Abstract
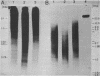
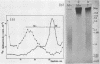
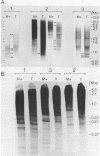
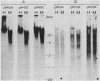
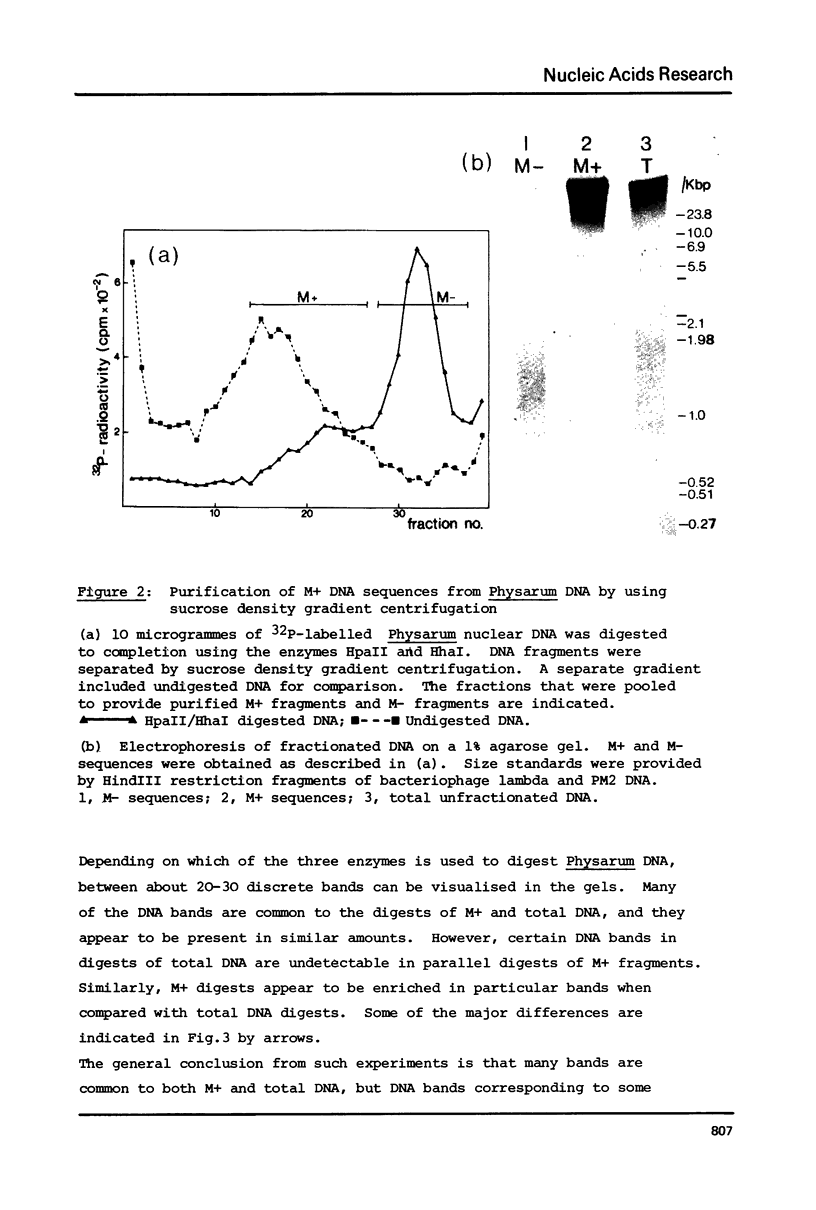
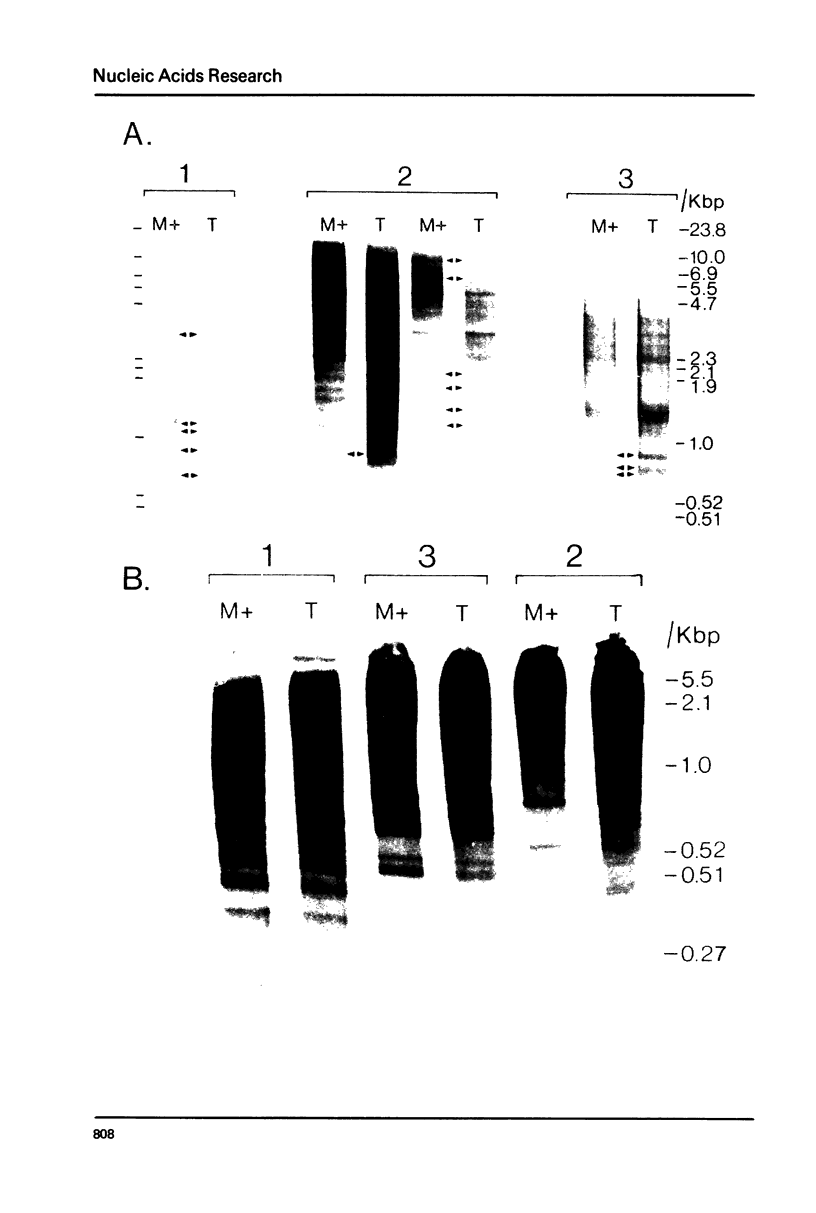
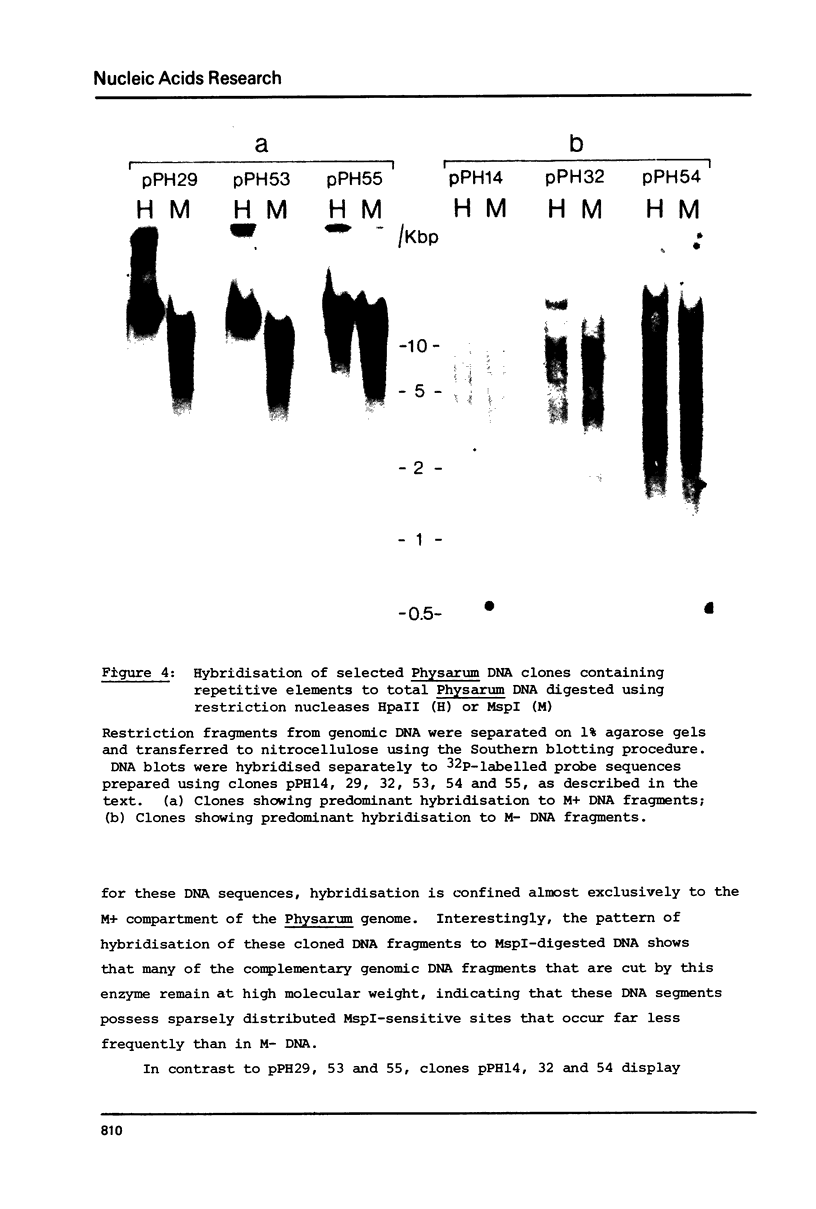
Nuclear DNA from the slime mould Physarum polycephalum is digested by the restriction endonuclease HpaII to generate a high molecular weight and a low molecular weight component. These are referred to as the M+ and the M- compartment, respectively. Sequences that are present in the M+ compartment are cleaved by MspI, the restriction enzyme isoschizomer of HpaII, thus showing that the recognition sequences for these enzymes in M+ DNA contain methylated CpG doublets. The distribution of repetitive sequences in the M+ and M- DNA compartments was investigated by comparison of the 'fingerprint' patterns of total Physarum DNA and isolated M+ DNA after digestion using different restriction endonucleases, and by probing for the presence of specific repetitive sequences in Southern blots of M+ and M- DNA by the use of cloned DNA segments. Both types of experiment indicate that many repetitive sequences are shared by both compartments, though some repetitive sequences appear to be considerably enriched, or are present exclusively, either in M+ DNA or in M- DNA.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams R. L., McKay E. L., Douglas J. T., Burdon R. H. Methylation of nucleosomal and nuclease sensitive DNA. Nucleic Acids Res. 1977 Sep;4(9):3097–3108. doi: 10.1093/nar/4.9.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. P. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980 Apr 11;8(7):1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. P., Taggart M. H., Smith B. A. Methylated and unmethylated DNA compartments in the sea urchin genome. Cell. 1979 Aug;17(4):889–901. doi: 10.1016/0092-8674(79)90329-5. [DOI] [PubMed] [Google Scholar]
