < 0.05. (d) Effect of dopamine agonist signaling on GluA2-3′ UTR luciferase construct activity (wild type or mutant), in vitro. Wild-type or mutant GluA2-3′ UTR luciferase construct was transfected into 13DIV hippocampal cells and treated with 10 nM SKF-38393 on 18DIV for 1 h at 37°C. After treatment, cells were washed and reincubated at 37°C for an additional 24 h, after which time the luciferase activity was measured. Data are shown relative to untreated cells transfected with either the wild-type or mutant construct, respectively. Bar plots show mean ± SD (n = 5 for wild-type construct; n = 3 for mutant construct); *, P < 0.0002. (e) Expression of miR-181a in various regions of the mouse brain in response to chronic treatment with psychotropic drugs. C57BL6 mice (four or five per group) were exposed to saline, cocaine (10 mg/kg), or amphetamine (5 mg/kg) for 5 days (1 injection/day) and sacrificed 4 h after the last injection. Total RNA (including small RNA species) was analyzed by qRT-PCR. Data were normalized to snoRNA 202, and data are shown relative to saline-treated mice. Hip, hippocampus; PFCx, prefrontal cortex; SLF, sublimbic forebrain, VMB, ventral midbrain. Bar plots show mean ± SD (n = 4 or 5 per group). *, P < 0.05; +, 0.1 > P > 0.05.
We next examined whether the increased levels of miR-181a in response to dopamine agonist signaling affect GluA2 expression. For this experiment, either wild-type or mutant GluA2-3′ UTR fused to the luciferase gene was transfected into 13DIV hippocampal cells and treated with 10 nM SKF-38393 at 18DIV and luciferase activity was measured 24 h later. The agonist treatment led to a significant decrease in the activity of the wild-type, but not of the mutant, GluA2-3′ UTR construct (Fig. 6d). From this, it can be deduced that the induction of miR-181a by dopamine agonist signaling leads to the generation of functional miR-181a that is potentially capable of downregulating GluA2 expression.
Upregulation of miR-181a in chronically drug-treated mice.
It is well established that chronic exposure to addictive psychotropic drugs can lead to neuroplastic changes in the brain (46, 63). Additionally, several recent studies have further demonstrated changes in miRNA abundance due to treatment with psychotropic drugs that are known to engage dopamine signaling (8, 9, 18, 22, 33). To monitor whether miR-181a expression is altered in vivo in response to chronic exposure to psychotropic drugs, C57BL/6 mice (four to five per group) were exposed to saline, cocaine (10 mg/kg), or amphetamine (5 mg/kg) for 5 days (1 injection/day) and sacrificed 4 h after the last injection. For each animal, the hippocampus, prefrontal cortex, sublimbic forebrain (nucleus accumbens region), and ventral midbrain were carefully microdissected and total RNA (including small RNA species) was extracted for the screening of miR-181a expression by qRT-PCR. Chronic drug exposure led to a significant induction of miR-181a expression in a drug-dependent and brain region-specific manner (Fig. 6e; data not shown for the induction of miR-181b and c). Of the two psychotropic drugs tested, amphetamine showed the most significant induction of miR-181a, which was observed in multiple regions of the brain, with the hippocampus showing the greatest degree of induction. In contrast, cocaine induced the expression of miR-181a most robustly in the prefrontal cortex. Taken together, these results demonstrate that drugs of abuse that are known to engage dopamine signaling potently induce miR-181a expression in relevant brain areas in vivo, notably the hippocampus and prefrontal cortex.
DISCUSSION
It is now well accepted that the intake of drugs of abuse can lead to fundamental changes in neuronal processes that regulate synaptic plasticity (46, 63). These changes can manifest in long-lasting neuroadaptations in several key regions of the mesolimbic dopamine system that may, in turn, contribute to behavioral changes characterized by altered processing of contextual information. However, the exact molecular mechanisms governing these neuroadaptations are far from clear and are the subject of intense scientific inquiry.
