Abstract
The past few years of research in human evolutionary genetics have provided novel insights and questions regarding how human adaptations to recent selective pressures have taken place. Here, we review the advances most relevant to understanding human evolution in response to pathogen-induced selective pressures. Key insights come from theoretical models of adaptive evolution, particularly those that consider spatially structured populations, and from empirical population genomic studies of adaptive evolution in humans. We also review the CCR5-Δ32 HIV resistance allele as a case study of pathogen resistance in humans. Taken together, the results make clear that the human response to pathogen-induced selection pressures depends on a complex interplay between the age of the pathogen, the genetic basis of potential resistance phenotypes, and how population structure impacts the adaptive process in humans.
Keywords: human adaptation, pathogen resistance, population structure, CCR5-Δ32
1. Introduction
A major aspect of human evolutionary biology has been the emergence of novel traits in humans—for example, the evolution of bipedalism, language capability, expanded brain size, manual dexterity and behavioural complexity. At the same time, much of human evolution has probably been less overt and largely in response to the myriads of pathogens that have faced humans in the past and continue to confront humans today. Considerations of the impacts of pathogen-driven evolution date back to Haldane's well-known paper suggesting the importance of infectious disease as a selective agent and driver of evolution [1,2]. Decades of research since Haldane's paper have made clear infectious disease should be an especially strong pressure on recent evolution in humans [3–5]. Most importantly, since the advent of agriculture in the Neolithic era, the increased density of human populations and rate of contact with animal reservoirs have heightened the rate of emergence of novel pathogens [6]. The mortality associated with pathogen epidemics is obvious and has been dramatic even over short time-scales of human history, so any heritable variation that might affect pathogen resistance is expected to have been under natural selection to some degree.
Variants that affect susceptibility to pathogens somehow perturb human physiology in a way that is relevant to the outcome of disease, and so discovering these variants and understanding their mechanisms is a potential trove of medical insight [7,8]. Protective alleles provide a hint for a potential treatment pathway; ideally, a therapeutic intervention can be designed that mimics the physiological perturbations brought about by the resistance allele. As an example, the CCR5-Δ32 allele (explained further below), which confers resistance to HIV in homozygous carriers [9–13], is now forming the basis of trial therapies based on stem cell transplantation. A recent paper [14] reports on an infected patient who received blood stem cells from a Δ32/Δ32 homozygous donor. After transplantation, this patient reconstituted his T cells to normal levels, and HIV viral levels were undetectable despite the patient not taking anti-retroviral therapies for a period of more than 3.5 years.
Discovering resistance variants is possible using the tools of genetic epidemiology, such as genome-wide association studies (GWAS) or linkage mapping in family-based designs [15–18]. The effectiveness of such approaches depends crucially on the spectrum of frequencies at which disease resistance alleles are found in populations. Human geneticists have an evolving paradigm for the genetic basis of complex disease traits (such as diabetes, cancer susceptibility and heart disease) that increasingly suggests the genetic basis of these traits may be a large number of rare variants of modest effect [19]. This paradigm is motivated by the so-called ‘missing-heritability’ problem, which points towards an insufficient number of common variants being discovered by recent large-scale GWAS [20], and evolutionary considerations which recognize that disease-causing alleles may have been under negative selection pressures, pushing their frequencies downward [21].
Given the unique positive selection pressures expected for disease resistance alleles, it is not clear what one should expect for the spectrum of allele frequencies for resistance alleles. For example, should resistance alleles be found as common variants (greater than 5% frequency in populations) and hence be amenable to mapping using genome-wide association approaches or should they be much more rare (less than 5% frequency) and hence investigated using re-sequencing-based approaches? In addition, it is relevant to ask whether they will be found on a single haplotype or several haplotypes.
The goal of this paper is to review some recent theoretical and empirical results that might be insightful with respect to the frequency and geographical distribution of pathogen-resistance alleles in humans. In particular, the past few years of research in human evolutionary genetics have provided novel perspectives and questions regarding how humans have adapted at a genetic level to recent selective pressures, such as pathogens. Key insights come from theoretical models of adaptive evolution, particularly those that consider spatially structured populations, and from empirical population genomic studies of adaptive evolution in humans.
2. Models of adaptive evolution: hard and soft sweeps
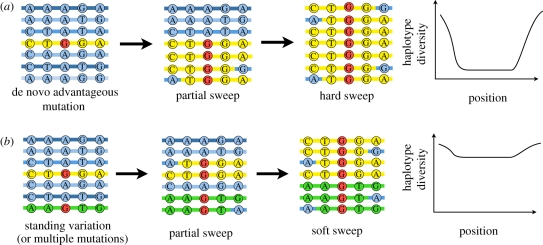
In evolutionary genetic studies of natural selection, two major outcomes of natural selection are now distinguished, hard sweeps and soft sweeps [22]. Hard sweeps are the outcome of a single instance of an advantageous mutation arising and spreading through a population. Because it arises from a single instance of mutation, the advantageous variant necessarily begins on a single chromosome, which defines a unique ancestral advantageous haplotype (figure 1). As this ancestral haplotype increases in frequency, or ‘sweeps’ through the population, it brings along neutral or nearly neutral genetic variants found on the ancestral haplotype. This impact on linked variation owing to selection has been understood for some time and was first called genetic hitchhiking by Maynard Smith & Haigh [23].
Figure 1.
