Abstract
The use of simple hosts such as Dictyostelium discoideum in the study of host pathogen interactions offers a number of advantages and has steadily increased in recent years. Infection-specific genes can often only be studied in a very limited way in man and even in the mouse model their analysis is usually expensive, time consuming and technically challenging or sometimes even impossible. In contrast, their functional analysis in D. discoideum and other simple model organisms is often easier, faster and cheaper. Because host-pathogen interactions necessarily involve two organisms, it is desirable to be able to genetically manipulate both the pathogen and its host. Particularly suited are those hosts, like D. discoideum, whose genome sequence is known and annotated and for which excellent genetic and cell biological tools are available in order to dissect the complex crosstalk between host and pathogen. The review focusses on host-pathogen interactions of D. discoideum with Legionella pneumophila, mycobacteria, and Salmonella typhimurium which replicate intracellularly.
Keywords: Amoeba, Dictyostelium discoideum, drug targets, functional analysis, infection, Legionella pneumophila, model organism, Mycobacteria, pathogen, Salmonella typhimurium, social amoeba, virulence factor.
INTRODUCTION
D. discoideum is a fascinating member of the amoebozoa, whose natural habitat is deciduous forest soil and decaying leaves, where the amoebae feed on bacteria and yeast and grow as separate, independent, single cells. Upon depletion of food, the cells undergo aggregation and cell differentiation, giving rise to a multi-cellular organism made up of different cell types [1]. The organism offers unique advantages for studying fundamental cellular processes with powerful molecular genetic, biochemical, and cell biological tools [2]. These processes include cell motility, chemotaxis, cytokinesis, signal transduction, and several aspects of development [3-6]. Additional advantages of D. discoideum are easy cultivation allowing large scale cultures and biochemical studies, the amenability to genetic and cell biological analysis and the availability of the genome sequence [2, 7-9]. As a soil amoeba and a phagocyte D. discoideum can be a natural host of opportunistic bacteria and may thus have developed strategies to avoid invasion by given pathogens or to counteract their intracellular survival and replication [10-12]. It has already been shown for a number of intracellular bacterial pathogens that they are resistant to free-living amoeba, such as Acanthamoeba castellanii [13]. A. castellanii occupies the same natural niche as e.g. L. pneumophila and mycobacteria where selection of virulence traits occurs (see also Sandström et al., this issue) [14, 15]. The organism might therefore be considered a closer model than D. discoideum to test their virulence, however, D. discoideum offers the advantage that mutants can easily be generated [2].
Phagocytosis is a very complex, evolutionarily conserved mechanism that is used by higher eukaryotes to clear dead cells and cell debris and to counter the constant threat posed by pathogens. For this purpose they harbour specialized cells such as macrophages, neutrophils or dendritic cells that have the ability to rapidly and efficiently internalize a variety of organisms and particles and degrade them. These cells represent professional phagocytes that are important for innate and adaptive immunity in metazoa. For lower eukaryotes like D. discoideum phagocytosis is a means to internalize bacteria that are used as food source. The ingested microorganism is trapped in a phagosome and, via the phagolysosomal pathway, is ultimately delivered to a lysosome where it is degraded by a cocktail of hydrolytic enzymes [10, 11, 16]. Efficient phagocytosis relies on signalling processes, a functioning cytoskeleton, in particular actin and actin-binding proteins, and vesicle trafficking and fusion. Pathogens, on the other hand, have evolved several means to interfere with these processes. They either block maturation of the phagosome, manipulate its identity and use it as a replication niche or escape from it into the cytosol [17].
In this review we first provide an introduction to D. discoideum as a model host for a number of bacterial pathogens followed by a brief description of L. pneumophila, mycobacteria, and S. typhimurium, bacterial pathogens that have been used to study host-pathogen interactions with D. discoideum. We then discuss host cell processes that are important for the uptake of the pathogen, the establishment of the replication niche and host defence. We finally address the potential of D. discoideum for drug screening.