In this study, we explored the role of miRNAs in contributing to the neuroadaptations caused by drugs of abuse due to their extensive reach into gene-regulatory mechanisms in the brain, most notably their widespread role at synapses of the hippocampus and cortex (48, 50, 53). Moreover, several recent studies have now identified a role for miRNAs in the rewarding effects of a number of addictive drugs of abuse (8, 9, 18, 19, 20, 22, 33, 44). In our study, we identified a total of 20 miRNAs that were significantly enriched in nucleus accumbens synaptoneurosomes, with 9 of them showing ≥2-fold enrichment (Fig. 1a and 1b; Table 1). miR-181a, an miRNA that was previously implicated in immune function (10), displayed a particularly robust and reproducible enrichment in synaptoneurosomes as judged by both Northern blotting and qRT-PCR analysis (Fig. 1c and d). miR-181a was also identified to be associated with Ago2 in the striatum and induced by cocaine in Drd2-expressing neurons, suggesting a likely role in cocaine addiction (47). Additionally, a somatodendritic gradient has been shown for miR-181a with over 2-fold enrichment in dendritic regions, further corroborating the detection of this miRNA at synaptic sites (31). Interestingly, a significant number of other miRNAs from our screen were also identified to be associated with Ago2 in the striatum, were induced by cocaine in Drd2-expressing neurons, and possessed a somatodendritic gradient. miRNAs falling into each of these categories are summarized in Table S2 in the supplemental material. In addition to the validation of miR-181a, we also verified the enrichment of two other miRNAs, miR-139-5p and miR-328, in our synaptoneurosome preparations (Fig. 1c). Similarly to miR-181a, these two miRNAs were also previously found to be associated with striatal Ago2 and favor a somatodendritic gradient distribution (see Table S2). The functional characterizations of these miRNAs are under way in our lab.
Among the computationally predicted targets for miR-181a, there was an apparent bias toward genes involved in various brain functions (Fig. 2a and 2b; see Tables S4 and S5 in the supplemental material). One of the target genes that we specifically focused on was the transcript encoding the GluA2 subunit of AMPA-Rs (Fig. 2c). A potential interaction between miR-181 family member miR-181b and GluA2 was previously investigated within the context of schizophrenia, where miR-181b showed elevated levels in the gray matter of the superior temporal gyrus (4). However, this study used a glioblastoma cell culture model and did not address the functional consequences with regard to synapse morphology and physiology. In our work, we observed a specific reduction in GluA2 surface expression and mEPSC frequency upon miR-181a expression, demonstrating for the first time an important functional role for miR-181a at the synapse. The regulation of AMPA-Rs is of particular interest for understanding the molecular mechanisms contributing to neuroadaptations caused by drugs of abuse, and also for synaptic plasticity in general, since the expression and subunit composition of AMPA-Rs at synapses are critical determinants of excitatory synaptic strength (6, 61). Interestingly, reduced GluA2 surface expression upon miR-181a overexpression correlates with a significant reduction in mEPSC frequency (Fig. 5b and c). The effects on frequency are most easily explained by a reduction in functional postsynaptic specializations. Consistent with this, dendrites of miR-181a overexpressing neurons have fewer and smaller spines (Fig. 4e and f). Spine defects could be a direct consequence of decreased GluA2 surface expression, since GluA2 was shown to promote spine growth via its extracellular N-terminal domain (43). However, although we monitor synapse morphology and physiology exclusively in cells where miR-181a was expressed postsynaptically, we cannot formally rule out the possibility that changes in mEPSC frequency are due to a retrograde signal that alters presynaptic function (e.g., probability of vesicle release) of synaptically connected nontransfected neurons. Furthermore, presynaptic changes occurring in termini that synapse onto the neuron they originate from (so-called autapses) could account for frequency alterations. A detailed electrophysiological characterization will be required to distinguish between these various possibilities. Surprisingly, the reduction in mEPSC frequency was accompanied by only a very subtle reduction in amplitude (Fig. 5d and e). Perhaps miR-181a triggers a selective removal of GluA2 from a small subset of synapses that subsequently become silent, whereas the majority of synapses would preserve their GluA2 content and therefore keep their original strength (21). This could be important for synapse-specific modifications, for example during long-term depression (LTD).