Hard and soft sweeps. (a) Hard sweeps. A de novo advantageous mutation (red G allele) arises on a single haplotype (marked in yellow). The advantageous allele increases in frequency and neutral or nearly neutral genetic variants at nearby locations also increase in frequency (genetic hitchhiking). As selection proceeds, recombination shuffles alleles off the ancestral haplotype (marked in yellow), and as a result after the completion of hard sweeps (fixation of the advantageous mutation), there is a trough in diversity around the selected locus with a sizeable reduction in genetic diversity at the location of the selected locus. (b) Soft sweeps. The advantageous mutation (red G allele) is found on multiple haplotypes (marked in yellow and green). After the completion of a soft sweep, the advantageous allele is fixed, but its less likely nearby neutral variants are also fixed. This leads to a small reduction in diversity at locations near the selected locus.
Recombination plays an important role in determining the chromosomal extent of genetic hitchhiking during a hard sweep. As one moves along the chromosome in either direction away from the advantageous mutation, the chromosomes carrying the advantageous mutation will show less and less of the ancestral haplotype because of the shuffling effects of recombination (figure 1). As a result, after the advantageous allele reaches fixation (a frequency of 100%), one finds the regions around the selected locus have a complete lack of genetic diversity (even at neutral sites) and as one looks further from the selected locus variation is restored to background levels. This trough of diversity leaves a strong observable pattern in the genome, and is one hallmark of a ‘hard’ sweep as opposed to a ‘soft’ sweep. Regions with an excess of rare variants might indicate regions where a hard sweep has finished and new mutations are entering the population. Regions where one haplotype has low diversity relative to all others might indicate regions partway through the hard sweep process—so-called partial sweeps. The first generation of selection scan methods was built to capture these signatures (e.g. Tajima's D [24], integrated haplotype score (iHS) [25], cross-population extended haplotype homozygosity (XP-EHH) [26], the composite likelihood ratio test [27], composite of multiple signals test [28]).
In contrast, models of soft sweeps are defined by the occurrence of the advantageous mutation on several haplotypes [29]. This can occur via two major routes—when selection first begins to act, the advantageous mutation may be pre-existing in the population on multiple haplotypes or while selection is taking place on the first instance of an advantageous mutation, additional mutations may arise on different haplotypes and likewise begin to spread. Pennings and Hermison [29–31] have explored models of soft sweeps in depth. Major factors influencing the rate are the total mutation rate and selection coefficient in favour of the advantageous allele and the effective population size.
After the completion of a soft sweep at a single locus, the advantageous allele is fixed in the population, but neutral variants at nearby locations are often not fixed (figure 1). This process leads to much less obvious signatures of selection—the signature in single nucleotide polymorphism (SNP) data is weak because one does not expect a big reduction in diversity at nearby loci. One might have some hope in full sequence data by looking for putatively functional variants that have changed in frequency dramatically (for example, relative to the allele frequency in populations that have not experienced the selective pressure). The prospects of detecting selection with current methods are worse if the soft sweep occurs because advantageous mutations are distributed across multiple loci [22]. In this case of polygenic adaptation, there is not necessarily a strong shift in allele frequency at any single locus.
A major concern in the study of human adaptive evolution has been that soft sweeps may play a larger role than hard sweeps, and that our ability to detect loci that have undergone recent adaptive evolution may be severely compromised as a result [22,32]. Recent empirical studies bear on this issue but before we consider them, we will review models of adaptive evolution that include a spatial component.
3. Spatial models of adaptive evolution
The models for recent adaptive evolution just discussed are typically conceptualized as processes that take place in single, randomly mating populations. Because humans have spread out across the globe and have limited dispersal, it is problematic to try to understand human evolution without considering population structure. Indeed, numerous recent studies show that humans have detectable spatial structure, not just at global scales, but even within sub-continental geographical areas (for a review, see [33]). The spatial structure patterns observed in humans are probably due to locally restricted mating and the outcome of human spatial expansion events and subsequent large-scale migrations. Hence, we need to consider adaptive evolution models in a spatial context.
One of the most classic and influential models of adaptive evolution in a spatial context is Fisher's deterministic ‘wave of advance’ model of selection and dispersal [34]. The wave of advance model is a continuous time, continuous space, partial differential equation model that describes the change in allele frequencies at a location in terms of dispersal and selection. In this model, when a new advantageous mutation arises in a population, it increases in frequency due to selection and spreads outwards from its origin to neighbouring locations due to dispersal. If the selective advantage of the allele is identical everywhere, then the frequency of an advantageous allele is more concentrated around its geographical origin relative to neutral alleles of the same age. This leads to highly differentiated advantageous alleles relative to neutral alleles and suggests that by looking for loci with extreme levels of divergence among populations (e.g. by finding loci with high values of the fixation index, FST), one can discover loci that have recently undergone adaptive evolution [35].
Fisher also described the speed of the expansion wave, that is, how quickly a new mutation will spread across space. The speed of the wave front depends on the average dispersal distance and additive selection coefficient. The wave is faster if dispersal is strong or selection is strong. This wave speed plays a crucial role in determining how long it will take an adaptive allele to spread between populations.
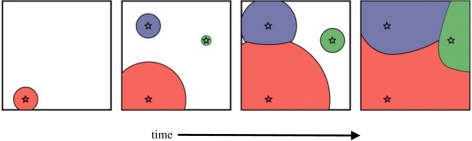
In a recent paper, Ralph & Coop [36] explore the consequences of the speed at which an advantageous allele spreads for the genetic architecture of a trait. In particular, they consider cases in which adaptation to a novel selection pressure (such as a novel pathogen) might take place by ‘multiple’ mutations of similar phenotypic consequence that arise independently at the same locus or different loci (parallel mutations). Suppose a new advantageous mutation arises in the population, and spreads outwards according to Fisher's wave-of-advance model. In the areas where the first mutation has not yet reached, a second allele conferring similar selective advantage might arise and begin spreading outward (figure 2). Ralph & Coop studied the dynamics of this process, and show how if a mutation arises in a location the first mutation has not yet reached and spreads from there, then the two waves will meet at some point and a geographical tiling of the habitat by different advantageous haplotypes will form. This spatial tessellation will dissipate over time due to mixing by migration.