D. discoideum, a Versatile Model to Study Host Pathogen Interactions
Although Depraitère and Darmon [18] described as early as 1978 that a few bacteria were pathogenic for D. discoideum, the system emerged as an experimental model for bacterial infections only ten years ago, when two groups demonstrated that D. discoideum could be used as host for L. pneumophila [19, 20]. Following these two reports, the number of pathogens for which D. discoideum has been shown to be a suitable host has increased steadily, the last entry being S. typhimurium (Table 1). In recent years it became clear that the basic mechanisms of host pathogen interactions are conserved between lower and higher eukaryotes [10, 21, 22]. Moreover, unicellular eukaryotes probably constitute a reservoir in which different pathogenic bacteria survive in the wild and where they develop novel virulence factors that are subsequently effective against animals or humans. Consequently, D. discoideum has become an attractive model system for investigating the infection with human pathogens [10-12, 23, 24]. D. discoideum cells are very suitable for cell biological assays and imaging, therefore, they have been used to study the dynamics of bacterial uptake, intracellular traffic of the pathogen-containing vacuole and. eventually, bacterial exit. However, the major contribution of D. discoideum infection studies resides in the identification of host cell factors that affect infection. To study these factors a large number of D. discoideum mutants are available from the Dictyostelium stock center (http://dictybase.org/ StockCenter/StockCenter.html), additional genes of interest can be tagged and easily disrupted and also untargeted mutational screens can be carried out [2, 25]. The immense value of the last approach was recently documented by Ralph Isberg’s lab where several new host cell factors that are important for infection with L. pneumophila were discovered and analysed [26]. As shown in Table 2, the list of genes favouring resistance or susceptibility to infection is increasing steadily. In addition, the bacterial side of the coin can easily be studied using D. discoideum as a screening host for wild-type or mutagenised pathogenic bacteria followed by a plaque assay (Table 3) [27, 28]. This approach that will, however, not be discussed in this review, works for pathogenic bacteria that have been already proven to infect D. discoideum and allows the fast detection of bacterial virulence genes. For recent reviews see Steinert and Heuner [29] and Weber et al. [30].
Table 1.
Bacteria that have been Successfully Used to Infect D. discoideum
| Bacterial Pathogen | References |
|---|---|
| Legionella pneumophila | [20, 49] |
| Mycobacterium avium, M. marinum, M. tuberculosis | [76, 121, 176] |
| Pseudomonas aeruginosa | [27, 76, 177] |
| Vibrio cholerae | [178] |
| Klebsiella pneumoniae | [179] |
| Neisseria meningitidis | [180] |
| Burkholderia cenocepacia | [181] |
| Salmonella typhimurium | [77] |
Table 2.
Host Cell Factors that Affect D. discoideum-Pathogen Interactions
| Host Cell Factor | Approach | Effects on Infection | Pathogen | References | |
|---|---|---|---|---|---|
| Uptake | Growth | ||||
| F Actin | inhibitors | down | normal | L.p. | [47] |
| α-actinin/ABP120 | knockout | down | down | L.p. | [173] |
| Coronin A | knockout | down | normal | L.p. | [173] |
| up | up* | M.m. | [176] | ||
| Coronin B | Knockout | up | normal | L.p. | [182] |
| overexpression | down | normal | |||
| Myosin1(A/B) | knockout | normal | up | L.p | [49] |
| Profilin I/II | knockout | normal | up | L.p. | [20] |
| Daip1 | knockout | down | normal | L.p. | [173] |
| Villidin | knockout | down | down | L.p | [173] |
| Lim C/D | knockout | down | down | L.p | [173] |
| Comitin | knockout | down | up | L.p. | [183] |
| Calnexin | knockout | down | down | L.p | [173] |
| Calreticulin | knockout | down | down | L.p. | [173] |
| Gβ subunit | knockout | down | down | L.p. | [173] |
| RacH | knockout | down | up | M.m., | [121] |
| down | up | L.p. | [47] | ||
| PLC | inhibitors | down | normal | L.p. | [47] |
| Calcium level | inhibitors | down | n.t. | L.p. | [173] |
| PI3K1/2 | knockout | normal | up | L.p. | [125] |
| PI3K1-5 | knockout | down | up | L.p. | [47] |
| PI3K1-5/PTEN | knockout | down | up | L.p. | [47] |
| PTEN | knockout | down | down | L.p. | [47] |
| Dd5P4 (OCRL1) | knockout | down | up | L.p | [30] |