Intriguingly, other GluA receptors (in particular GluA1) lack canonical miR-181a binding sites in their 3′ UTRs, suggesting that miR-181a selectively targets GluA2 for translational repression. Therefore, we speculate that miR-181a could also contribute to alterations in the subunit composition of AMPA-Rs by selectively reducing the expression of GluA2 subunits. Specifically, miR-181a may block local synthesis of GluA2 but not of GluA1, possibly leading to an increase in calcium-permeable GluA2-lacking (Cp) AMPA-Rs at synapses. Cp-AMPA-Rs are transiently inserted into synapses during several forms of Hebbian and homeostatic plasticity (40), where they seem to play an important role in the induction phase of plasticity. A relative increase in Cp-AMPA-Rs upon miR-181a expression could also compensate for the loss of GluA2-containing AMPA-Rs, providing an additional explanation why we observed only subtle reductions of mEPSC amplitudes under these conditions (Fig. 5d and e).
In the context of addiction, drugs of abuse are known to influence AMPA-R assembly in order to provoke abnormal plasticity that contributes to reward-seeking behavior(s) (65, 66). For example, cocaine is known to alter postsynaptic AMPA-R components by promoting the exchange of GluA2-containing AMPA-Rs (i.e., GluA1/2 or GluA2/3 receptors) for GluA2-lacking AMPA-Rs (i.e., GluA1/1 or GluA1/3) (1, 3, 39). Furthermore, GluA2-lacking AMPA-Rs are required for drug-craving behavior following prolonged cocaine withdrawal (11). As such, the regulation of GluA2 expression by miR-181a could be part of a homeostatic plasticity mechanism in response to increased dopamine signaling. Indeed, we found that the expression of miR-181a (and other members of this family such as miR-181b and -c) is upregulated by dopamine signaling in cell culture (Fig. 6a to d) and by psychotropic drugs in mouse models of chronic drug addiction (Fig. 6e and data not shown for miR-181b and -c). Moreover, dopamine agonist treatment reduced the expression of the GluA2 reporter in an miR-181a binding site-dependent manner (Fig. 6d). Consistent with our results, Schuman and colleagues recently observed dopamine-dependent increases in local GluA1, but not GluA2, synthesis in dendrites of hippocampal neurons (54). Together, these results are consistent with the idea that aberrantly expressed miR-181a, in response to drugs of abuse/dopamine signaling, can lead to perturbation in AMPA-R subunit assembly by preferentially targeting the GluA2 subunit for posttranscriptional downregulation. In turn, this might contribute to some of the altered neuroadaptations associated with drug-afflicted areas of the brain. In this regard, it will be very interesting to test the potential involvement of miR-181a in drug-induced alterations in plasticity and behavior in animal models in future studies.
Since the 3′ UTR of mRNAs is usually targeted by multiple microRNAs (2), miR-181a is likely not the only posttranscriptional regulator of GluA2-expression. Interestingly, the highly conserved and exclusively neuronal miR-124 (30) also has a conserved binding site in the vicinity of the binding site for miR-181a (Fig. 2c). Moreover, miR-124 has also been shown to respond to cocaine treatment (downregulation) (8, 9), which might translate into increased expression of target gene(s), like GluA2. However, in our work, we found that miR-124 was not enriched at MSN synapses (Fig. 1c), and we did not see significant alterations in the expression pattern of this miRNA upon dopamine signaling (Fig. 6b). Clearly, more work is needed to elucidate the exact contribution of other posttranscriptional regulators, at the level of both miRNAs and RNA binding proteins (RBPs), in the regulation of GluA2 expression.
A finding from this work that needs more clarification is the mechanism by which miR-181a is induced in response to dopamine signaling in different brain areas. There are two possibilities by which this may occur: (i) increased transcriptional output of the gene encoding miR-181a or (ii) increased processing of miR-181a from its precursor molecule (pre-miRNA). In our study, we found some preliminary experimental support for the latter. Specifically, by microarray screening, we were able to detect one of the miR-181a precursors, pre-miR-181a-2 (but not pre-miR-181a-1) in the synaptoneurosomes prepared from the nucleus accumbens tissue (see Table S6 in the supplemental material). This could indicate synapto-dendritic transport and/or local processing of pre-miR-181a-2 in response to dopamine signaling. Consistent with this model, proteins of the miRNA processing machinery (e.g., Dicer, eIF2C2) have been found within the vicinity of dendrites close to synapses (35, 36).
