Abstract
Dentin sialophosphoprotein (DSPP) mutations cause dentin dysplasia type II (DD-II) and dentinogenesis imperfecta (DGI) types II and III. We identified two kindreds with DGI-II exhibiting vertical bands of hypoplastic enamel. Both families had a previously reported DSPP mutation that segregated with the disease phenotype. Oral photographs and dental radiographs of four affected and one unaffected participant in one family and the proband in the second family were used to document the dental phenotypes. We aligned the 33 unique allelic DSPP sequences showing variable patterns of insertions and deletions (indels), generated a merged DPP sequence that includes sequences from all DSPP length haplotypes, and mapped the known DSPP mutations in this context. Analyses of the DSPP sequence changes and their likely effects on protein expression, as well as published findings of the dental phenotype in Dspp null mice support the hypothesis that all DSPP mutations cause pathology through dominant-negative effects. Noting that Dspp is transiently expressed by preameloblasts during formation of the dentino-enamel junction, we hypothesize that DSPP dominant-negative effects potentially cause cellular pathology in preameloblasts that, in turn, causes enamel defects. We conclude that enamel defects can be part of the dental phenotype caused by DSPP mutations, although DSPP is not critical for dental enamel formation.
Keywords: enamel, dentin, DSPP, dentinogenesis imperfecta, genetics
The principal organic components in forming enamel are amelogenin, ameloblastin and enamelin (1). In tooth dentin they are type I collagen and dentin sialophosphoprotein (Dspp) (2). During formation of the dentino-enamel junction, when the initial dentin and enamel layers mineralize, there is transient expression of enamel proteins by odontoblasts and dentin proteins by preameloblasts (3). Dspp is only transiently expressed by preameloblasts, but is secreted by odontoblasts throughout dentin formation (4, 5). Despite the transient expression of dentin and enamel proteins by opposing cells during formation of the DEJ, it is clear that enamel proteins such as amelogenin and enamelin are not critical for dentinogenesis and Dspp is not critical for amelogenesis. Patients with amelogenesis imperfecta (AI) caused by mutations in AMELX or ENAM do not exhibit a dentin phenotype (6). In addition, there is a loss of selection to maintain the integrity of enamel protein genes in vertebrates that have lost the ability to form dental enamel during evolution (7). Dspp is not expressed by secretory or maturation stage ameloblasts and enamel volume and density are normal in Dspp null mice (8). In humans, mutations in DSPP cause inherited dentin malformations, such as dentin dysplasia type II (DD-II), and dentinogenesis imperfecta types II and III (DGI-II and DGI-III). Dental enamel often undergoes rapid attrition in patients with dentinogenesis imperfecta (9, 10), which is generally ascribed to a lack of support by the defective underlying dentin.
To date, 35 different DSPP mutations have been shown to cause inherited dentin defects in humans (11). All show an autosomal dominant pattern of transmission, which means that only one DSPP allele is defective. This contrasts with the recessive pattern of transmission observed in mice following total disruption of the Dspp gene (12). Tell-tale characteristics of the reported human mutations led to the hypothesis that all of the disease-causing DSPP mutations lead to the production of a mutant protein that damages the expressing cell and/or the extracellular matrix (13). If this compelling hypothesis is true, DSPP mutations cause inherited dentin defects through a dominant-negative mechanism, and not by a loss of function or a reduction in the amount of normal DSPP protein (haploinsufficiency), which is the pathogenic mechanism at work in the Dspp knockout mice. We use the term “dominant-negative” to indicate the phenotype is caused by the pathological effects of the mutant protein, although the actual pathological mechanism might be better described as a “gain of function”.
DSPP 5-prime Mutations
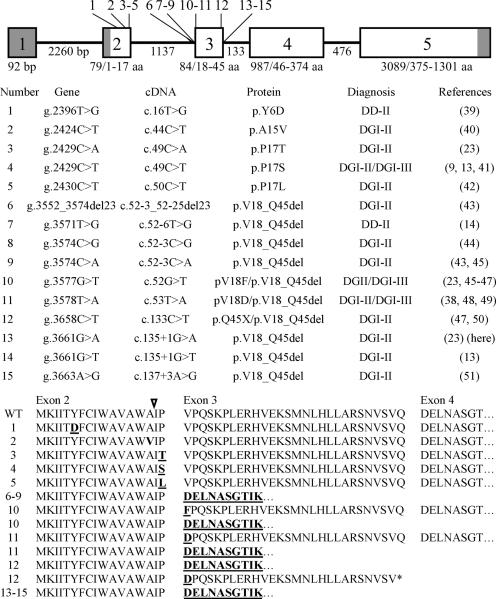
Fifteen different disease-causing mutations have been reported in the 5' end of DSPP (Fig. 1). Five of the mutations are in exon 2, which encodes the signal peptide (amino acids 1 to 15) plus two amino acids from the N-terminus of DSPP (amino acids 16 and 17). The first two mutations are within the signal peptide and must cause a problem with protein targeting, as the sequence of the secreted protein is identical to the wild-type. The next three mutations alter the amino acids in the +2 and +3 positions relative to the signal peptide cleavage site. The remaining ten mutations cluster around the splice boundaries of exon 3. Where experimental evidence is available, the result of a defective splicing junction was the skipping of exon 3, rather than intron retention (14, 15). All of the DSPP 5-prime disease-causing mutations (either directly or by skipping of exon 3) alter the context of the signal peptide cleavage site. Six of the 15 mutations have been reported more than once, so many of the DSPP 5' mutations that potentially cause inherited dentin defects may have been identified. None of the DSPP 5' mutations alter the protein at the many conserved sites that are involved in potentially important posttranslational modifications (16), suggesting that heterozygous loss of function mutations in DSPP do not cause inherited dentin defects. A reasonable explanation for the dominant-negative effects is that the 5' mutations interfere with protein targeting (secretion) and cause cell pathology (13).
Figure 1.