Figure 2.
Parallel adaptation in a spatially distributed population. Multiple (independent) advantageous mutations (origins marked with red, blue and green stars) arise and spread out over specific areas (marked with coloured circles). The waves eventually meet and form a tiling of the habitat with unique advantageous haplotypes. Adapted from Ralph & Coop [36].
Further, they found conditions that lead to parallel adaptation in different subsets of the range. Their results could be expressed in terms of a characteristic length, such that when the habitat size is greater than the characteristic length, adaptation via parallel mutations is plausible. This characteristic length can be defined as the distance travelled by an unobstructed spreading wave before it is expected that another successful mutation would have arisen within the area so far enclosed. The characteristic length is determined by the dispersal parameter, the mutation rate and the population density, with the plausibility of parallel mutations decreasing with dispersal rate and increasing with mutation rate and population density. However, the characteristic length depends only weakly on the selection coefficient, as selection both hastens the spread of an allele and conversely increases the chances that a new mutation escapes loss due to drift and begins to spread.
For humans, Ralph & Coop suggest that human population genetics parameters make it plausible that adaptation via parallel mutations will be common for habitats greater than 3000 km across (for reference, this is the rough spatial scale of Eurasia). Therefore, when implementing selection scans, we have to keep in mind that globally we might expect to observe soft sweep patterns (multiple advantageous haplotypes), while locally we might see hard sweep patterns (a single advantageous haplotype).
4. Insights from recent genomic studies of human adaptive evolution
Numerous population genomic studies of human adaptive evolution have been conducted in the past 5 years, and several reviews document major features of these studies [37–41]. Here, we highlight a few results from studies of selection carried out with the Human Genome Diversity Cell Line Panel (HGDP) sample genotyped by Li et al. [42]. The sample has a uniquely broad geographical representation in that it contains 938 individuals from 53 populations sampled throughout the world. Here, we first focus on two papers based on studying signatures of selection from SNP data obtained with the Illumina 650 K platform [43,44].
These studies investigated selection using several established population genetic methods, including computation of statistics regarding haplotype structure (iHS, XP-EHH) [25,43] and allele frequency differentiation (FST) [45], identifying outliers in the distribution of these statistics, and comparing the results to simulations. Among the outliers were variants in loci related to immune system function, presumably which evolved in response to pathogens. Two non-synonymous variants that had extreme allele frequency differentiation among the populations compared were found in the TLR6 and DPP3 genes [43]. TLR6 encodes a toll-like receptor involved in innate immunity that is highly differentiated between European populations and neighbouring populations. DPP3 is most differentiated between Amerindian populations and other HGDP populations and is highly expressed in lymphoblasts. Further evidence for selection on immune function genes was found via a cluster of SNPs in the human leucocyte antigen (HLA) region that differentiate the Oroqen from Han populations and SNPs in a cluster of interleukin receptors that differentiate the Oroqen from the Han. These SNPs provide some evidence that by scanning population genomic variation for signatures of selection, variants that might affect pathogen resistance can be found. A further impressive example of this is given by recent work that has identified signatures of selection that can be linked to Lassa fever [26,46].
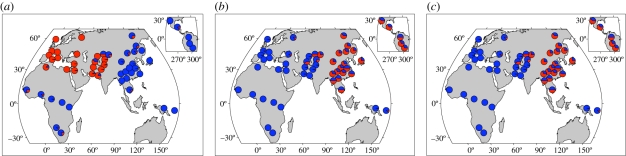
Beyond detecting loci that putatively have undergone adaptive evolution, a major conclusion of these studies was the importance of geography in structuring adaptive evolution in humans. These results are best evidenced by considering the signatures of selection observed at skin pigmentation loci. Previous studies have noted that skin pigmentation has been an important phenotype that has undergone adaptive evolution in the recent past (see the review by Jablonski & Chaplin [47]). In the HGDP selection scans, some of the most extreme signatures of selection fell in known skin pigmentation loci. KITLG, SLC24A5 and MC1R all showed signatures of selection with haplotypes that have increased in frequency as a result of lightening of skin pigmentation [44]. The KITLG-selected haplotype is found across Eurasia and the Americas, the SLC24A5-selected haplotype is found only in Western Eurasia, and the MC1R haplotype is found only in Eastern Eurasia and the Americas (figure 3). Overall, one observes that adaptive haplotypes that lead to lighter skin pigmentation are geographically localized.
Figure 3.
Skin pigmentation sweep signatures in the HGDP samples. The proportion of red within each pie denotes the frequency of the recently arisen advantageous haplotype at (a) the SLC24A5 locus, (b) the MC1R locus and (c) the KITLG locus. Adapted from Coop et al. [44].
Presumably the same selective pressure is acting across Northern Eurasia to lighten skin pigmentation, but the genetic basis for lightened pigmentation is different in different populations. This observation loosely fits with the parallel mutation model of Ralph & Coop [36] in which multiple adaptive haplotypes arise and spread in response to the same selective pressure. The geographical extent of the spread is perhaps associated with age of the mutation. The KITLG mutation may have occurred early enough to spread out across all out-of-Africa populations, but SLC24A5 and MC1R mutations are relatively younger so that they are geographically localized.
