Summary
Activated RAS promotes dimerization of members of the RAF kinase family1-3. ATP-competitive RAF inhibitors activate ERK signaling4-7 by transactivating RAF dimers4. In melanomas with mutant BRAF(V600E), levels of RAS activation are low and these drugs bind to BRAF(V600E) monomers and inhibit their activity. This tumor-specific inhibition of ERK signaling results in a broad therapeutic index and RAF inhibitors have remarkable clinical activity in patients with melanomas that harbor mutant BRAF(V600E)8. However, resistance invariably develops. Here, we identify a novel resistance mechanism. We find that a subset of cells resistant to vemurafenib (PLX4032, RG7204) express a 61kd variant form of BRAF(V600E) that lacks exons 4-8, a region that encompasses the RAS-binding domain. p61BRAF(V600E) exhibits enhanced dimerization in cells with low levels of RAS activation, as compared to full length BRAF(V600E). In cells in which p61BRAF(V600E) is expressed endogenously or ectopically, ERK signaling is resistant to the RAF inhibitor. Moreover, a mutation that abolishes the dimerization of p61BRAF(V600E) restores its sensitivity to vemurafenib. Finally, we identified BRAF(V600E) splicing variants lacking the RAS-binding domain in the tumors of six of 19 patients with acquired resistance to vemurafenib. These data support the model that inhibition of ERK signaling by RAF inhibitors is dependent on levels of RAS-GTP too low to support RAF dimerization and identify a novel mechanism of acquired resistance in patients: expression of splicing isoforms of BRAF(V600E) that dimerize in a RAS-independent manner.
RAF inhibitors have remarkable clinical activity in mutant BRAF melanomas that is limited by acquisition of drug resistance8. To identify novel mechanisms of resistance, we generated cell lines resistant to vemurafenib by exposing the BRAF-mutant (V600E) melanoma cell line SKMEL-239 to a high dose of drug (2μM). At this concentration, vemurafenib effectively inhibited ERK signaling and induced cell cycle arrest and cell death (Fig. 1a-c, Supplementary Fig. 2a and data not shown (DNS)). Five independent vemurafenib-resistant cell populations were generated after approximately 2 months of continuous drug exposure (Fig. 1a). We chose this approach rather than one of gradual adaptation to increasing concentrations of drug since it more closely represents the clinical situation8.
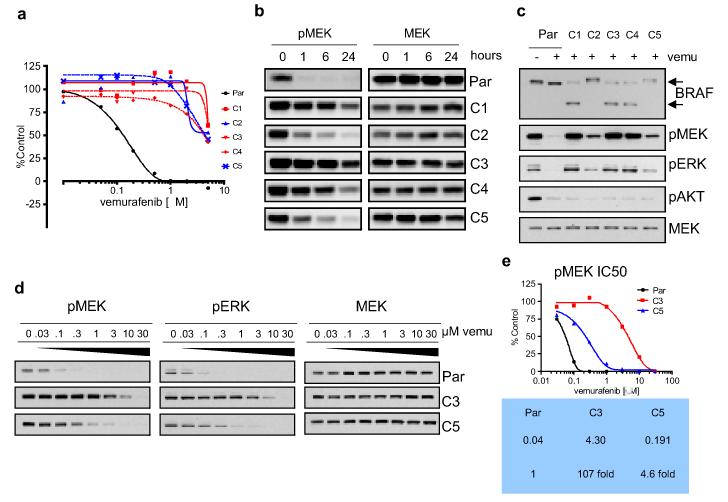
Figure 1. Resistance to the RAF inhibitor vemurafenib (PLX4032) is associated with failure of the drug to inhibit ERK signaling.
a. Vemurafenib IC50 curves (at 5 days) for the SKMEL-239 parental cell line and five vemurafenib-resistant clones. b. Effects of 2μM vemurafenib on ERK signaling in parental (Par) and resistant clones (C1-5). c. Western blot for components of the ERK and AKT signaling pathways in parental and resistant clones (2μM PLX4032/24 hours). d. Dose-response of pMEK and pERK downregulation at 1 hour to increasing concentrations of vemurafenib in parental and two representative resistant clones (C3 and C5). e. Graphic representation of the chemiluminescent signal intensities from 1d and IC50s for inhibition of MEK phosphorylation by vemurafenib in the parental and C3 and C5 clones.