Mutations in the 5-prime end of DSPP causing inherited dentin defects. Top: The intron/exon structure of DSPP. The five DSPP exons are indicated by numbered boxes; the four introns are the lines connecting the exons. The numbers below each intron show the number of basepairs (bp) in that intron. The numbers below each exon show the number of bp in that exon followed by the range of amino acids encoded by it. Locations of the 15 reported DSPP 5' mutations are indicated by numbered lines, starting at the 5' end of DSPP. The first five mutations are in exon 2. We hypothesize that these mutations directly interfere with signal peptide function or cleavage. The other ten mutations cluster around the splice boundaries of exon 3. Skipping of exon 3 might affect cleavage or functioning of the DSPP signal peptide of (amino acids 1 to 15). Signal peptide malfunction is presumed to interfere with protein secretion and cause cell pathology. Middle: List of the 15 reported DSPP 5' mutations and their dental phenotypes. Note that six of the fourteen DSPP 5' mutations have been reported more than once and that the diagnoses of DGI-II or DGI-III have both been used to describe the phenotype of persons with the same DSPP mutation. Bottom: Wild-type (WT) and mutant predicted amino acid DSPP sequences. Changed amino acids are in bold and underlined; the signal peptide cleavage site is indicated by and arrow.
DSPP 3-prime Mutations
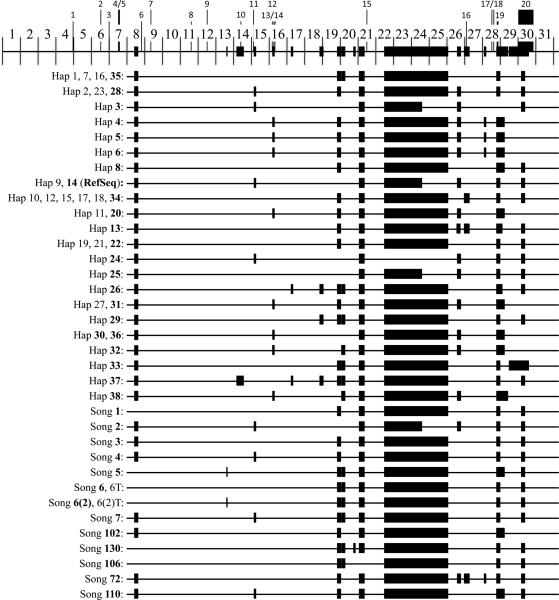
Twenty different disease-causing mutations have been reported in the 3' end of DSPP, within the coding region for dentin phosphoprotein (DPP), the C-terminal cleavage product of DSPP (17–19). Dpp is the most acidic protein known, having an isoelectric point near 1.1 (20). Its sequence is highly repetitive and the length of the repeat region varies greatly among different mammalian species and even within species, which is reflected in the size of the Dpp protein (21). The first studies to successfully characterize the DPP coding region in patients with inherited dentin defects reported therein 22 (13) and 13 (22) different patterns of insertions and deletions (indels). McKnight et al. (13) identified 22 DPP indel patterns in 188 normal human chromosomes, while Song et al. (22) reported 13 DPP indel patterns from 220 normal human chromosomes. Only two of the DPP indel patterns were identical between the two studies, so 33 unique indel patterns are known for human DPP coding region, with the deduced amino acid sequences ranging from 770 to 902 amino acids. The DPP region of the DSPP reference sequence (GB# NM_014208.3) has 839 amino acids. We carefully aligned the DPP coding region from each of the 33 haplotypes having novel indel patterns and noted the positions of the 20 disease-causing mutations in the DSPP 3-prime region (Supporting Fig. S1). This allowed us to construct a merged DPP coding region that includes all of the sequences found within the 33 known DPP length haplotypes (Fig. S2). The currently known DPP length haplotypes and the locations of the 20 disease-causing mutations in the merged DPP coding region are charted in Fig. 2. The 20 3-prime disease-causing mutations and their associated phenotypes are listed in Fig. 3.
Figure 2.
Human DPP length haplotypes and positions of indels and disease-causing mutations in merged human DPP. Top: The merged DPP sequence (from Fig. S2) is shown divided into a little over 31 equal, numbered segments corresponding to 90 bp (30 aa) of DPP. The thin bar represents universal segments found on all DPP length haplotypes; the thick bars are variable regions found only on some DPP length haplotypes. The numbered lines above the merged DPP map mark the locations of the twenty DSPP 3-prime disease-causing mutations (listed in Fig. 3). Note that most indels are located in the downstream half of the DPP code. Bottom: Maps showing positions of the indels (thick bars) on each of the 33 novel length haplotypes. The first seven segments of DPP show no length polymorphisms and are not included in the diagrams for each length haplotype. There can be multiple DPP haplotypes for each length haplotype because of nucleotide substitutions (polymorphism that don't effect length). The 22 length haplotypes from McKnight et al. (13) and 13 length haplotypes from Song et al. (22) are shown.
Figure 3.
Top: DSPP intron/exon structure with the DPP region expanded to show the positions of indels (thick bars) and sites of 20 disease-causing frame shift mutations (numbered). Bottom: List of the 20 reported DSPP 3-prime mutations, their designations, and their associated dental phenotypes. All twenty disease-causing mutations shift the downstream sequence into the −1 reading frame.
Because the DPP coding region is so repetitive, an unusual thing happens when a genetic alteration causes a shift in its reading frame. A deletion or insertion that causes a shift to the −2 reading frame (i.e., a deletion of two nucleotides or the insertion of four) will find an in-frame translation termination codon after introducing only a few extraneous amino acids (Fig. S3). A deletion or insertion that causes a shift to the −1 reading frame however, continues without hitting a translation termination signal until downstream of the natural DPP stop codon. Removing the first nucleotide at the beginning of the DPP coding region in the DSPP reference sequence replaces DPP with an 850 amino acid missense protein that is comprised of 35% alanine, 23% valine, 20% threonine, and 12% isoleucine. Any −1 reading frame shift starting 47 or fewer codons before the start of DPP in exon 5 or within the DPP coding region itself will terminate translation at the same stop codon downstream of the normal DSPP termination codon. The length of the missense peptide in each case is a function of the location of the frame-shift and which DPP length haplotype it occurs in. It turns out that all 20 of the disease-causing mutations in the DPP coding region shift the reading frame to the −1 frame. Mutations that shift all or part of the DPP coding region into the −2 reading frame terminate translation without producing much of a missense peptide. Such changes reduce the amount of DPP protein (haploinsufficiency) or generate a truncated DPP protein (loss of function), but these types of mutations have not been observed, presumably because they do not cause inherited dentin defects. This analysis supports the hypothesis that disease-causing mutations in DSPP are the result of dominant-negative effects and that haploinsufficiency secondary to the loss of a single DSPP allele does not cause disease in the heterozygous condition.