A similar pattern is observed in one of the most canonical examples of adaptive evolution in humans, selection for lactase persistence. Evidence of a partial selective sweep in European populations has been detected at the loci that control lactase persistence [48]. The partial nature of the sweep is probably the result of the recent age of onset of the selective pressure, here believed to be due to the advent of dairy culture and the digestion of dairy products throughout an individual's lifetime. Recent work on lactase persistence in Africa and the Middle East has shown how a different set of mutations contributes to lactase persistence in these populations [49]. This is another example of how multiple, geographically localized mutations have been involved in human adaptation to recent selective pressures.
While these examples do not prove a rule, they are suggestive that a model of parallel mutation, in which there are hard sweeps locally and soft sweeps globally, may be typical for recent human adaptive evolution. Further support for the importance of parallel mutations across human populations is provided by another recent analysis of the HGDP data [50]. In this study, the authors found evidence of an excess of non-synonymous variants that have undergone parallel divergence relative to synonymous variants.
A recent analysis of the 1000 Genomes Project data also suggests the plausibility that hard sweeps are rare across humans [51]. Under models of hard sweeps, one expects a reduction in genetic diversity around locations where amino acid substitutions have taken place. In the analysis of the 1000 Genomes pilot data, Hernandez et al. [51] found the trough in diversity around non-synonymous sites was not lower than that around synonymous substitutions (which serve as a form of internal control for other factors than recent selection affecting diversity). The results suggest that hard sweeps must be uncommon relative to soft sweeps, or if they are common, the selective pressures associated with the hard sweeps must be quite weak.
5. Case study: the CCR5-Δ32 HIV-resistance allele
Before synthesizing the results surveyed thus far, we will consider one case study of a pathogen resistance allele in detail. One of the most well-studied pathogen resistance variants is the Δ32 mutation found in the C–C motif chemokine receptor 5, CCR5. This 32 bp deletion inactivates the expression of CCR5 on the cell surface of T cells and as a result confers a complete resistance to infection from T-cell trophic HIV strains in homozygous carriers [9–13]. In heterozygote individuals, infection is still possible but there is a delay of onset of AIDS following HIV infection [10]. CCR5 is not alone in carrying variants that delay the onset of AIDS [15] and the variation in AIDS mortality due to heritable factors is exerting a selective pressure on HIV resistance variants [52].
Nearly all of the known resistance variants predate the emergence of AIDS—simple considerations of allele frequency make this obvious: novel mutations that have occurred since the emergence of AIDS would be found at vanishingly small frequencies (for example, in a single patient), but many of the variants are found at polymorphic frequencies (greater than 5% frequency in at least some populations). Because the HIV resistance variants predate the emergence of AIDS as a selective pressure, the selection due to AIDS is acting on standing variation and is expected in the long term to produce soft sweep signatures of selection.
While selection from standing variation would appear to be the rule in the response to AIDS, the history of that standing variation has been an interesting question. It has been posited that prior to the onset of AIDS, the Δ32 allele may have conferred a selective advantage to an ancestral pathogen [12,53]. In this sense, it may have undergone a partial sweep due to resistance to an ancestral pathogen and now is getting further swept due to its protective role in responding to a novel pathogen. To play with the hitchhiking motif, one might think of this as a ‘pony express’ model—the selective agent changes but the increase in allele frequency keeps moving onward.
The motivation for this elaborate model came from the surprising observation that haplotypes carrying the Δ32 allele were typically paired with a particular configuration of microsatellite alleles [53]. This linkage disequilibrium is indicative of allele age, and in an early study the age of the Δ32 allele was estimated as approximately 1300 AD [53]. Such a young age was at odds with a relatively polymorphic frequency in Europe of 10 per cent on average [54–56]. The best explanation was a positive selection pressure, and this ancestral selective advantage for carriers of the allele was estimated to be between 5 and 35 per cent [53,57]. Further support came from what was judged to be an elevated level of population differentiation, perhaps resulting from localized positive selection. The Δ32 allele was found at frequencies reaching approximately 16 per cent in Northern Europe and decreasing as one moves southwards to Southern Europe and eastwards into Eurasia [54–56]. In essence, the Δ32 allele was judged to show evidence of a partial sweep in Europe prior to HIV (though the sweep terminology was not in use then).
This conclusion led to subsequent debates about the ancestral selective pressure. Given the estimated allele age and the concentration of allele frequency in Europe, the suggestion was put forward that Δ32 conferred resistance to the bubonic plaque and, in particular, the impact of the Black Death epidemic in Europe [53]. Subsequent works suggested smallpox as a candidate, based on factors such as the retroviral nature of both smallpox and HIV, the more sustained impact of smallpox on European populations, and the age structure of smallpox deaths disproportionately affecting younger age classes and hence leading to more selective impact [58].
Within this context, one of us investigated models of a geographically localized signature due to a partial sweep, by fitting a Fisher's wave of advance model to the allele frequency data [59]. This approach can only infer the relative strength of dispersal to selection. To get an absolute sense of the strength of selection, one needs an allele age estimate, and given the allele age estimates available at the time, the indication from the data was that the additive selection coefficient could be greater than 10 per cent and that the allele had dispersed rapidly (average dispersal distances more than 100 km per generation) during its sweep.