Resistance of SKMEL-239 cells to vemurafenib was associated with decreased sensitivity of ERK signaling to the drug (Fig. 1b, c, Supplementary Fig. 2b). Analysis revealed the presence of two distinct classes of resistant clones. In the first, exemplified by the C3 clone, the IC50 for pMEK inhibition was more than 100-fold higher than that of the parental cell line (Fig. 1d, e). Despite a similar degree of resistance to the anti-proliferative and pro-apoptotic effects of the drug, the second class of clones, exemplified by clone C5, demonstrated only a modest increase in pMEK IC50 (4.5-fold higher than the parental cell line). All five resistant clones retained sensitivity to the MEK inhibitor PD03259019, albeit at slightly higher doses (Supplementary Fig. 3a, b).
Analysis of DNA and cDNA derived from the five resistant clones showed that all retained expression of BRAF(V600E) (Supplementary Fig. 4a, b). We did not detect mutation in BRAF at the gatekeeper site10, RAS mutation, upregulation of receptor tyrosine kinase activity or COT overexpression (Supplementary Fig. 5a, b and DNS). Analysis of BRAF protein expression showed that each of the resistant clones expressed a 90kd band that co-migrated with the band observed in parental cells. In the C1, C3 and C4 clones, a new band was also identified, at an approximate molecular weight of 61kd (p61BRAF(V600E), Fig. 1c, Supplementary Fig. 2b). No band of this size was detected in parental SKMEL-239 cells or in a panel of 22 other melanoma cell lines (Supplementary Fig. 6).
PCR analysis of cDNA revealed the expected single transcript of 2.3kb, representing full-length BRAF in parental cells and two transcripts of 2.3kb and 1.7kb in C3 cells. The 1.7kb product was a BRAF transcript that contained the V600E mutation and an in-frame deletion of exons 4-8 (Fig. 2a and Supplementary Fig. 7). This 1.7kb transcript is predicted to encode a protein of 554 amino acids (M.W. 61kd), consistent with the lower BRAF band detected by immunoblotting. Exons 4-8 include domains critical for RAF activation, most notably, the RAS-binding domain (RBD) and the cysteine-rich domain (CRD)3. Analogous deletions in wild-type BRAF and CRAF promote RAF dimerization and render RAS activity dispensable for this process1,4. The 61kd BRAF variant identified in C3 was also detected in clones C1 and C4 by qPCR, with a primer that anneals specifically to the exons 3/9 junction (Supplementary Fig. 8). Inspection of the BRAF locus on chromosome 7q34 by array CGH data suggested no evidence of an intragenic somatic deletion within the BRAF gene.
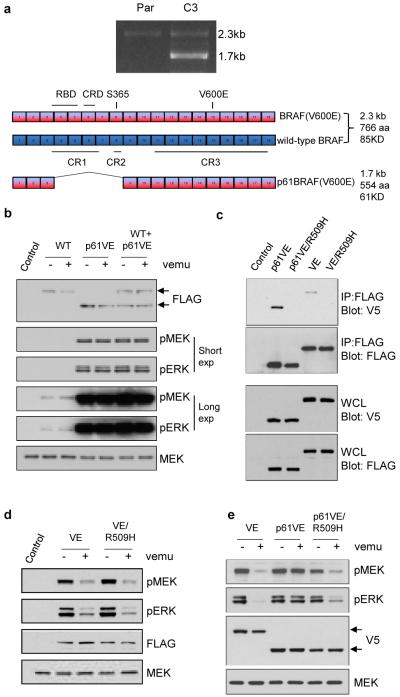
Figure 2. A BRAF(V600E) variant that lacks exons 4-8 is resistant to the RAF inhibitor vemurafenib.