We have identified two DGI-II kindreds that show dentin defects typical of this condition, but they also display pathological malformations of their dental enamel. We have characterized the dental condition in these families and identified the same DSPP mutation in both, which was previously reported to cause DGI (23), but without documenting the dental phenotype. We hypothesize that transient expression of the mutated DSPP protein may have caused a dominant-negative effect in preameloblasts that resulted in the observed enamel malformations.
Materials and methods
The study protocol and subject consents were reviewed and approved by the Institution Review Board at the University of Michigan and appropriate informed consent was obtained from all subjects. Two families were recruited. Medical and dental histories were obtained and pedigrees constructed that span four generations. Oral photographs and radiographs were taken for five members of family 1 (III:7, III:8, IV:1, IV:2, IV:3) and the proband of family 2 (IV:2). Copies of previous dental radiographs were obtained for members IV:1, IV:2, and IV:3 of family 1 and members II:4, III:7, III:8, and IV:3 of family 2. Dental phenotypes were assessed by oral examination and by inspection of dental radiographs. In total 15 persons were recruited: 11 affected and 4 unaffected. The subjects ranged in age from 9 through adult.
DNA Isolation
Ten ml of peripheral whole blood was obtained from participating family members. In family 1 we were able to obtain DNA from five affected (II:6; III:7; IV:1, age 18; IV:2, age 17; and IV:3, age 16) and two unaffected (I:2 and III:8) members. In family 2 we were able to obtain DNA from six affected (II:3; III:4; III:7; IV:1; IV:2, age 13; and IV:3, age 9) and two unaffected (II:4, III:8) members. Genomic DNA was isolated using the QIAamp DNA Blood Maxi Kit (Qiagen, Valencia, CA, USA). Ages of adults were not specified.
Polymerase Chain Reaction (PCR)
PCR amplifications were done using the Platinum® PCR SuperMix (Invitrogen, Carlsbad, CA, USA). The reactions had a 5 min denaturation at 94 °C, followed by 35 cycles each with denaturation at 94 °C for 90 s, primer annealing at 56–59 °C for 60 s, and product extension at 72 °C for 90 s. In the final cycle the 72 °C extension was for 7 min. PCR amplification products were purified by QIAquick PCR Purification Kit and protocol (Qiagen).
Exons 2 through 4 were amplified along with their adjoining intron sequences. Exon 5 was characterized up to the repetitive region. Exon 2 was amplified with forward 5'gatgtccccataaccacacc and reverse 5'ctccatgacttctgggcatt primers to generate a 596 bp product (annealing 56 °C; extension 72 °C/ 90 s). Exons 3 and 4 were amplified together as a 1490 bp product using the forward 5'caagccctgtaagaagccact and reverse 5'acatggatgcttgtcatggt primers (annealing 59 °C; extension 72 °C/90 s). The 5' part of exon 5 was amplified using the forward 5'cctatggcaacttttcccagt and reverse 5' tgtcattgtcatcattcccatt primers (annealing 56 °C; extension 70 °C/90 s) to generate a 589 bp product.
DNA Sequence Analysis
DNA sequencing was performed at the University of Michigan DNA sequencing core. The original PCR primers used to generate each amplimer were used for sequencing reaction. In addition, for the larger exon 3–4 amplification product, two sense primers 5'-ggaccatgggaaagaagatg and 5'-gcatccagggacaagtaagc, and three antisense oligonucleotides 5'-cattcccttctcccttgtga, 5'-cctcgtttctacaggaattctca and 5'-tggaggtcttgtctccatca were used for sequencing reactions.
Alignment of 33 DPP indel patterns
Haplotype sequences from McKnight et al. (13) were imported from GenBank and provided the haplotype sequences starting with DSPP nucleotide 2572 encoding Arg396. Each sequence can be obtained by searching the National Center for Biotechnology Information (NCBI) database with “DSPP haplotype #”. For the purpose of this analysis, the reference sequence (NM_014208.3) was used to complete the DPP sequences 5-prime to this position, with the exception of Clone 37, where the short sequence 5' to nucleotide 2572 was obtained from Hap37A to ensure correct positioning of indel 1. The Song et al. (22) haplotype sequences were not available in GenBank. These sequences were obtained by altering the DSPP cDNA reference sequence (NM_014208.3) according to Tables 2 and 3 in Song et al. (22). Song et al. reported 15 haplotypes that showed 13 different indel patterns. The Song et al. haplotype designations are in bold. One Song haplotype from each of the 13 groups was used in the alignment: 1 (803 aa), 2 (839 aa), 3 (788 aa), 4 (788 aa), 5 (790 aa), [6, 6T, (797 aa)], [6(2), 6(2)T (796 aa)], 7 (782 aa), 102 (791 aa), 130 (794 aa), 106 (806 aa), 72 (785 aa), 110 (782 aa). Song haplotype 2 has the same indel pattern as McKnight haplotypes 9 and 14 as well as the reference sequence. Song haplotype 3 has the same indel pattern as McKnight haplotypes 19, 21, and 22.