- Bird A. P., Taggart M. H. Variable patterns of total DNA and rDNA methylation in animals. Nucleic Acids Res. 1980 Apr 11;8(7):1485–1497. doi: 10.1093/nar/8.7.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm T. L., Drahovsky D. Verteilung und enzymatische Hypermethylierung invers repetitiver DNA-Sequenzen in verschiedenen Mäuse- und menschlichen Zellen. Z Naturforsch C. 1979 Aug;34(7-8):558–564. [PubMed] [Google Scholar]
- Browne M. J., Cato A. C., Burdon R. H. The distribution of modified and non-modified C-G doublets in BHK-21 cell DNA. FEBS Lett. 1978 Jul 1;91(1):69–73. doi: 10.1016/0014-5793(78)80019-2. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E. H., Britten R. J. Organization, transcription, and regulation in the animal genome. Q Rev Biol. 1973 Dec;48(4):565–613. doi: 10.1086/407817. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Hough B. R., Amenson C. S., Britten R. J. General interspersion of repetitive with non-repetitive sequence elements in the DNA of Xenopus. J Mol Biol. 1973 Jun 15;77(1):1–23. doi: 10.1016/0022-2836(73)90359-8. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Drahovsky D., Boehm T. L. Enzymatic DNA methylation in higher eukaryotes. Int J Biochem. 1980;12(4):523–528. doi: 10.1016/0020-711x(80)90002-6. [DOI] [PubMed] [Google Scholar]
- Drahovsky D., Boehm T. L., Kreis W. Distribution pattern and enzymic hypermethylation of inverted repetitive DNA sequences in P815 mastocytoma cells. Biochim Biophys Acta. 1979 Jun 20;563(1):28–35. doi: 10.1016/0005-2787(79)90004-2. [DOI] [PubMed] [Google Scholar]
- Hardman N., Jack P. L., Brown A. J., McLachlan A. Distribution of inverted repeat sequences in nuclear DNA from Physarum polycephalum. Eur J Biochem. 1979 Feb 15;94(1):179–187. doi: 10.1111/j.1432-1033.1979.tb12884.x. [DOI] [PubMed] [Google Scholar]
- Hardman N., Jack P. L., Fergie R. C., Gerrie L. M. Sequence organisation in nuclear DNA from Physarum polycephalum. Interspersion of repetitive and single-copy sequences. Eur J Biochem. 1980 Jan;103(2):247–257. doi: 10.1111/j.1432-1033.1980.tb04309.x. [DOI] [PubMed] [Google Scholar]