| Phg1 | knockout | normal | up | K.p. | [179] |
| Nramp1 | knockout | normal | up | L.p., M.a | [105] |
| overexpression | normal | down | |||
| VacB (flotillin) | knockout | normal | down | L.p.,M.m. | [121] |
| RtoA | knockout | normal | down | L.p. | [184] |
| Kil1 | knockout | normal | up | K.p. | [179] |
| overexpression | normal | down | |||
| TirA | knockout | n.t. | up | L.p. | [185] |
| Rnl, hsp60 | KO/antisense | normal | up | L.p. | [39] |
| AMPK | overexpression | normal | up | L.p. | [39] |
| antisense | normal | normal | |||
| ATG1, 6, 7 | knockout | normal | up | S.t. | [77] |
| ATG9 | knockout | down | up | L.p. | [150] |
| DupA | knockout | n.t. | down | L.p., M.m | [26] |
Pathogen uptake and intracellular growth in D. discoideum mutants or upon treatment of wild type cells with inhibitors (F actin: cytochalasin A, latrunculin A; phospholipase C: U73122; intracellular calcium levels: BAPTA-AM, Thapsigargin). Effects ("up" or "down") on bacterial uptake or intracellular growth are relative to AX2 control cells, in the case of inhibitors, or to the parental strain, in the case of mutants. n.t.: not tested.
Solomon et al. 2003 reported an enhanced initial rate of replication until day 4 in comparison to the AX2 control cells. L.p., L. pneumophila; K.p., K. pneumoniae; M.a, M. avium; M.m., M. marinum; S.t., S. typhimurium.
Table 3.
D. discoideum as Screening Host for Microbial Genes
| Gene | Pathogen | References |
|---|---|---|
| dotH, dotI, dotO | L. pneumophila | [19] |
| lepA, lepB | L. pneumophila | [186, 187] |
| sdhA | L. pneumophila | [188] |
| vipD | L. pneumophila | [189] |
| lqs, rpoS, letA | L. pneumophila | [190-192] |
| sidJ | L. pneumophila | [193] |
| enhC | L. pneumophila | [194] |
| sidC, sdcA, sidM | L. pneumophila | [126] |
| legC3 | L. pneumophila | [195] |
| rpoS | L. pneumophila | [196] |
| ankB | L. pneumophila | [197] |
| lpnE | L.pneumophila | [30] |
| vas | V. cholerae | [178] |
| lasR, rhl, pscJ, exoU | P. aeruginosa | [27, 177] |
| trpD, pchH, pchI | P. aeruginosa | [198] |
| Rd1 | M. marinum | [122] |
In the following section we will concentrate on infection studies with L. pneumophila, Mycobacteria and S. typhimurium, whereas for the other pathogens listed in Table 1 the interested reader is referred to a recent excellent review by Margaret Clarke [12].
PATHOGENS THAT INFECT D. DISCOIDEUM
L. pneumophila
In August 1976 a large outbreak of severe pneumonia affected attendees of a convention of war veterans in Philadelphia, USA. The outbreak was caused by a previously unrecognized bacterium and of 182 reported cases 29 were fatal. In early 1977 the causative agent of the “Legionnaires’ disease”, was nailed down and named L. pneumophila [31]. The bacterium is Gram-negative and now known as a facultative intracellular parasite. Meanwhile, it is clear that L. pneumophila is a significant cause of pneumonia. The majority of cases of Legionnaires’ disease are caused by L. pneumophila serogroup 1, but other serogroups and other species are also pathogenic [32-35]. L. pneumophila infection of alveolar human macrophages usually occurs through inhalation of contaminated aerosols produced by water systems such as air-conditioning units or showers [35]. Upon cell entry the L. pneumophila containing vacuole (LCV) is formed but does not enter the endo-lysosomal pathway [36-38]. Instead, a series of alternative docking events take place, including transient recruitment of mitochondria after about 1 hour [39] followed by association of ribosomes after about 4 hours. Then L. pneumophila proliferates, becomes acid tolerant and produces a flagellum. After 16 to 20 hours the LCV fuses with lysosomes. Finally, necrosis of the host cell is triggered, which leads to the release of the bacteria [40, 41]. A role of the mitochondria in the infection process is supported by two recent papers with D. discoideum as host. In mitochondrially diseased cells L. pneumophila could replicate better than in wild-type cells and this was suppressed by inhibiting the expression of the catalytic subunit of the AMP-activated protein kinase (AMPK), the central cellular energy sensor. Conversely, overexpression of the AMPK catalytic subunit enhanced the intracellular growth of L. pneumophila [39]. Interestingly, this protein is upregulated in mitochondrial diseases and also upon infection with L. pneumophila. By which mechanism AMPK facilitates infection remains unclear.