a. PCR analysis of BRAF in cDNA from parental (P) and C3 cells. Sequencing of the 1.7kb product expressed in the C3 clones but not in parental cells revealed an in frame deletion of five exons (4-8) in cis with the V600E mutation. Abbreviations: CR1: Conserved Region 1, CR2: Conserved Region 2, CR3: Conserved Region 3, RBD: RAS-binding domain, CRD: Cystine-Rich Domain. b. Full length wild-type BRAF and the 1.7kb/61kd splice variant of BRAF(V600E) were expressed in 293H cells. The effect of vemurafenib (2μM for 1 hour) on ERK signaling in the presence of p61BRAF(V600E) was analyzed by western blot for pMEK and pERK. c. To compare levels of dimerization, 293H cells co-expressing FLAG tagged and V5-tagged p61BRAF(V600E), full length BRAF(V600E) and the corresponding dimerization-deficient mutants p61BRAF(V600E/R509H) and BRAF(V600E/R509H) were lysed followed by immunoprecipitation with FLAG antibody. Western blotting with V5 or FLAG antibodies was performed as indicated. d. Comparison of MEK/ERK activation and sensitivity of ERK signaling to vemurafenib (2μM for 1 hour) in 293H cells expressing either Flag-tagged BRAF(V600E) or the dimerization mutant Flag-tagged BRAF(V600E/R509H). e. Constructs expressing V5-tagged BRAF(V600E), p61BRAF(V600E) or the dimerization mutant p61BRAF(V600E/R509H) were transfected into 293H cells and treated with DMSO or 2μM vemurafenib for 1 hour.
The 1.7kb transcript was cloned into an expression vector and expressed in 293H cells, alone or together with full-length wild-type BRAF. ERK signaling was resistant to vemurafenib when p61BRAF(V600E) was ectopically expressed (Fig 2b). Expression of p61BRAF(V600E) in parental SKMEL-239 cells or in HT-29 (BRAF(V600E)) cells also resulted in failure of vemurafenib to effectively inhibit ERK signaling (Supplementary Fig. 9a, b). To test whether ERK signaling in C3 cells was dependent on p61BRAF(V600E), we designed siRNAs directed against either the 3/9 splice junction or a region within the exon 4-8 deletion to selectively suppress the expression of p61BRAF(V600E) or full-length BRAF, respectively. In parental cells, ERK signaling was inhibited by knockdown of full-length BRAF(V600E) (Supplementary Fig. 10a). In C3 cells, phosphorylation of MEK, cyclin D1 expression and cell growth were inhibited upon knockdown of p61BRAF(V600E) but not of full-length wild type BRAF, ARAF or CRAF (Supplementary Fig. 10b, c). Moreover, in C3 cells in which the expression of full-length BRAF or CRAF was knocked down, ERK signaling remained resistant to vemurafenib (Supplementary Fig. 10d).
Vemurafenib inhibits the kinase activity of RAF immunoprecipitated from cells, but activates intracellular RAF in BRAF wild-type cells4. This suggests that the conditions required for transactivation in vivo are not recapitulated in the in vitro assay. We tested whether p61BRAF(V600E) is also sensitive to this inhibitor in vitro. Although the in vitro activity of p61BRAF(V600E) was slightly higher than full-length BRAF(V600E), similar concentrations of vemurafenib caused their inhibition in vitro (Supplementary Fig. 11). These data indicate that resistance of p61BRAF(V600E) to vemurafenib is not due to its inability to bind the inhibitor.