- Hardman N., Jack P. L. Periodic organisation of foldback sequences in Physarum polycephalum nuclear DNA. Nucleic Acids Res. 1978 Jul;5(7):2405–2423. doi: 10.1093/nar/5.7.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houck C. M., Rinehart F. P., Schmid C. W. A ubiquitous family of repeated DNA sequences in the human genome. J Mol Biol. 1979 Aug 15;132(3):289–306. doi: 10.1016/0022-2836(79)90261-4. [DOI] [PubMed] [Google Scholar]
- Jelinek W. R., Toomey T. P., Leinwand L., Duncan C. H., Biro P. A., Choudary P. V., Weissman S. M., Rubin C. M., Houck C. M., Deininger P. L. Ubiquitous, interspersed repeated sequences in mammalian genomes. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1398–1402. doi: 10.1073/pnas.77.3.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaina B., Hinz R. Analysis of the Vicia faba genome by use of restriction endonucleases. Eur J Biochem. 1979 May 2;96(1):159–166. doi: 10.1111/j.1432-1033.1979.tb13025.x. [DOI] [PubMed] [Google Scholar]
- Katz L., Kingsbury D. T., Helinski D. R. Stimulation by cyclic adenosine monophosphate of plasmid deoxyribonucleic acid replication and catabolite repression of the plasmid deoxyribonucleic acid-protein relaxation complex. J Bacteriol. 1973 May;114(2):577–591. doi: 10.1128/jb.114.2.577-591.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel A. R., Firtel R. A. A family of short, interspersed repeat sequences at the 5' end of a set of Dictyostelium single-copy mRNAs. Cell. 1979 Apr;16(4):787–796. doi: 10.1016/0092-8674(79)90094-1. [DOI] [PubMed] [Google Scholar]
- Lapeyre J. N., Becker F. F. Analysis of highly repeated DNA sequences of rat with EcoR1 endonuclease. Biochim Biophys Acta. 1980 Mar 28;607(1):23–35. doi: 10.1016/0005-2787(80)90217-8. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. Novel classes of mouse repeated DNAs. Nucleic Acids Res. 1980 Aug 11;8(15):3247–3258. doi: 10.1093/nar/8.15.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippsen P., Streeck R. E., Zachau H. G. Defined fragments of calf, human, and rat DNA produced by restriction nucleases. Eur J Biochem. 1974 Jun 15;45(2):479–488. doi: 10.1111/j.1432-1033.1974.tb03573.x. [DOI] [PubMed] [Google Scholar]