Results
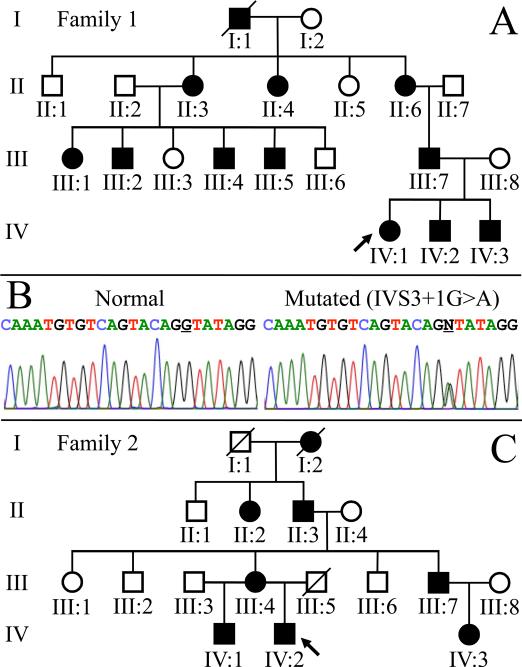
Two apparently unrelated Caucasian families of Northern European ancestry with inherited defects of dentin were examined at a University of Michigan dental clinic. There was no history of any unusual bone brittleness or unexplained hearing loss in either family. Affection status (the presence of inherited dentin defects) was oral ascertained by routine examination and pedigrees of the two kindreds going back four generations were constructed following interviews with multiple family members (Fig. 4). Mutational analyses of DSPP 5-prime region identified the same disease-causing mutation in both families: IVS3+1G>A; c.135+1G>A. This transition mutation changes the first nucleotide of intron 3, which is absolutely required for mRNA splicing. The mutation was identified in all ten of the affected members that donated samples for mutation analyses. The enamel surfaces of the probands, particularly in the anteriors, appeared shriveled, with vertical lines of hypoplastic enamel Fig. 5.
Figure 4.
A & C: Pedigrees of two Caucasian families with dentinogenesis imperfecta type II (DGI-II). The diseases of these two families segregated as an autosomal dominant trait. B: DNA sequencing chromatograms identified the same reported disease-causing mutation (IVS3+1G>A) in both families. This mutation changes the first nucleotide of intron 3 and is predicted to disturb normal proper mRNA splicing, presumably by causing the skipping of exon 3 during RNA splicing.
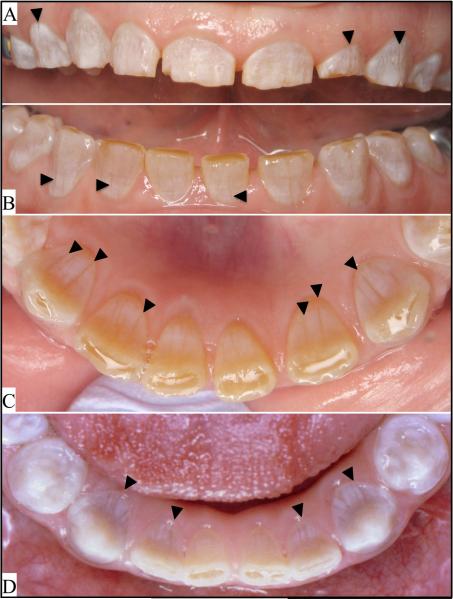
Figure 5.
A–C: Oral photos of the proband of family 1, and D: the proband of family 2. Arrowheads mark the most conspicuous vertical bands of hypoplastic enamel. More complete oral photographic surveys of the probands are provided in Figs. 4S and 9S.
Dental phenotyping: Family 1
In family 1, the proband was an 18 yr old female (IV:1) exhibiting translucent and chalky-colored crowns with vertical grooves of hypoplastic enamel and spaces between the maxillary and mandibular anterior teeth (Fig. S4). The enamel was more opaque on radiographs than dentin. The pulp chambers and many of the root canals were already obliterated. The roots of many teeth were noticeably shorter than normal, particularly in the anteriors. Some of the molar crowns had a bulbous morphology. Attrition was apparent along the incisal edges of the anteriors and the buccal cusp tips of the mandibular cuspids and premolars.
The proband's youngest brother (IV:3) at age 16 already had extensive attrition of all anterior teeth, and detectable attrition on the buccal cusp tips of the mandibular premolars (Fig. S5). The crowns were translucent or chalky, with some vertical grooves still evident in cervical areas that had not yet abraded, such as the buccals of #12 and #13. The pulp chambers and root canals were obliterated and the molar and premolar crowns appeared bulbous. The enamel was more radiopaque than dentin. The probands other brother (IV:2) at age 17 had extensive attrition of the anterior teeth, and lesser attrition on the buccal cusp tips of the mandibular premolars and second molars (Fig. S6), presumably worsened by failure of the crowns on three of the first molars. Like his siblings, the crowns were translucent and chalky, the pulp chambers and root canals were obliterated, the molar and premolar crowns were bulbous (which appeared to play a role in the impaction of the lower left third molar), and the enamel was more radiopaque than dentin. Vertical grooves were evident on the buccal cervical areas of the premolars, which were not yet destroyed by attrition. The father (III:7) had a very similar dentition, although the crowns were mostly translucent rather than chalky (Fig. S7). Attrition was most severe in the maxillary anteriors, but was not as bad as his sons' condition. The radiographs showed obliterated pulp chambers and root canals, bulbous crowns and enamel that was more radiopaque then dentin. The enamel surfaces had a polished appearance. The mother (III:8) was not affected and had a normal and healthy dentition (Fig. S8). The expression of the DGI-II phenotype in the four affected members of this family was remarkably consistent.
Dental phenotyping: Family 2
The proband of family 2 (IV:2) at age 12 was the youngest affected person to be characterized. Like family 1, the proband of family 2 had classic DGI-II (Fig. S9). The crowns alternated between chalky and translucent in color and had vertical grooves in the enamel. The pulp chambers and most of the root canals in the incisors and first molars had obliterated. The crowns of the posterior teeth were bulbous. The enamel was more radiopaque than dentin. Attrition was evident on the incisal edges of the anterior teeth. The affected mother had maxillary and mandibular complete dentures. Taking the differences in age between the subjects in the two families, the dental phenotypes of these patients with the IVS3+1G>A; c.143+1G>A mutation in DSPP were very similar.