It has been shown that the N-terminus of RAF negatively regulates the C-terminal catalytic domain11 and that truncation of the N-terminus results in constitutive dimerization of the protein in the absence of activated RAS1. To ask whether deletion of exons 4-8 promotes dimerization of p61BRAF(V600E), we co-expressed two constructs encoding the same protein (either p61BRAF(V600E) or full-length BRAF(V600E)) but with different tags (Flag or V5) and then immunoblotted for V5 after immunoprecipating FLAG. As shown in Fig. 2c, dimerization of p61BRAF(V600E) was significantly elevated compared to that of full-length BRAF(V600E). The R509 residue is within the BRAF dimerization interface. Mutation of this residue to a histidine significantly diminishes dimerization of wild-type BRAF and results in loss of its catalytic activity in cells4,12. However, full length BRAF(V600E/R509H) expressed in 293H cells retained its ability to fully activate ERK signaling and remained sensitive to vemurafenib (Fig. 2d). Moreover, BRAF(V600E/R509H) fully activated ERK signaling when expressed in either BRAF-null or ARAF/CRAF-null MEFs (Supplementary Fig. 12a, b). These results show that, in contrast to wild-type BRAF, BRAF(V600E) can signal as a monomer and that active RAS and dimerization are not necessary for its activation.
Our model implies that, in tumors with BRAF(V600E), elevation of RAS-GTP or alterations that cause increased RAF dimerization in the absence of RAS activation will confer resistance to RAF inhibitors4,13. To test whether resistance mediated by p61BRAF(V600E) was the result of elevated dimer formation, we introduced the R509H dimerization-deficient mutation into p61BRAF(V600E). In 293H cells expressing p61BRAF(V600E), phosphorylation of ERK was elevated and was insensitive to vemurafenib (Fig. 2e). ERK activity was also elevated in cells expressing p61BRAF(V600E/R509H), but to a slightly lesser degree. p61BRAF(V600E/R509H) showed impaired dimerization, confirming that the R509H mutation located within the dimerization interface disrupts the formation of p61RAF(V600E) dimers (Fig. 2c and Supplementary Figure 13). Finally, this monomeric p61BRAF(V600E/R509H) was sensitive to RAF inhibitors; in cells ectopically expressing this mutant, ERK signaling was inhibited by vemurafenib (Fig. 2e). Thus, the R509H mutation by impairing dimerization of p61BRAF(V600E) restores sensitivity to the RAF inhibitor.
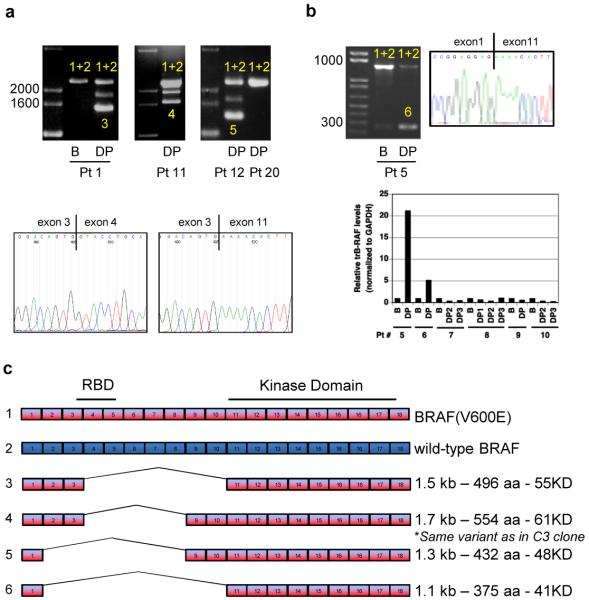
To determine whether BRAF variants can account for clinical resistance to RAF inhibitors, we analyzed tumors from nineteen melanoma patients with acquired resistance to vemurafenib. PCR analysis of cDNA from pre-treatment samples showed a single band of the expected size (2.3kb) which was sequenced and confirmed to include both BRAF(V600E) and wild-type BRAF transcripts (Fig. 3A, B and DNS). We identified two PCR products in six of the post-treatment progression samples, including three with matching pre-treatment samples. The shorter PCR products encoded BRAF(V600E) transcripts lacking exons 4-10 (pt 1), exons 4-8, (pt 11, identical to the variant identified in the resistant cell lines), exons 2-8 (pt 12), or exons 2-10 (pts 5, 6 and 19) (Fig. 3a-c, Supplementary Table 1). Mutations in NRAS, were identified in 4 of 19 progression samples and were found to be mutually exclusive with BRAF splicing variants. BRAF splicing variants were not detected in two samples derived from patients with intrinsic resistance (pt 20 shown) or in 27 additional melanomas resected from vemurafenib-naïve patients (Fig. 3a, Supplementary Table 1 and DNS). Finally, we did not detect significant levels of BRAF splicing variants in melanoma cell lines and tumors that have not been exposed to RAF inhibitors. However, the assay employed cannot exclude the possibility that such variants may be expressed in small amounts prior to drug selection and subsequently selected under conditions of continuous treatment with the drug.