At the same time, a second study [60] reinvestigated evidence for selection at CCR5 and found an important problem in the previous studies. The recombination rates estimated between the microsatellites near CCR5 and the locus itself were much smaller than previously estimated. In turn, the association between Δ32 and the microsatellite alleles was not necessarily unexpected given its frequency—what was once a clear hitchhiking signature became compatible with selective neutrality. This highlights one of the pitfalls of population genetic inference—some approaches rely on parameters inferred from external data sets and if uncertainty in such parameters is poorly accounted for, downstream analyses can be misled. Sabeti et al. also found CCR5 was not an outlier from other loci in the genome with respect to haplotype homozygosity (a statistic sensitive to partial selective sweeps) and with the revised map estimated the age to be more than 5000 years old [60].
Further corroboration for an older age came from ancient DNA studies that found Δ32 in samples as old as 3000 years old, clearly incompatible with previous younger allele age estimates [61]. In the light of the older allele age and weaker selection coefficient, the geographical analysis of Novembre et al. [59] produces dispersal estimates that are more in line with dispersal rates estimated from historical records (1–75 km per generation) [62]. In sum, it does not seem to be possible to reject a model in which Δ32 was a neutral variant existing within Europe prior to the advent of AIDS. Positive selection prior to the emergence of the AIDS epidemic may be relevant for other HIV-resistance variants though, as suggested recently by Klimentidis et al. [63] in particular for an HIV-resistance variant in the HLA region.
6. Synthesis
To bring together these various insights, it is best to stratify our considerations by the age of the pathogen. For young pathogens, such as HIV, evolution from standing variation may be typical, much like in the CCR5-Δ32 example. For older pathogens, evolution via novel, and perhaps multiple geographically localized mutations may be more typical. Some of these localized mutations may have arisen after the selective pressure and hence have partial hard sweep signatures that may be detectable using existing selection scan methods (iHS, for example). The identification of sweep signals associated with the Lassa fever example is exemplary of a hard sweep localized in Africa, and the lactase persistence and skin pigmentation examples described above are instructive as well.
For young pathogens, if resistance variants exist they almost necessarily derive from standing variation and so it is worth noting a few points about the nature of standing variation in humans. Recent studies have made clear that relative to expectations for constant-sized populations human sequence variation shows a strong excess of rare variants, most probably owing to population growth [64,65]. When designing mapping studies to discover novel resistance variants, we must keep these expectations in consideration. Approaches that will have the highest chances for success will focus on rare variants (for example, via re-sequencing) and will carry out mapping studies in multiple, local populations that are directly experiencing the pathogen. Also, because selection signatures may be relatively weak, there is strong incentive for doing direct phenotype-mapping approaches, such as genome-wide association or re-sequencing-based approaches.
One relatively novel selection signature method that may work well for discovering pathogen-resistance alleles is the approach of correlating allele frequencies with environmental variables that are indicative of selection pressure. This method does not rely directly on the expectations of a hard sweep model and so may have more power in soft sweep scenarios. Di Rienzo and co-workers [66] have had success using this approach for finding variants associated with climate adaptations and metabolism [67] and diet and subsistence shifts in humans. An important feature of the methods they applied was that they account for background demography [68].
One aspect of human population genetics that we have not described in this review is the potential impact of allele surfing [69–74], which is the outcome of serial founder effects and can be misleading for selection scans as it can make neutral variants appear as if they have swept to high frequency. Another important aspect we have not discussed is how different forms of balancing selection may impact variation at pathogen-resistance loci. This is particularly relevant for variants that exist in the HLA region, where there are high levels of genetic diversity and evidence for balancing selection. For a relevant review, see [75]. We have instead focused on the impact of directional selection for pathogen resistance. That said, during the initial spread of an allele that exhibits overdominance (a mechanism of balancing selection) there may be sweep-like signatures that can be detected, and distinguishing directional from balancing selection may be difficult.
Finally, it is worth noting that an appreciation for the impact of soft sweeps and polygenic adaptation is relatively new in human evolutionary genetics, and that researchers will inevitably step up to the challenge of finding methods that are more sensitive for detecting regions of the genome that have experienced such soft sweeps. The next few years, empowered in part by rapidly growing sequence data, will hopefully see exciting developments that will address some of the challenges highlighted here.
Acknowledgements
For this work, J.N. was supported by a Searle Scholars Award and NSF award EF-0928987. E.H. was supported by NIH training grant 5T32AI007370.