Taken together, our work identifies mammalian AMPA-Rs as targets of neuronal miRNAs, with important implications for both physiological and pathological plasticity mechanisms. Furthermore, our results suggest that perturbations in the GluA2 subunit composition of AMPA-Rs in response to drug-induced expression of the nucleus accumbens-enriched miRNA miR-181a may lead to maladaptive changes in synaptic transmission and plasticity in brain regions critical to the drug reward pathway. miR-181a could therefore represent a novel and promising target for therapeutic intervention in situations of excessive dopamine signaling, including but not limited to drug abuse.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the excellent technical assistance of Tatjana Wüst. We thank Roberto Fiore for cloning the GluA2-3′ UTR. We also thank Roberto Fiore and Sharof Khudayberdiev for critical reading of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB488 to G.M.S.), the Human Frontier Science Program (Career Development Award to G.M.S.), and the National Institute on Drug Abuse (1R21DA025102-01 to G.M.S.). G.M.S. is an EMBO Young Investigator.
We declare that we have no conflict of interest.
Footnotes
Published ahead of print 5 December 2011
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Argilli E, Sibley DR, Malenka RC, England PM, Bonci A. 2008. Mechanism and time course of cocaine-induced long-term potentiation in the ventral tegmental area. J. Neurosci. 28:9092–9100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 [DOI] [PubMed] [Google Scholar]
- 3. Bellone C, Luscher C. 2006. Cocaine triggered AMPA receptor redistribution is reversed in vivo by mGluR-dependent long-term depression. Nat. Neurosci. 9:636–641 [DOI] [PubMed] [Google Scholar]
- 4. Beveridge NJ, et al. 2008. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum. Mol. Genet. 17:1156–1168 [DOI] [PubMed] [Google Scholar]
- 5. Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193 [DOI] [PubMed] [Google Scholar]
- 6. Burnashev N, Monyer H, Seeburg PH, Sakmann B. 1992. Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron 8:189–198 [DOI] [PubMed] [Google Scholar]
- 7. Bussey KJ, et al. 2003. MatchMiner: a tool for batch navigation among gene and gene product identifiers. Genome Biol. 4:R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chandrasekar V, Dreyer JL. 2009. microRNAs miR-124, let-7d and miR-181a regulate cocaine-induced plasticity. Mol. Cell. Neurosci. 42:350–362 [DOI] [PubMed] [Google Scholar]
- 9. Chandrasekar V, Dreyer JL. 2011. Regulation of MiR-124, Let-7d, and MiR-181a in the accumbens affects the expression, extinction, and reinstatement of cocaine-induced conditioned place preference. Neuropsychopharmacology 36:1149–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen CZ, Li L, Lodish HF, Bartel DP. 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science 303:83–86 [DOI] [PubMed] [Google Scholar]
- 11. Conrad KL, et al. 2008. Formation of accumbens GluR2-lacking AMPA receptors mediates incubation of cocaine craving. Nature 454:118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cougot N, et al. 2008. Dendrites of mammalian neurons contain specialized P-body-like structures that respond to neuronal activation. J. Neurosci. 28:13793–13804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Di Chiara G. 2002. Nucleus accumbens shell and core dopamine: differential role in behavior and addiction. Behav. Brain Res. 137:75–114 [DOI] [PubMed] [Google Scholar]
- 14. Fiore R, et al. 2009. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 28:697–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gao C, Sun X, Wolf ME. 2006. Activation of D1 dopamine receptors increases surface expression of AMPA receptors and facilitates their synaptic incorporation in cultured hippocampal neurons. J. Neurochem. 98:1664–1677 [DOI] [PubMed] [Google Scholar]