- Pollock J. M., Jr, Swihart M., Taylor J. H. Methylation of DNA in early development: 5-methyl cytosine content of DNA in sea urchin sperm and embryos. Nucleic Acids Res. 1978 Dec;5(12):4855–4861. doi: 10.1093/nar/5.12.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly J. G., Braun R., Thomas C. A., Jr Methjylation in Physarum DNA. FEBS Lett. 1980 Jul 28;116(2):181–184. doi: 10.1016/0014-5793(80)80638-7. [DOI] [PubMed] [Google Scholar]
- Schneiderman M. H., Billen D. Methylation rapidly reannealing DNA during the cell cycle of Chinese hamster cells. Biochim Biophys Acta. 1973 May 18;308(3):352–360. doi: 10.1016/0005-2787(73)90327-4. [DOI] [PubMed] [Google Scholar]
- Solage A., Cedar H. Organization of 5-methylcytosine in chromosomal DNA. Biochemistry. 1978 Jul 11;17(14):2934–2938. doi: 10.1021/bi00607a036. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Vanyushin B. F., Mazin A. L., Vasilyev V. K., Belozersky A. N. The content of 5-methylcytosine in animal DNA: the species and tissue specificity. Biochim Biophys Acta. 1973 Mar 28;299(3):397–403. doi: 10.1016/0005-2787(73)90264-5. [DOI] [PubMed] [Google Scholar]
- Vanyushin B. F., Tkacheva S. G., Belozersky A. N. Rare bases in animal DNA. Nature. 1970 Mar 7;225(5236):948–949. doi: 10.1038/225948a0. [DOI] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. DNA methylation at a CCGG sequence in the large intron of the rabbit beta-globin gene: tissue-specific variations. Nucleic Acids Res. 1978 Dec;5(12):4631–4634. doi: 10.1093/nar/5.12.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker P. A., Hardman N. Methylation of nuclear DNA in Physarum polycephalum. Biochem J. 1980 Dec 1;191(3):859–862. doi: 10.1042/bj1910859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ploeg L. H., Flavell R. A. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980 Apr;19(4):947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]