Figure 3. Identification of splice variants of BRAF(V600E) in human tumors resistant to vemurafenib.
a. PCR analysis of cDNA derived from tumor samples from patients treated with vemurafenib. In samples with only one band (full-length BRAF), both BRAF(V600E) and wild-type BRAF (1+2) were detected. In resistant tumor samples expressing shorter transcripts, the shorter transcript was a splicing variant of BRAF(V600E) (3, 4, 5). The figure shows samples from three patients with acquired resistance to PLX4032: baseline (B) and disease progression (DP) samples from patient 1 and post-treatment samples from patients 11 and 12. A tumor sample from a patient with de novo resistance to vemurafenib (patient 20) is also shown. The intermediate band in samples expressing splicing variants (Pts 1, 11, 12) is an artifact of the PCR reaction resulting from switching between two very similar templates. Representative Sanger sequencing traces showing the junction between exons 3 and 11 in the DP sample from patient 1 compared to the full-length transcript derived from the baseline pre-treatment sample. b. As in A, baseline (B) and disease progression (DP) samples from a patient with an exon 2-10 deletion. RNA/cDNA levels of the exon 2-10 deletion were determined by qPCR using an exon 1/11 junction primer. The data are shown as the average of duplicates and expressed as relative levels between patient-matched samples c. Exon organization of the splicing variants found in tumors from six patients that initially responded and then progressed on vemurafenib.
In tumors from patients that have been analyzed, resistance to vemurafenib is frequently associated with inability of the drug to inhibit ERK signaling14. Our model suggests that this can be due to increased RAF dimer formation in the cell4. This can happen in at least two ways: increasing RAS-GTP levels and induction of RAS-independent dimerization (Supplementary Figure 1a, b). NRAS mutation has recently been reported in resistant tumors15. Other mechanisms of resistance to RAF inhibitors in model systems and in patients have also been reported, including activation of receptor tyrosine kinases, COT overexpression and MEK1 mutation 15-18. We now report a lesion in patient tumors that results in RAF inhibitor resistance by inducing increased, RAS-independent dimerization.
Expression of BRAF(V600E) splicing variants is the first resistance mechanism identified that involves a structural change in BRAF. In each case, the alternative splicing forms identified in the cell lines and patients were in frame and confined to the mutant BRAF allele. This suggests that generation of the splicing variants is likely due to a mutation or epigenetic change that affects BRAF splicing and not to a loss of global splicing fidelity19. The identification of BRAF variants lacking the RAS-binding domain in six of nineteen patients with acquired resistance suggests that this mechanism is clinically important. Acquired resistance mediated by BRAF(V600E) splicing variants is due to insensitivity of the enzyme to RAF inhibitors. These tumors should retain sensitivity to inhibitors of downstream components of the pathway such as MEK, which was indeed the case (Supplementary Fig. 3). Therefore, MEK inhibitors used in combination with vemurafenib could delay or prevent resistance by this mechanism.