- 16. Grimson A, et al. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27:91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He Y, Yang C, Kirkmire CM, Wang ZJ. 2010. Regulation of opioid tolerance by let-7 family microRNA targeting the mu opioid receptor. J. Neurosci. 30:10251–10258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hollander JA, et al. 2010. Striatal microRNA controls cocaine intake through CREB signalling. Nature 466:197–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang W, Li MD. 2009. Differential allelic expression of dopamine D1 receptor gene (DRD1) is modulated by microRNA miR-504. Biol. Psychiatry 65:702–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang W, Li MD. 2009. Nicotine modulates expression of miR-140*, which targets the 3′-untranslated region of dynamin 1 gene (Dnm1). Int. J. Neuropsychopharmacol. 12:537–546 [DOI] [PubMed] [Google Scholar]
- 21. Huang YH, et al. 2009. In vivo cocaine experience generates silent synapses. Neuron 63:40–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Im HI, Hollander JA, Bali P, Kenny PJ. 2010. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat. Neurosci. 13:1120–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Job C, Eberwine J. 2001. Localization and translation of mRNA in dendrites and axons. Nat. Rev. Neurosci. 2:889–898 [DOI] [PubMed] [Google Scholar]
- 24. John B, et al. 2004. Human MicroRNA targets. PLoS Biol. 2:e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ju W, et al. 2004. Activity-dependent regulation of dendritic synthesis and trafficking of AMPA receptors. Nat. Neurosci. 7:244–253 [DOI] [PubMed] [Google Scholar]
- 26. Kacharmina JE, Job C, Crino P, Eberwine J. 2000. Stimulation of glutamate receptor protein synthesis and membrane insertion within isolated neuronal dendrites. Proc. Natl. Acad. Sci. U. S. A. 97:11545–11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Karr J, et al. 2009. Regulation of glutamate receptor subunit availability by microRNAs. J. Cell Biol. 185:685–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kauer JA, Malenka RC. 2007. Synaptic plasticity and addiction. Nat. Rev. Neurosci. 8:844–858 [DOI] [PubMed] [Google Scholar]
- 29. Krek A, et al. 2005. Combinatorial microRNA target predictions. Nat. Genet. 37:495–500 [DOI] [PubMed] [Google Scholar]
- 30. Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. 2003. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 9:1274–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kye MJ, et al. 2007. Somatodendritic microRNAs identified by laser capture and multiplex RT-PCR. RNA 13:1224–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Le Moal M, Simon H. 1991. Mesocorticolimbic dopaminergic network: functional and regulatory roles. Physiol. Rev. 71:155–234 [DOI] [PubMed] [Google Scholar]
- 33. Lippi G, et al. 2011. Targeting of the Arpc3 actin nucleation factor by microRNA-29a/b regulates dendritic spine morphology. J. Cell Biol. 194:889–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 35. Lugli G, Larson J, Martone ME, Jones Y, Smalheiser NR. 2005. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J. Neurochem. 94:896–905 [DOI] [PubMed] [Google Scholar]
- 36. Lugli G, Torvik VI, Larson J, Smalheiser NR. 2008. Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J. Neurochem. 106:650–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Luscher C, et al. 1999. Role of AMPA receptor cycling in synaptic transmission and plasticity. Neuron 24:649–658 [DOI] [PubMed] [Google Scholar]
- 38. Malenka RC, Nicoll RA. 1999. Long-term potentiation—a decade of progress? Science 285:1870–1874 [DOI] [PubMed] [Google Scholar]
- 39. Mameli M, Bellone C, Brown MT, Luscher C. 2011. Cocaine inverts rules for synaptic plasticity of glutamate transmission in the ventral tegmental area. Nat. Neurosci. 14:414–416 [DOI] [PubMed] [Google Scholar]
- 40. Man HY. 2011. GluA2-lacking, calcium-permeable AMPA receptors—inducers of plasticity? Curr. Opin. Neurobiol. 21:291–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miranda RC, et al. 2010. MicroRNAs: master regulators of ethanol abuse and toxicity? Alcohol. Clin. Exp. Res. 34:575–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nestler EJ. 2001. Molecular neurobiology of addiction. Am. J. Addict. 10:201–217 [DOI] [PubMed] [Google Scholar]
- 43. Passafaro M, Nakagawa T, Sala C, Sheng M. 2003. Induction of dendritic spines by an extracellular domain of AMPA receptor subunit GluR2. Nature 424:677–681 [DOI] [PubMed] [Google Scholar]