Zhang and Kuspa [42] found a decrease of mitochondrial mRNAs already 4h post infection and cleavage of the large subunit of the mitochondrial rRNA into two distinct fragments suggesting that L. pneumophila specifically disrupts mitochondrial protein synthesis in D. discoideum during the course of infection. Cleavage was particularly pronounced 24 hours post infection and may be correlated with cell death [42].
The pathogenicity of L. pneumophila is determined by a number of virulence factors, among them the 24 dot/icm (defect in organelle trafficking/intracellular multiplication) gene products that are responsible for the formation of a type IV secretion system. A large number of effector proteins are transported into the cytoplasm of the host cell and are responsible for the modified phagosome maturation that allows survival of L. pneumophila [40, 43]. Genome sequencing of three clinical L. pneumophila isolates has revealed new putative virulence factors, among them many eukaryotic-like proteins that are likely to be implicated in different steps of the L. pneumophila life cycle [44]. So far no transmission of L. pneumophila among humans has been observed and it is assumed that freshwater amoebae and not human alveolar macrophages are the natural host of L. pneumophila [40, 45].
To study the infection process of L. pneumophila, guinea pigs, different protozoa, monocytes and other human cells have been used, while the suitability of D. discoideum was only recognized much later [19, 20, 46]. The infection and replication processes of macrophages and D. discoideum with L. pneumophila appear very similar. Recent evidence suggests that uptake of L. pneumophila into D. discoideum occurs by macropinocytosis [47], whereas in macrophages macropinocytosis as well as phagocytosis have been described [48]. However, infection in D. discoideum proceeds slower than in macrophages and host cell lysis occurs only after more than 48 hours [49, 50]. Meanwhile a wealth of information about host cell and bacterial factors that are important in the infection process has been obtained with D. discoideum as the model system (Tables 2 and 3, and see below).
Mycobacterium tuberculosis and Mycobacterium marinum
There are around 100 different species of Mycobacteria which is the only genus in the family of Mycobacteriaceae [51]. Mycobacteria have a rod-like appearance and are usually considered Gram-positive. The grouping is based on the lack of an outer cell membrane, though, due to their characteristic cell wall, they do not retain the crystal violet in Gram staining well. Their cell wall is hydrophobic, waxy and thicker than in many other bacteria. It is composed of the hydrophobic mycolate layer and a peptide-glycan layer held together by arabinogalactan. Mycobacteria live in water and in the soil, are aerobic, and acid-fast [51]. Several members from the Mycobacteria group, including M. tuberculosis are human pathogens [35] and cause tuberculosis and other granulomatous lesions: Tuberculosis kills nearly 3 million people annually [52]. Virulence depends among other factors on the region of difference (RD) 1 locus, which encodes components of a type seven secretion system (ESX-1 system) and essential secreted effectors like CFP-10, ESAT-6 [53] and on two large families of proteins, PE and PPE, which could provide antigenic variation to the pathogen in order to evade the host immune response [54-56]. M. marinum is a close relative of M. tuberculosis and infects amphibians, fishes and also humans [57]. In 1954 M. marinum was identified as being responsible for the cutaneous granulomatous lesions of 80 persons who had used the same swimming pool. Therefore, the disease is called swimming pool or fish tank granuloma [58]. M. tuberculosis and M. marinum share common mechanisms of pathogenicity and the pathologies and lesions they cause are almost indistinguishable [59]. Since M. tuberculosis is a biosafety level 3 human pathogen, its study is labor intensive and carries the risk of accidental exposure. Therefore, mycobacterial models like M. marinum, Mycobacterium bovis (BCG strain) or Mycobacterium avium are increasingly used to understand M. tuberculosis virulence [60]. On the host side, the mouse is the most commonly used model, however, M. tuberculosis is not a natural pathogen of mice and the course of tuberculosis differs from the human disease. In recent years, zebrafish, D. melanogaster, C. elegans and D. discoideum have been firmly established as surrogate hosts [61].