Supplementary Material
Methods Summary.
Vemurafenib20 (PLX4032) was obtained from Plexxikon Inc. PD0325901 was synthesized in the MSKCC Organic Synthesis Core Facility by O. Ouerfelli. Flag-tagged BRAF constructs have been described previously4. All other plasmids were created using standard cloning methods, with pcDNA3.1 (Invitrogen) as a vector. Mutations were introduced using the site-directed Mutagenesis Kit (Stratagene). The C1-5 vemurafenib-resistant cells were generated by continuous exposure of parental SKMEL-239 cells to 2μM of drug until the emergence of resistant colonies. Single cell cloning was then performed prior to biological characterization.
For cDNA preparation, the Superscript III First-Strand Synthesis kit (Invitrogen) was used. Sanger sequencing of the products was performed by Genewiz. For qPCR analysis, cDNA synthesis was carried out with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qPCR was performed with the iQ SYBR Green RT-PCR Super Mix (BioRad) and the C1000 Thermal Cycler (BioRad). The comparative Ct method was employed to quantify transcripts and delta Ct was measured in triplicate. Sequences of all primers used are available upon request.
Melanoma tumor specimens from patients treated with vemurafenib were collected on an IRB-approved protocols and were flash frozen immediately after resection or biopsy. To determine tumor content, 5 μm sections were cut, stained with hematoxylin and eosin, and scored by a pathologist. If the specimen had >70% tumor content (excluding necrosis), the remainder of the frozen tumor was homogenized using a Bullet Blender (Next Advance, Inc.) with 0.9-2 mm stainless steel beads for 5 min at a speed setting of 10. RNA was then extracted from the tumor homogenate using the RNeasy Mini Kit (Invitrogen) and quantified.
Acknowledgements
We are grateful to Manuela Baccarini and Catrin Pritchard for the RAF knock-out MEFs. We would like to thank Kevan Shokat, Chao Zhang, Sarat Chandarlapaty, Christine Pratilas and Kenneth Robzyk for useful discussions, Cailian Liu for her technical expertise and Tony J. Riley from the Media Services, MSKCC. We also thank Donald Hucks of the Vanderbilt Innovative Translational Research Shared Resource (ITR) for technical assistance, Pamela L. Lyle for scoring the tumor sections, and Thinle Chodon for assistance with tumor procurement. The ITR is supported by the Vanderbilt-Ingram Cancer Center and the TJ Martell Foundation. This work has been funded by the National Institutes of Health (R.S.L., N.R., D.B.S), the Beene Foundation (D.B.S), the Melanoma Research Alliance (K.T.F., J.A.S. P.B.C., R.S.L., N.R., D.B.S.) and the STARR Foundation (N.R., D.B.S). R.S.L. was supported by the Burroughs Wellcome Fund, American Skin Association, Joint Center for Translational Medicine, Sidney Kimmel Foundation, and Stand Up to Cancer. T.M. was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), NCI, Center for Cancer Research and P.B.C was supported in part by the Danny Federici Melanoma Fund. P.I.P. was supported by T32 CACA062948-15.
Appendix
Methods
Compounds
Vemurafenib (PLX4032) was obtained from Plexxikon Inc. PD0325901 was synthesized in the MSKCC Organic Synthesis Core Facility by O. Ouerfelli. Drugs were dissolved in DMSO and stored at −20°C.
Cell proliferation and cell cycle analysis
All melanoma cell lines were generated by A. Houghton (MSKCC) or obtained from ATCC. 293H cells were obtained from Invitrogen. Cells were maintained in DMEM (293H and MEFs), or RPMI (all other cell lines) supplemented with 2mM glutamine, antibiotics and 10% fetal bovine serum. We confirmed by DNA fingerprinting21 that all PLX4032-resistant, SKMEL-239 clones were derived from the same patient, thus excluding the possibility of cross contamination (Supplemental Table 2). For proliferation assays, cells were plated in 6 well plates and 24 hours later were treated with varying concentrations of inhibitors as indicated. IC50 values were calculated using Graph Pad Prism v.5. For cell cycle and apoptosis studies, cells were seeded in 6 well dishes the day prior to drug treatment. For analysis, both adherent and floating cells were harvested and stained with ethidium bromide as described previously 22.