Discussion
We characterized the dental phenotype of two families with a classic DGI-II phenotype and observed enamel defects in the form of vertical grooves in both probands. Both families had the same DSPP transition mutation (IVS3+1G>A). This mutation might cause the retention of intron 2, or the skipping of exon 3. Retention of intron 2 would lead to premature translation termination and delete all but two amino acids of the DSPP secreted protein. Such a transcript would likely be degraded by nonsense mediated decay (24), potentially causing haploinsufficiency but not dominant-negative effects. Skipping of exon 3 would delete amino acids 18 through 45 and potentially alter the context of the signal peptide cleavage site after Ala15. This is the most likely predicted scenario and has experimental support (14, 15). Ten of the disease-causing DSPP 5' mutations localize along the splice junctions of exon 3. We hypothesize that during RNA splicing of transcripts expressed from the mutant DSPP allele, exon 3 is skipped all or part of the time, and the resulting protein is not targeted properly and causes cell pathology.
DSPP has a signal peptide and is translated into the endoplasmic reticulum (ER) for secretion. Following translocation into the ER, the DSPP signal peptide is cleaved by the ER signal peptidase complex (SPC). Secretory proteins have a distinct secondary structure requirement at their cleavage site for processing by ER SPC (25–28). The flanking amino acid residues around the signal peptide cleavage site significantly influence this cleavage process (28). The DSPP IVS3+1G>A mutation, which alters the consensus sequence of a splice donor site, presumably leads to skipping of exon 3. Although exon 3 skipping of DSPP does not cause a frameshift or premature termination, it significantly changes the C-terminal flanking amino acid residues of signal peptide cleavage site, which could disturb signal peptide cleavage and interfere with normal DSPP folding and secretion. Mutations in or near the signal peptide coding region of other genes are known to cause diseases by inducing ER stress and cell pathology (29–32). Different proteins have different specific requirements for targeting. Recently it has been shown that a proline in the +2 position, which is specifically altered in three of the DSPP 5' mutations, is critical for the export of nucleobindin 1 (33). A mouse amelogenin missense mutation (p.Y64H) induced extensive ER stress and ameloblast cell pathology (34).
During enamel formation, DSPP is transiently expressed in preameloblasts. To explain the enamel defects in our patients, we hypothesize that the DSPP IVS3+1G>A mutation interferes with DSPP secretion and causes ER stress and cell pathology in pre-ameloblasts leading to developmental enamel defects. However, this pathogenic concept does not suggest that DSPP plays an important role in enamel formation. Any genetic or environmental aberration that induces ameloblast pathology will potentially cause developmental enamel defects. DSPP is transiently expressed at a sensitive transition period in ameloblast differentiation when the basal lamina is being degraded, enamel proteins are starting to be expressed, and cell-matrix interactions are in a state of flux (1). Pathology of secretory stage ameloblasts could explain the thin enamel. The vertical grooves might relate to buckling of the ameloblast layer during transition/early maturation because of ameloblast over-crowding on the pathologically small enamel surface. The scarcity of reports of enamel defects in persons with DSPP related dentin malformations might be due to the rapid attrition of enamel in DGI patients, so the enamel defects are seldom observed.
The finding that disease-causing mutations cluster at the borders of exon 3 and that improper splicing can potentially be caused by mutations at the edges of the exon (i.e. in the coding sequence) or 6 nucleotides into the intron suggests that this exon might be particularly susceptible to skipping, despite the fact that inclusion of this exon is critical to avoid pathological consequences. Perhaps this exon has undergone recent changes during evolution. DSPP belongs to the secretory calcium binding phosphoprotein (SCPPs) family of genes that all evolved from SPARCL1 (secreted protein, acidic, cysteine-rich related) (35). In humans there are seventeen SCPP genes in the enamel/milk/saliva group and five in the evolutionarily older dentin/bone group, which includes DSPP (36). DSPP exon 3 is unusual in that it is longer than the exon 3 of its homologues and does not encode a Golgi casein kinase phosphorylation site.
We assembled data concerning natural and pathological variations in DSPP. Alignment of the 33 known DPP indel patterns (Fig. S1) allowed us to produce a merged sequence and to chart the 20 frame-shift mutations (Fig. 2). Such a merged sequence should prove helpful in localizing the positions of indels in yet to be discovered length variations in the DPP code. As only two of the DPP indel patterns were the same in the two studies that reported them, there are likely to be many more the DPP indel patterns in the population as a whole.
Recently reported DSPP mutations and their associated phenotypes strengthen our previously stated hypothesis that autosomal dominant dentin defects caused by DSPP mutations should be considered a single disease, with a continuous spectrum of phenotypes that can be subclassified according to their severity into DD-II, DGI-II, and DGI-III, with DD-II being the least, and DGI-III being the most severe (37). Patients with the fifteen disease-causing 5' DSPP mutations have been diagnosed as having DD-II, DGI-II, and DGI-III. The p.P17S, p.PV18F and p.V18D mutations have been identified in persons diagnosed with DGI-II and DGI-III. The twenty disease-causing 5' DSPP −1 frame shift mutations have been identified in patients diagnosed with DD-II and DGI-II. All of the 35 characterized disease-causing DSPP showed a dominant pattern of inheritance. The pathogenesis in all cases (which is currently based upon logic and not scientific demonstration) seems to occur by a similar mechanism: dominant-negative effects induced by the mutant DSPP protein.
In this study we provided a detailed summary and analysis of the normal polymorphic structures of DSPP and the 35 DSPP mutations that cause DD-II, DGI-II and DGI-III. Current evidence supports the interpretation that DSPP mutations cause these dominant forms of inherited dentin defects by a dominant-negative mechanism, and that the three disease designations reflect varying levels of severity of a common disease. We characterized the dental phenotypes and the mutation that causes them in two DGI-II families. The dental phenotypes in these two families were similar, and both showed linear vertical defects in the enamel. We interpreted these enamel defects as likely being developmental in origin, rather than being caused by damage to the weakened structure during or following eruption. In a recent case, a 5' DSPP mutation (c.53T>A, p.V18D) caused enamel defects that were evident on radiographs even before tooth eruption (38).