References
- 1.Haldane J. B. S. 1949. The rate of mutation of human genes. Hereditas 35(Suppl. 1), 267–272 10.1111/j.1601-5223.1949.tb03339.x (doi:10.1111/j.1601-5223.1949.tb03339.x) [DOI] [Google Scholar]
- 2.Lederberg J. J. B. S. 1999. Haldane (1949) on infectious disease and evolution. Genetics 153, 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zinkernagel R. M., Hengartner H., Stitz L. 1985. On the role of viruses in the evolution of immune-responses. Br. Med. Bull. 41, 92–97 [DOI] [PubMed] [Google Scholar]
- 4.Howard J. C. 1991. Immunology: disease and evolution. Nature 352, 565–567 10.1038/352565a0 (doi:10.1038/352565a0) [DOI] [PubMed] [Google Scholar]
- 5.Hill A. V. S., Motulsky A. G. 1999. Genetic variation and human diseases: the role of natural selection. In Evolution in health and disease (ed. Stearns S. C.), pp. 50–61 New York, NY: Oxford University Press [Google Scholar]
- 6.Ewald P. W. 1994. Evolution of infectious disease. New York, NY: Oxford University Press [Google Scholar]
- 7.Kaslow R. A., McNicholl J., Hill A. V. S. 2008. Genetic susceptibility to infectious diseases. New York, NY: Oxford University Press [Google Scholar]
- 8.Hill A. V. S. 2012. Evolution, revolution and heresy in the genetics of infectious disease susceptibility. Phil. Trans. R. Soc. B 367, 840–849 10.1098/rstb.2011.0275 (doi:10.1098/rstb.2011.0275) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Y., et al. 1996. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat. Med. 2, 1240–1243 10.1038/nm1196-1240 (doi:10.1038/nm1196-1240) [DOI] [PubMed] [Google Scholar]
- 10.Dean M., et al. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science 273, 1856–1862 10.1126/science.273.5283.1856 (doi:10.1126/science.273.5283.1856) [DOI] [PubMed] [Google Scholar]
- 11.Biti R., Ffrench R., Young J., Bennetts B., Stewart G., Liang T. 1997. HIV-1 infection in an individual homozygous for the CCR5 deletion allele. Nat. Med. 3, 252–253 10.1038/nm0397-252 (doi:10.1038/nm0397-252) [DOI] [PubMed] [Google Scholar]
- 12.O'Brien T. R., Winkler C., Dean M., Nelson J. A., Carrington M., Michael N. L., White G. C. 1997. HIV-1 infection in a man homozygous for CCR5 Δ32. Lancet 349, 1219. 10.1016/S0140-6736(97)24017-1 (doi:10.1016/S0140-6736(97)24017-1) [DOI] [PubMed] [Google Scholar]
- 13.Zimmerman P. A., et al. 1997. Inherited resistance to HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: studies in populations with contrasting clinical phenotypes, defined racial background, and quantified risk. Mol. Med. 3, 23–36 [PMC free article] [PubMed] [Google Scholar]
- 14.Allers K., Hutter G., Hofmann J., Loddenkemper C., Rieger K., Thiel E., Schneider T. 2011. Evidence for the cure of HIV infection by CCR5Δ32/Δ32 stem cell transplantation. Blood 117, 2791–2799 10.1182/blood-2010-09-309591 (doi:10.1182/blood-2010-09-309591) [DOI] [PubMed] [Google Scholar]
- 15.Hill A. V. 2006. Aspects of genetic susceptibility to human infectious diseases. Annu. Rev. Genet. 40, 469–486 10.1146/annurev.genet.40.110405.090546 (doi:10.1146/annurev.genet.40.110405.090546) [DOI] [PubMed] [Google Scholar]
- 16.Fellay J., et al. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317, 944–947 10.1126/science.1143767 (doi:10.1126/science.1143767) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fellay J., et al. 2009. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 5, e1000791. 10.1371/journal.pgen.1000791 (doi:10.1371/journal.pgen.1000791) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Limou S., et al. 2009. Genomewide association study of an AIDS-nonprogression cohort emphasizes the role played by HLA genes (ANRS Genomewide Association Study 02). J. Infect. Dis. 199, 419–426 10.1086/596067 (doi:10.1086/596067) [DOI] [PubMed] [Google Scholar]
- 19.Schork N. J., Murray S. S., Frazer K. A., Topol E. J. 2009. Common versus rare allele hypotheses for complex diseases. Curr. Opin. Genet. Dev. 19, 212–219 10.1016/j.gde.2009.04.010 (doi:10.1016/j.gde.2009.04.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarthy M. I. 2009. Exploring the unknown: assumptions about allelic architecture and strategies for susceptibility variant discovery. Genome Med. 1, 66. 10.1186/gm66 (doi:10.1186/gm66) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pritchard J. K. 2001. Are rare variants responsible for susceptibility to complex diseases? Am. J. Hum. Genet. 69, 124–137 10.1086/321272 (doi:10.1086/321272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pritchard J. K., Pickrell J. K., Coop G. 2010. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 20, R208–R215 10.1016/j.cub.2009.11.055 (doi:10.1016/j.cub.2009.11.055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maynard Smith J. M., Haigh J. 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23, 23–35 10.1017/S0016672300014634 (doi:10.1017/S0016672300014634) [DOI] [PubMed] [Google Scholar]
- 24.Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voight B. F., Kudaravalli S., Wen X., Pritchard J. K. 2006. A map of recent positive selection in the human genome. PLoS Biol. 4, e72. 10.1371/journal.pbio.0040072 (doi:10.1371/journal.pbio.0040072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabeti P. C., et al. 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449, 913–918 10.1038/nature06250 (doi:10.1038/nature06250) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen R., Williamson S., Kim Y., Hubisz M. J., Clark A. G., Bustamante C. 2005. Genomic scans for selective sweeps using SNP data. Genome Res. 15, 1566–1575 10.1101/gr.4252305 (doi:10.1101/gr.4252305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grossman S. R., et al. 2010. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science 327, 883–886 10.1126/science.1183863 (doi:10.1126/science.1183863) [DOI] [PubMed] [Google Scholar]