- 44. Pietrzykowski AZ, et al. 2008. Posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron 59:274–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rao A, Steward O. 1991. Evidence that protein constituents of postsynaptic membrane specializations are locally synthesized: analysis of proteins synthesized within synaptosomes. J. Neurosci. 11:2881–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Robinson TE, Kolb B. 2004. Structural plasticity associated with exposure to drugs of abuse. Neuropharmacology 47(Suppl. 1):33–46 [DOI] [PubMed] [Google Scholar]
- 47. Schaefer A, et al. 2010. Argonaute 2 in dopamine 2 receptor-expressing neurons regulates cocaine addiction. J. Exp. Med. 207:1843–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schratt G. 2009. microRNAs at the synapse. Nat. Rev. Neurosci. 10:842–849 [DOI] [PubMed] [Google Scholar]
- 49. Schratt GM, Nigh EA, Chen WG, Hu L, Greenberg ME. 2004. BDNF regulates the translation of a select group of mRNAs by a mammalian target of rapamycin-phosphatidylinositol 3-kinase-dependent pathway during neuronal development. J. Neurosci. 24:7366–7377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schratt GM, et al. 2006. A brain-specific microRNA regulates dendritic spine development. Nature 439:283–289 [DOI] [PubMed] [Google Scholar]
- 51. Schuman EM, Dynes JL, Steward O. 2006. Synaptic regulation of translation of dendritic mRNAs. J. Neurosci. 26:7143–7146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sibley DR, Leff SE, Creese I. 1982. Interactions of novel dopaminergic ligands with D-1 and D-2 dopamine receptors. Life Sci. 31:637–645 [DOI] [PubMed] [Google Scholar]
- 53. Siegel G, et al. 2009. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat. Cell Biol. 11:705–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Smith WB, Starck SR, Roberts RW, Schuman EM. 2005. Dopaminergic stimulation of local protein synthesis enhances surface expression of GluR1 and synaptic transmission in hippocampal neurons. Neuron 45:765–779 [DOI] [PubMed] [Google Scholar]
- 55. Reference deleted.
- 56. Sprengel R, et al. 2001. Glutamate receptor channel signatures. Trends Pharmacol. Sci. 22:7–10 [DOI] [PubMed] [Google Scholar]
- 57. Steward O, Schuman EM. 2001. Protein synthesis at synaptic sites on dendrites. Annu. Rev. Neurosci. 24:299–325 [DOI] [PubMed] [Google Scholar]
- 58. Sun X, Wolf ME. 2009. Nucleus accumbens neurons exhibit synaptic scaling that is occluded by repeated dopamine pre-exposure. Eur. J. Neurosci. 30:539–550 [DOI] [PubMed] [Google Scholar]
- 59. Sutton MA, Schuman EM. 2005. Local translational control in dendrites and its role in long-term synaptic plasticity. J. Neurobiol. 64:116–131 [DOI] [PubMed] [Google Scholar]
- 60. Sutton MA, Wall NR, Aakalu GN, Schuman EM. 2004. Regulation of dendritic protein synthesis by miniature synaptic events. Science 304:1979–1983 [DOI] [PubMed] [Google Scholar]
- 61. Swanson GT, Kamboj SK, Cull-Candy SG. 1997. Single-channel properties of recombinant AMPA receptors depend on RNA editing, splice variation, and subunit composition. J. Neurosci. 17:58–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tada T, Sheng M. 2006. Molecular mechanisms of dendritic spine morphogenesis. Curr. Opin. Neurobiol. 16:95–101 [DOI] [PubMed] [Google Scholar]
- 63. Tang J, Dani JA. 2009. Dopamine enables in vivo synaptic plasticity associated with the addictive drug nicotine. Neuron. 63:673–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tusher VG, Tibshirani R, Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98:5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wolf ME. 2010. The Bermuda Triangle of cocaine-induced neuroadaptations. Trends Neurosci. 33:391–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wolf ME, Mangiavacchi S, Sun X. 2003. Mechanisms by which dopamine receptors may influence synaptic plasticity. Ann. N. Y. Acad. Sci. 1003:241–249 [DOI] [PubMed] [Google Scholar]
- 67. Zhou R, et al. 2009. Evidence for selective microRNAs and their effectors as common long-term targets for the actions of mood stabilizers. Neuropsychopharmacology 34:1395–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ziviani E, et al. 2011. Ryanodine receptor-2 upregulation and nicotine-mediated plasticity. EMBO J. 30:194–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.