If enamel defects can occur when DSPP is mutated, is DSPP necessary for proper dental enamel formation? Because Dspp is not expressed by ameloblasts during dental enamel formation and no enamel defects are observed in Dspp null mice, the answer is almost certainly no. No dental phenotype is observed in Dspp heterozygous mice (where only one Dspp allele is deleted) (8). In contrast, all of the 35 reported disease-causing DSPP mutations show a dominant pattern of inheritance. Analyses of these mutations strongly suggest that the resulting dental phenotype is not caused by a loss of Dspp function, but is due to poorly understood pathological mechanisms that we have referred to simply as “dominant-negative effects”. As Dspp is transiently expressed by preameloblasts, we propose that enamel defects associated with DSPP mutations are probably the result of cell pathology induced by this transient expression.
Supplementary Material
Acknowledgments
We thank the family for their participation, and the Pediatric Dental Clinic at the University of Michigan for their cooperation. This investigation was supported in part by USPHS Research Grant DE015846 from the National Institute of Dental and Craniofacial Research (NIDCR), National Institutes of Health (NIH), Bethesda, MD 29892, and a Science Research Center grant to the Bone Metabolism Research Center (2010-0001741) funded by the Ministry of Education, Science, and Technology (MEST) of the Republic of Korea.
References
- 1.Simmer JP, Papagerakis P, Smith CE, Fisher DC, Rountrey AN, Zheng L, Hu JC. Regulation of dental enamel shape and hardness. J Dent Res. 2010;89:1024–1038. doi: 10.1177/0022034510375829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nanci A. Dentin-Pulp Complex. In: Nanci A, editor. Ten Cate's Oral Histology Development, Structure, and Function. Mosby; St. Louis, MO, USA: 2008. pp. 191–238. [Google Scholar]
- 3.Nagano T, Oida S, Ando H, Gomi K, Arai T, Fukae M. Relative levels of mRNA encoding enamel proteins in enamel organ epithelia and odontoblasts. J Dent Res. 2003;82:982–986. doi: 10.1177/154405910308201209. [DOI] [PubMed] [Google Scholar]
- 4.Bronckers AL, D'Souza RN, Butler WT, Lyaruu DM, van Dijk S, Gay S, Woltgens JH. Dentin sialoprotein: biosynthesis and developmental appearance in rat tooth germs in comparison with amelogenins, osteocalcin and collagen type-I. Cell Tissue Res. 1993;272:237–247. doi: 10.1007/BF00302729. [DOI] [PubMed] [Google Scholar]
- 5.Begue-Kirn C, Krebsbach PH, Bartlett JD, Butler WT. Dentin sialoprotein, dentin phosphoprotein, enamelysin and ameloblastin: tooth-specific molecules that are distinctively expressed during murine dental differentiation. Eur J Oral Sci. 1998;106:963–970. doi: 10.1046/j.0909-8836.1998.eos106510.x. [DOI] [PubMed] [Google Scholar]
- 6.Wright JT, Hart TC, Hart PS, Simmons D, Suggs C, Daley B, Simmer J, Hu J, Bartlett JD, Li Y, Yuan ZA, Seow WK, Gibson CW. Human and mouse enamel phenotypes resulting from mutation or altered expression of AMEL, ENAM, MMP20 and KLK4. Cells Tissues Organs. 2009;189:224–229. doi: 10.1159/000151378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meredith RW, Gatesy J, Cheng J, Springer MS. Pseudogenization of the tooth gene enamelysin (MMP20) in the common ancestor of extant baleen whales. Proc Biol Sci. 2011;278:993–1002. doi: 10.1098/rspb.2010.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki S, Sreenath T, Haruyama N, Honeycutt C, Terse A, Cho A, Kohler T, Muller R, Goldberg M, Kulkarni AB. Dentin sialoprotein and dentin phosphoprotein have distinct roles in dentin mineralization. Matrix Biol. 2009;28:221–229. doi: 10.1016/j.matbio.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hart PS, Hart TC. Disorders of human dentin. Cells Tissues Organs. 2007;186:70–77. doi: 10.1159/000102682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim JW, Simmer JP. Hereditary dentin defects. J Dent Res. 2007;86:392–399. doi: 10.1177/154405910708600502. [DOI] [PubMed] [Google Scholar]
- 11.Nieminen P, Papagiannoulis-Lascarides L, Waltimo-Siren J, Ollila P, Karjalainen S, Arte S, Veerkamp J, Walton VT, Kustner EC, Siltanen T, Holappa H, Lukinmaa PL, Alaluusua S. Frameshift mutations in dentin phosphoprotein and dependence of dentin disease phenotype on mutation location. J Bone Miner Res. 2011;26:873–880. doi: 10.1002/jbmr.276. [DOI] [PubMed] [Google Scholar]
- 12.Sreenath T, Thyagarajan T, Hall B, Longenecker G, D'Souza R, Hong S, Wright JT, MacDougall M, Sauk J, Kulkarni AB. Dentin sialophosphoprotein knockout mouse teeth display widened predentin zone and develop defective dentin mineralization similar to human dentinogenesis imperfecta type III. J Biol Chem. 2003;278:24874–24880. doi: 10.1074/jbc.M303908200. [DOI] [PubMed] [Google Scholar]