- 29.Hermisson J., Pennings P. S. 2005. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics 169, 2335–2352 10.1534/genetics.104.036947 (doi:10.1534/genetics.104.036947) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pennings P. S., Hermisson J. 2006. Soft sweeps II: molecular population genetics of adaptation from recurrent mutation or migration. Mol. Biol. Evol. 23, 1076–1084 10.1093/molbev/msj117 (doi:10.1093/molbev/msj117) [DOI] [PubMed] [Google Scholar]
- 31.Pennings P. S., Hermisson J. 2006. Soft sweeps III: the signature of positive selection from recurrent mutation. PLoS Genet. 2, e186. 10.1371/journal.pgen.0020186 (doi:10.1371/journal.pgen.0020186) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pritchard J. K., Di Rienzo A. 2010. Adaptation: not by sweeps alone. Nat. Rev. Genet. 11, 665–667 10.1038/nrg2880 (doi:10.1038/nrg2880) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novembre J., Ramachandran S. 2011. Perspectives on human population structure at the cusp of the sequencing era. Annu. Rev. Genomics Hum. Genet. 12, 245–274 10.1146/annurev-genom-090810-183123 (doi:10.1146/annurev-genom-090810-183123) [DOI] [PubMed] [Google Scholar]
- 34.Fisher R. 1937. The wave of advance of advantageous genes. Ann. Eugen. 7, 355–369 10.1111/j.1469-1809.1937.tb02153.x (doi:10.1111/j.1469-1809.1937.tb02153.x) [DOI] [Google Scholar]
- 35.Lewontin R. C., Krakauer J. 1973. Distribution of gene frequency as a test of the theory of the selective neutrality of polymorphisms. Genetics 74, 175–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ralph P., Coop G. 2010. Parallel adaptation: one or many waves of advance of an advantageous allele? Genetics 186, 647–668 10.1534/genetics.110.119594 (doi:10.1534/genetics.110.119594) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akey J. M. 2009. Constructing genomic maps of positive selection in humans: where do we go from here? Genome Res. 19, 711–722 10.1101/gr.086652.108 (doi:10.1101/gr.086652.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Enattah N. S., et al. 2008. Independent introduction of two lactase-persistence alleles into human populations reflects different history of adaptation to milk culture. Am. J. Hum. Genet. 82, 57–72 10.1016/j.ajhg.2007.09.012 (doi:10.1016/j.ajhg.2007.09.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biswas S., Akey J. M. 2006. Genomic insights into positive selection. Trends Genet. 22, 437–446 10.1016/j.tig.2006.06.005 (doi:10.1016/j.tig.2006.06.005) [DOI] [PubMed] [Google Scholar]
- 40.Nielsen R., Hellmann I., Hubisz M., Bustamante C., Clark A. G. 2007. Recent and ongoing selection in the human genome. Nat. Rev. Genet. 8, 857–868 10.1038/nrg2187 (doi:10.1038/nrg2187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabeti P. C., et al. 2006. Positive natural selection in the human lineage. Science 312, 1614–1620 10.1126/science.1124309 (doi:10.1126/science.1124309) [DOI] [PubMed] [Google Scholar]
- 42.Li J. Z., et al. 2008. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319, 1100–1104 10.1126/science.1153717 (doi:10.1126/science.1153717) [DOI] [PubMed] [Google Scholar]
- 43.Pickrell J. K., et al. 2009. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 19, 826–837 10.1101/gr.087577.108 (doi:10.1101/gr.087577.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coop G., et al. 2009. The role of geography in human adaptation. PLoS Genet. 5, e1000500. 10.1371/journal.pgen.1000500 (doi:10.1371/journal.pgen.1000500) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holsinger K. E., Weir B. S. 2009. Genetics in geographically structured populations: defining, estimating and interpreting FST. Nat. Rev. Genet. 10, 639–650 10.1038/nrg2611 (doi:10.1038/nrg2611) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andersen K. G., Shylakhter I., Tabrizi S., Grossman S. R., Happi C. T., Sabeti P. C. 2012. Genome-wide scans provide evidence for positive selection of genes implicated in Lassa fever. Phil. Trans. R. Soc. B 367, 868–877 10.1098/rstb.2011.0299 (doi:10.1098/rstb.2011.0299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jablonski N. G., Chaplin G. 2012. Human skin pigmentation, migration, and disease susceptibility. Phil. Trans. R. Soc. B 367, 785–792 10.1098/rstb.2011.0308 (doi:10.1098/rstb.2011.0308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bersaglieri T., Sabeti P. C., Patterson N., Vanderploeg T., Schaffner S. F., Drake J. A., Rhodes M., Reich D. E., Hirschhorn J. N. 2004. Genetic signatures of strong recent positive selection at the lactase gene. Am. J. Hum. Genet. 74, 1111–1120 10.1086/421051 (doi:10.1086/421051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tishkoff S. A., et al. 2007. Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 39, 31–40 10.1038/ng1946 (doi:10.1038/ng1946) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tennessen J. A., Akey J. M. 2011. Parallel adaptive divergence among geographically diverse human populations. PLoS Genet. 7, e1002127. 10.1371/journal.pgen.1002127 (doi:10.1371/journal.pgen.1002127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hernandez R. D., Kelley J. L., Elyashiv E., Melton S. C., Auton A., McVean G. 1000 Genomes Project, Sella G., Przeworski M. 2011. Classic selective sweeps were rare in recent human evolution. Science 331, 920–924 10.1126/science.1198878 (doi:10.1126/science.1198878) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schliekelman P., Garner C., Slatkin M. 2001. Natural selection and resistance to HIV. Nature 411, 545–546 10.1038/35079176 (doi:10.1038/35079176) [DOI] [PubMed] [Google Scholar]