Western blotting and receptor tyrosine kinase (RTK) arrays
Western blot analysis was performed as previously described9. The following antibodies were used: p217/p221-MEK (pMEK), p202/p204-ERK (pERK), p338CRAF, p473AKT, MEK (Cell Signaling), V5 tag (Invitrogen), ARAF, BRAF, cyclin D1, p27, COT (Santa Cruz), CRAF (BD Biosciences), Flag tag, β-actin (Sigma). For immunoprecipitations of tagged proteins: anti-Flag M2 affinity gel (Sigma). The Human Phospho-RTK array Kit (R&D Systems) was utilized to detect kinase activation within a panel of RTKs. Briefly, cells were plated in 10 cm dishes and harvested after 24 hours. Following lysis, 500 μg of lysate was applied to a membrane-anchored RTK array and incubated at 4°C for 24 hours. Membranes were exposed to chemiluminescent reagents and images captured using the ImageQuant LAS 4000 instrument (GE HealthCare).
Plasmids/Trasfections
Flag-tagged BRAF constructs have been described previously4. All other plasmids were created using standard cloning methods, with pcDNA3.1 (Invitrogen) as a vector. Mutations were introduced using the site-directed Mutagenesis Kit (Stratagene). For transfection studies, cells were seeded into 35mm or 100mm plates and transfected the following day using Lipofectamine 2000 (Invitrogen). Cells were collected 24 hours later for subsequent analysis.
Immunoprecipitations and kinase assays
Cells were lysed in lysis buffer (50mM Tris, pH7.5, 1% NP40, 150mM NaCl, 10% glycerol, 1mM EDTA) supplemented with protease and phosphatase inhibitor cocktail tablets (Roche). Immunoprecipitations were performed at 4°C for 4h, followed by three washes with lysis buffer and, in cases of subsequent kinase assay, one final wash with kinase buffer (25mM Tris, pH 7.5, 10mM MgCl2). Kinase assays were conducted in the presence of 200μM ATP at 30°C for 20min with inactive MEK(K97R) (Millipore) as a substrate. The kinase reaction was terminated by adding sample buffer and boiling. Kinase activity was determined by immunoblotting for pMEK.
siRNA knockdown
In order to selectively knock down p61BRAF(V600E) or full-length BRAF, siRNA duplexes were designed to target the junction between exons 3-9 (JC-1 and JC-2) or sequences within exons 4-8 (ex[4-8]-1 and ex[4-8]-2. The sequences are the following: JC-1: GGACAGUGGACUUGAUUAGUU, JC-2: AGGACAGUGGACUUGAUUAUU, ex[4-8]-1: ACUGAUAUUUCCUGGCUUAUU, ex[4-8]-2: CUGUCAAACAUGUGGUUAUUU. To knock down ARAF and CRAF we used siRNA pools. All siRNA duplexes were from Dharmacon and transfections were carried out with Lipofectamine 2000 (Invitrogen) at a final siRNA concentration of 50nM, according to the manufacturer’s instructions. 72 hours later, cells were either counted to estimate cell growth, or subjected to immunoblot analysis.
Footnotes
Contributions
P.I.P., M.J., T.G.G, A.R., R.S.L., N.R., and D.B.S. designed experiments and analyzed data. P.I.P, Y.P., M.J., X.K., C.N., G.M., H.S., M.A., B.T., M.T.G. M.S., K.D., and T.M. performed experiments and analyzed data. J.A.W, K.T.F, M.C.K., J.A.S., P.B.C., and R.S.L. provided tumors for analysis. All authors contributed to the writing of the paper.