- 13.Mcknight DA, Suzanne Hart P, Hart TC, Hartsfield JK, Wilson A, Wright JT, Fisher LW. A comprehensive analysis of normal variation and disease-causing mutations in the human DSPP gene. Hum Mutat. 2008;29:1392–1404. doi: 10.1002/humu.20783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee SK, Hu JC, Lee KE, Simmer JP, Kim JW. A dentin sialophosphoprotein mutation that partially disrupts a splice acceptor site causes type II dentin dysplasia. J Endod. 2008;34:1470–1473. doi: 10.1016/j.joen.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee KE, Lee SK, Jung SE, Lee Z, Kim JW. Functional splicing assay of DSPP mutations in hereditary dentin defects. Oral Dis. 2011 Oct;17(7):690–5. doi: 10.1111/j.1601-0825.2011.01825.x. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 16.Yamakoshi Y, Nagano T, Hu JC, Yamakoshi F, Simmer JP. Porcine dentin sialoprotein glycosylation and glycosaminoglycan attachments. BMC Biochem. 2011;12:1–6. doi: 10.1186/1471-2091-12-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y, Lu Y, Chen S, Prasad M, Wang X, Zhu Q, Zhang J, Ball H, Feng J, Butler WT, Qin C. Key proteolytic cleavage site and full-length form of DSPP. J Dent Res. 2010;89:498–503. doi: 10.1177/0022034510363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Marschall Z, Fisher LW. Dentin sialophosphoprotein (DSPP) is cleaved into its two natural dentin matrix products by three isoforms of bone morphogenetic protein-1 (BMP1) Matrix Biol. 2010;29:295–303. doi: 10.1016/j.matbio.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsuchiya S, Simmer JP, Hu JC, Richardson AS, Yamakoshi F, Yamakoshi Y. Astacin proteases cleave dentin sialophosphoprotein (Dspp) to generate dentin phosphoprotein (Dpp) J Bone Miner Res. 2011;26:220–228. doi: 10.1002/jbmr.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jonsson M, Fredriksson S, Jontell M, Linde A. Isoelectric focusing of the phosphoprotein of rat-incisor dentin in ampholine and acid pH gradients. Evidence for carrier ampholyte-protein complexes. J Chromatogr. 1978;157:234–242. doi: 10.1016/s0021-9673(00)92338-0. [DOI] [PubMed] [Google Scholar]
- 21.Yamakoshi Y, Lu Y, Hu JC, Kim JW, Iwata T, Kobayashi K, Nagano T, Yamakoshi F, Hu Y, Fukae M, Simmer JP. Porcine dentin sialophosphoprotein: Length polymorphisms, glycosylation, phosphorylation, and stability. J Biol Chem. 2008;283:14835–14844. doi: 10.1074/jbc.M800633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song YL, Wang CN, Fan MW, Su B, Bian Z. Dentin phosphoprotein frameshift mutations in hereditary dentin disorders and their variation patterns in normal human population. J Med Genet. 2008;45:457–464. doi: 10.1136/jmg.2007.056911. [DOI] [PubMed] [Google Scholar]
- 23.Xiao S, Yu C, Chou X, Yuan W, Wang Y, Bu L, Fu G, Qian M, Yang J, Shi Y, Hu L, Han B, Wang Z, Huang W, Liu J, Chen Z, Zhao G, Kong X. Dentinogenesis imperfecta 1 with or without progressive hearing loss is associated with distinct mutations in DSPP. Nat Genet. 2001;27:201–204. doi: 10.1038/84848. [DOI] [PubMed] [Google Scholar]
- 24.Bhuvanagiri M, Schlitter AM, Hentze MW, Kulozik AE. NMD: RNA biology meets human genetic medicine. Biochem. 2010;430:365–377. doi: 10.1042/BJ20100699. [DOI] [PubMed] [Google Scholar]
- 25.Duffaud G, Inouye M. Signal peptidases recognize a structural feature at the cleavage site of secretory proteins. J Biol Chem. 1988;263:10224–10228. [PubMed] [Google Scholar]
- 26.Paetzel M, Karla A, Strynadka NC, Dalbey RE. Signal peptidases. Chem Rev. 2002;102:4549–4580. doi: 10.1021/cr010166y. [DOI] [PubMed] [Google Scholar]
- 27.Hon LS, Zhang Y, Kaminker JS, Zhang Z. Computational prediction of the functional effects of amino acid substitutions in signal peptides using a model-based approach. Hum Mutat. 2009;30:99–106. doi: 10.1002/humu.20798. [DOI] [PubMed] [Google Scholar]
- 28.Choo KH, Ranganathan S. Flanking signal and mature peptide residues influence signal peptide cleavage. BMC Bioinformatics. 2008;9:S15. doi: 10.1186/1471-2105-9-S12-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes AE, Ralston SH, Marken J, Bell C, MacPherson H, Wallace RG, van Hul W, Whyte MP, Nakatsuka K, Hovy L, Anderson DM. Mutations in TNFRSF11A, affecting the signal peptide of RANK, cause familial expansile osteolysis. Nat Genet. 2000;24:45–48. doi: 10.1038/71667. [DOI] [PubMed] [Google Scholar]
- 30.Kutz WE, Wang LW, Dagoneau N, Odrcic KJ, Cormier-Daire V, Traboulsi EI, Apte SS. Functional analysis of an ADAMTS10 signal peptide mutation in Weill-Marchesani syndrome demonstrates a long-range effect on secretion of the full-length enzyme. Hum Mutat. 2008;29:1425–1434. doi: 10.1002/humu.20797. [DOI] [PubMed] [Google Scholar]
- 31.Vijayasarathy C, Sui R, Zeng Y, Yang G, Xu F, Caruso RC, Lewis RA, Ziccardi L, Sieving PA. Molecular mechanisms leading to null-protein product from retinoschisin (RS1) signal-sequence mutants in X-linked retinoschisis (XLRS) disease. Hum Mutat. 2010;31:1251–1260. doi: 10.1002/humu.21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bleyer AJ, Zivna M, Hulkova H, Hodanova K, Vyletal P, Sikora J, Zivny J, Sovova J, Hart TC, Adams JN, Elleder M, Kapp K, Haws R, Cornell LD, Kmoch S, Hart PS. Clinical and molecular characterization of a family with a dominant renin gene mutation and response to treatment with fludrocortisone. Clin. 2010;74:411–422. doi: 10.5414/cnp74411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsukumo Y, Tsukahara S, Saito S, Tsuruo T, Tomida A. A novel endoplasmic reticulum export signal: proline at the +2-position from the signal peptide cleavage site. J Biol Chem. 2009;284:27500–27510. doi: 10.1074/jbc.M109.021592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barron MJ, Brookes SJ, Kirkham J, Shore RC, Hunt C, Mironov A, Kingswell NJ, Maycock J, Shuttleworth CA, Dixon MJ. A mutation in the mouse Amelx tri-tyrosyl domain results in impaired secretion of amelogenin and phenocopies human X-linked amelogenesis imperfecta. Hum Mol Genet. 2010;19:1230–1247. doi: 10.1093/hmg/ddq001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawasaki K, Weiss KM. Mineralized tissue and vertebrate evolution: the secretory calcium-binding phosphoprotein gene cluster. Proc Natl Acad Sci U S A. 2003;100:4060–4065. doi: 10.1073/pnas.0638023100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawasaki K, Weiss KM. SCPP gene evolution and the dental mineralization continuum. J Dent Res. 2008;87:520–531. doi: 10.1177/154405910808700608. [DOI] [PubMed] [Google Scholar]