- 53.Stephens J. C., et al. 1998. Dating the origin of the CCR5-Δ32 AIDS-resistance allele by the coalescence of haplotypes. Am. J. Hum. Genet. 62, 1507–1515 10.1086/301867 (doi:10.1086/301867) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Samson M., et al. 1996. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382, 722–725 10.1038/382722a0 (doi:10.1038/382722a0) [DOI] [PubMed] [Google Scholar]
- 55.Martinson J. J., Chapman N. H., Rees D. C., Liu Y. T., Clegg J. B. 1997. Global distribution of the CCR5 gene 32-basepair deletion. Nat. Genet. 16, 100–103 10.1038/ng0597-100 (doi:10.1038/ng0597-100) [DOI] [PubMed] [Google Scholar]
- 56.Libert F., et al. 1998. The ΔCCR5 mutation conferring protection against HIV-1 in Caucasian populations has a single and recent origin in Northeastern Europe. Hum. Mol. Genet. 7, 399–406 10.1093/hmg/7.3.399 (doi:10.1093/hmg/7.3.399) [DOI] [PubMed] [Google Scholar]
- 57.Slatkin M. 2001. Simulating genealogies of selected alleles in a population of variable size. Genet. Res. 78, 49–57 [DOI] [PubMed] [Google Scholar]
- 58.Galvani A. P., Slatkin M. 2004. Intense selection in an age-structured population. Proc. R. Soc. Lond. B 271, 171–176 10.1098/rspb.2003.2573 (doi:10.1098/rspb.2003.2573) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Novembre J., Galvani A. P., Slatkin M. 2005. The geographic spread of the CCR5 Δ32 HIV-resistance allele. PLoS Biol. 3, e339. 10.1371/journal.pbio.0030339 (doi:10.1371/journal.pbio.0030339) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sabeti P. C., et al. 2005. The case for selection at CCR5-Δ32. PLoS Biol. 3, e378. 10.1371/journal.pbio.0030378 (doi:10.1371/journal.pbio.0030378) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hummel S., Schmidt D., Kremeyer B., Herrmann B., Oppermann M. 2005. Detection of the CCR5-Δ32 HIV resistance gene in Bronze Age skeletons. Genes Immun. 6, 371–374 10.1038/sj.gene.6364172 (doi:10.1038/sj.gene.6364172) [DOI] [PubMed] [Google Scholar]
- 62.Wijsman E. M., Cavalli-Sforza L. L. 1984. Migration and genetic population structure with special reference to humans. Annu. Rev. Ecol. Syst. 15, 279–301 10.1146/annurev.es.15.110184.001431 (doi:10.1146/annurev.es.15.110184.001431) [DOI] [Google Scholar]
- 63.Klimentidis Y. C., Aissani B., Shriver M. D., Allison D. B., Shrestha S. 2011. Natural selection among Eurasians at genomic regions associated with HIV-1 control. BMC Evol. Biol. 11, 173. 10.1186/1471-2148-11-173 (doi:10.1186/1471-2148-11-173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Coventry A., et al. 2010. Deep resequencing reveals excess rare recent variants consistent with explosive population growth. Nat. Commun. 1, 131. 10.1038/ncomms1130 (doi:10.1038/ncomms1130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Altshuler D. M., et al. 2010. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 10.1038/nature09298 (doi:10.1038/nature09298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hancock A. M., Alkorta-Aranburu G., Witonsky D. B., Di Rienzo A. 2010. Adaptations to new environments in humans: the role of subtle allele frequency shifts. Phil. Trans. R. Soc. B 365, 2459–2468 10.1098/rstb.2010.0032 (doi:10.1098/rstb.2010.0032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hancock A. M., Witonsky D. B., Gordon A. S., Eshel G., Pritchard J. K., Coop G., Rienzo A. D. 2008. Adaptations to climate in candidate genes for common metabolic disorders. PLoS Genet. 4, e32. 10.1371/journal.pgen.0040032 (doi:10.1371/journal.pgen.0040032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coop G., Witonsky D., Di Rienzo A., Pritchard J. K. 2010. Using environmental correlations to identify loci underlying local adaptation. Genetics 185, 1411–1423 10.1534/genetics.110.114819 (doi:10.1534/genetics.110.114819) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramachandran S., Deshpande O., Roseman C. C., Rosenberg N. A., Feldman M. W., Cavalli-Sforza L. L. 2005. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc. Natl Acad. Sci. USA 102, 15 942–15 947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prugnolle F., Manica A., Balloux F. 2005. Geography predicts neutral genetic diversity of human populations. Curr. Biol. 15, R159–R160 10.1016/j.cub.2005.02.038 (doi:10.1016/j.cub.2005.02.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edmonds C. A., Lillie A. S., Cavalli-Sforza L. L. 2004. Mutations arising in the wave front of an expanding population. Proc. Natl Acad. Sci. USA 101, 975–979 10.1073/pnas.0308064100 (doi:10.1073/pnas.0308064100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vlad M. O., Cavalli-Sforza L. L., Ross J. 2004. Enhanced (hydrodynamic) transport induced by population growth in reaction-diffusion systems with application to population genetics. Proc. Natl Acad. Sci. USA 101, 10 249–10 253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Klopfstein S., Currat M., Excoffier L. 2006. The fate of mutations surfing on the wave of a range expansion. Mol. Biol. Evol. 23, 482–490 10.1093/molbev/msj057 (doi:10.1093/molbev/msj057) [DOI] [PubMed] [Google Scholar]
- 74.Excoffier L., Ray N. 2008. Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol. Evol. 23, 347–351 10.1016/j.tree.2008.04.004 (doi:10.1016/j.tree.2008.04.004) [DOI] [PubMed] [Google Scholar]
- 75.Meyer D., Thomson G. 2001. How selection shapes variation of the human major histocompatibility complex: a review. Ann. Hum. Genet. 65(Pt 1), 1–26 10.1046/j.1469-1809.2001.6510001.x (doi:10.1046/j.1469-1809.2001.6510001.x) [DOI] [PubMed] [Google Scholar]