Competing financial interests
J.A.S., P.B.C., N.R. and D.B.S. have participated on an advisory board for Roche. J.A.S. and P.B.C. have received clinical trial research funding from Roche. P.B.C. has received research support and served on an advisory board for Genentech. A.R. has received honorarium serving on the advisory boards of Roche-Genentech. N.R. and D.B.S. have received research funding from Astra-Zeneca.
References
- 1.Weber CK, Slupsky JR, Kalmes HA, Rapp UR. Active Ras induces heterodimerization of cRaf and BRaf. Cancer Res. 2001;61:3595–3598. [PubMed] [Google Scholar]
- 2.Rushworth LK, Hindley AD, O’Neill E, Kolch W. Regulation and role of Raf-1/B-Raf heterodimerization. Mol Cell Biol. 2006;26:2262–2272. doi: 10.1128/MCB.26.6.2262-2272.2006. doi:26/6/2262 [pii] 10.1128/MCB.26.6.2262-2272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nat Rev Mol Cell Biol. 2004;5:875–885. doi: 10.1038/nrm1498. doi:nrm1498 [pii] 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- 4.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. doi:10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heidorn SJ, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. doi:S0092-8674(09)01626-2 [pii] 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatzivassiliou G, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. doi:nature08833 [pii] 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 7.Joseph EW, et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proc Natl Acad Sci U S A. 2010;107:14903–14908. doi: 10.1073/pnas.1008990107. doi:1008990107 [pii] 10.1073/pnas.1008990107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flaherty KT, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. doi:10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Solit DB, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–362. doi: 10.1038/nature04304. doi:nature04304 [pii] 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whittaker S, et al. Gatekeeper mutations mediate resistance to BRAF-targeted therapies. Sci Transl Med. 2010;2:35ra41. doi: 10.1126/scitranslmed.3000758. doi:2/35/35ra41 [pii] 10.1126/scitranslmed.3000758. [DOI] [PubMed] [Google Scholar]
- 11.Cutler RE, Jr., Stephens RM, Saracino MR, Morrison DK. Autoregulation of the Raf-1 serine/threonine kinase. Proc Natl Acad Sci U S A. 1998;95:9214–9219. doi: 10.1073/pnas.95.16.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajakulendran T, Sahmi M, Lefrancois M, Sicheri F, Therrien M. A dimerization- dependent mechanism drives RAF catalytic activation. Nature. 2009;461:542–545. doi: 10.1038/nature08314. doi:nature08314 [pii] 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 13.Poulikakos PI, Rosen N. Mutant BRAF melanomas--dependence and resistance. Cancer Cell. 2011;19:11–15. doi: 10.1016/j.ccr.2011.01.008. doi:10.1016/j.ccr.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 14.McArthur G, et al. Molecular analyses from a phase I trial of vemurafenib to study mechanism of action (MOA) and resistance in repeated biopsies from BRAF mutation-positive metastatic melanoma patients (pts) J Clin Oncol. 2011;29(suppl) abstr 8502. [Google Scholar]
- 15.Nazarian R, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. doi:10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johannessen CM, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. doi:10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villanueva J, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. doi:S1535-6108(10)00484-8 [pii] 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagle N, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. doi:10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre- mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. doi:S0092-8674(10)01378-4 [pii] 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bollag G, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. doi:nature09454 [pii] 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janakiraman M, et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res. 2010;70:5901–5911. doi: 10.1158/0008-5472.CAN-10-0192. doi:0008-5472.CAN-10-0192 [pii] 10.1158/0008-5472.CAN-10-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nusse M, Beisker W, Hoffmann C, Tarnok A. Flow cytometric analysis of G1- and G2/M- phase subpopulations in mammalian cell nuclei using side scatter and DNA content measurements. Cytometry. 1990;11:813–821. doi: 10.1002/cyto.990110707. doi:10.1002/cyto.990110707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.