- 37.Beattie ML, Kim JW, Gong SG, Murdoch-Kinch CA, Simmer JP, Hu JC. Phenotypic variation in dentinogenesis imperfecta/dentin dysplasia linked to 4q21. J Dent Res. 2006;85:329–333. doi: 10.1177/154405910608500409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee SK, Lee KE, Hwang YH, Kida M, Tsutsumi T, Ariga T, Park JC, Kim JW. Identification of the DSPP mutation in a new kindred and phenotype-genotype correlation. Oral Dis. 2011;17:314–319. doi: 10.1111/j.1601-0825.2010.01760.x. [DOI] [PubMed] [Google Scholar]
- 39.Rajpar MH, Koch MJ, Davies RM, Mellody KT, Kielty CM, Dixon MJ. Mutation of the signal peptide region of the bicistronic gene DSPP affects translocation to the endoplasmic reticulum and results in defective dentine biomineralization. Hum Mol Genet. 2002;11:2559–2565. doi: 10.1093/hmg/11.21.2559. [DOI] [PubMed] [Google Scholar]
- 40.Malmgren B, Lindskog S, Elgadi A, Norgren S. Clinical, histopathologic, and genetic investigation in two large families with dentinogenesis imperfecta type II. Hum Genet. 2004;114:491–498. doi: 10.1007/s00439-004-1084-z. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, Chen L, Liu J, Zhao Z, Qu E, Wang X, Chang W, Xu C, Wang QK, Liu M. A novel DSPP mutation is associated with type II dentinogenesis imperfecta in a Chinese family. BMC Med Genet. 2007;8:52. doi: 10.1186/1471-2350-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee S-K, Lee K-E, Cho I-K, Hyun H-K, Lee S-H, Kim J-W. IADR 2011. San Diego, CA, USA: Novel DSPP mutation and characterization of the mutational effect. Abstract 820. [Google Scholar]
- 43.Wang H, Hou Y, Cui Y, Huang Y, Shi Y, Xia X, Lu H, Wang Y, Li X. A novel splice site mutation in the dentin sialophosphoprotein gene in a Chinese family with dentinogenesis imperfecta type II. Mutat Res. 2009;662:22–27. doi: 10.1016/j.mrfmmm.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 44.Kim JW, Nam SH, Jang KT, Lee SH, Kim CC, Hahn SH, Hu JC, Simmer JP. A novel splice acceptor mutation in the DSPP gene causing dentinogenesis imperfecta type II. Hum Genet. 2004;115:248–254. doi: 10.1007/s00439-004-1143-5. [DOI] [PubMed] [Google Scholar]
- 45.Holappa H, Nieminen P, Tolva L, Lukinmaa PL, Alaluusua S. Splicing site mutations in dentin sialophosphoprotein causing dentinogenesis imperfecta type II. Eur J Oral Sci. 2006;114:381–384. doi: 10.1111/j.1600-0722.2006.00391.x. [DOI] [PubMed] [Google Scholar]
- 46.Kim JW, Hu JC, Lee JI, Moon SK, Kim YJ, Jang KT, Lee SH, Kim CC, Hahn SH, Simmer JP. Mutational hot spot in the DSPP gene causing dentinogenesis imperfecta type II. Hum Genet. 2005;116:186–191. doi: 10.1007/s00439-004-1223-6. [DOI] [PubMed] [Google Scholar]
- 47.Song Y, Wang C, Peng B, Ye X, Zhao G, Fan M, Fu Q, Bian Z. Phenotypes and genotypes in 2 DGI families with different DSPP mutations. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2006;102:360–374. doi: 10.1016/j.tripleo.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 48.Kida M, Tsutsumi T, Shindoh M, Ikeda H, Ariga T. De novo mutation in the DSPP gene associated with dentinogenesis imperfecta type II in a Japanese family. Eur J Oral Sci. 2009;117:691–694. doi: 10.1111/j.1600-0722.2009.00683.x. [DOI] [PubMed] [Google Scholar]
- 49.Lee SK, Lee KE, Jeon D, Lee G, Lee H, Shin CU, Jung YJ, Lee SH, Hahn SH, Kim JW. A novel mutation in the DSPP gene associated with dentinogenesis imperfecta type II. J Dent Res. 2009;88:51–55. doi: 10.1177/0022034508328168. [DOI] [PubMed] [Google Scholar]
- 50.Zhang X, Zhao J, Li C, Gao S, Qiu C, Liu P, Wu G, Qiang B, Lo WH, Shen Y. DSPP mutation in dentinogenesis imperfecta Shields type II. Nat Genet. 2001;27:151–152. doi: 10.1038/84765. [DOI] [PubMed] [Google Scholar]
- 51.Bai H, Agula H, Wu Q, Zhou W, Sun Y, Qi Y, Latu S, Chen Y, Mutu J, Qiu C. A novel DSPP mutation causes dentinogenesis imperfecta type II in a large Mongolian family. BMC Med Genet. 2010;11:23. doi: 10.1186/1471-2350-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee KE, Kang HY, Lee SK, Yoo SH, Lee JC, Hwang YH, Nam KH, Kim JS, Park JC, Kim JW. Novel dentin phosphoprotein frameshift mutations in dentinogenesis imperfecta type II. Clin Genet. 2011;79:378–384. doi: 10.1111/j.1399-0004.2010.01483.x. [DOI] [PubMed] [Google Scholar]
- 53.McKnight DA, Simmer JP, Hart PS, Hart TC, Fisher LW. Overlapping DSPP mutations cause dentin dysplasia and dentinogenesis imperfecta. J Dent Res. 2008;87:1108–1111. doi: 10.1177/154405910808701217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.