S. typhimurium
Salmonella enterica serovar Typhimurium is one of more than 2000 species of the Salmonella enterica genus, which are resident bacteria of the gut in vertebrates. Only a handful of them are etiological agents of gastroenteritis and the more severe typhoid fever. Typhoid fever, which is characterized by fever, intestinal perforation and hemorrhage, enlargement of mesenteric lymph nodes, spleen and liver, is caused mostly by S. enterica serovar Typhi, which is a human pathogen that does not cause disease in other animals.
S. typhimurium is spread in both animals and humans and is the major agent of food-borne (mainly meat and eggs) gastroenteritis, a disease characterized by diarrhea, abdominal pain, nausea, vomiting and fever. Acute enteritis may last for up to a week and resolves spontaneously, but the disease is a major economic problem. In contrast to S. typhi which is endemic in Asia, Africa and South America, S. typhimurium is widespread also in Europe and North America, with an estimate of 1.4 million cases of enterocolitis, including 550 annual deaths, in the USA alone [62].
Established animal model systems for S. typhimurium are the mouse and C. elegans. In the mouse, for which S. typhimurium is a natural pathogen, the symptoms resemble those of typhoid fever in humans, which include enlargement of mesenteric lymph nodes, spleen and liver and eventually sepsis. After colonization of the intestinal epithelium, the bacteria are internalized by resident macrophages in the submucosa and rapidly disseminate by infecting circulating macrophages, B and T cells and eventually colonizing resident phagocytes in liver and spleen [62].
S. typhimurium internalization occurs by phagocytosis or macropinocytosis. Phagocytosis is common in professional phagocytes, and is induced by binding to lipopolysaccharide, fimbriae or flagellin receptors. Macropinocytosis is, instead, a highly specific bacterium-induced process for entering non-professional phagocytes as well as phagocytes. The process is regulated by the type 3 secretion system (T3SS), a protein complex encoded in the SPI1 (Salmonella pathogenicity island 1) gene locus, that secretes several effectors in the cell, inducing re-organization of the actin cytoskeleton, with formation of massive localized membrane ruffles and macropinocytic cups [63-67]. The outcome of infection depends on the modality of uptake, with macropinocytosis leading preferentially to formation of a survival and replication niche, the Salmonella-containing vacuole (SCV), whereas bacteria taken up by phagocytosis are mostly transported to lysosomes. The SCV is initially characterized by acquisition of early endosomal markers, which are removed and substituted within 60 to 90 minutes by late endosomal and lysosomal markers [68, 69]. Maturation of the SCV and virulence are controlled by the SPI2 T3SS system, a second secretion system that secretes hundreds of proteins into the cytoplasm [67, 70-73]. Boucrot et al. showed that the SCV recruited the plus-end-directed motor kinesin and that this event was regulated by proteins translocated by the SPI2 T3SS, among them SifA [74]. Interestingly the early SCV migrated to the perinuclear area and escaped the fusion with lysosomes [75].
S. thyphimurium is phagocytosed by D. discoideum amoebae almost as well as E. coli B/r. An earlier report suggested that the bacterium was not pathogenic for D. discoideum [76]. Jia et al. [77] reported that D. discoideum knockout mutants for autophagy genes atg1, atg6 or atg7, in contrast to control cells, supported establishment of a replicative niche, suggesting that autophagy was required for S. typhimurium degradation. By using a DNA microarray approach, a different pattern of RNA expression was found, in comparison to non-pathogenic bacteria, suggestive of cells entering starvation, despite the fact that S. typhimurium was ingested. The starvation response of the cells and its potential subversion by S. typhimurium is under study (Sillo et al., unpublished results).
CRUCIAL HOST CELL PROCESSES
Phagocytosis and Macropinocytosis
Invasive bacteria exploit phagocytosis or macropinocytosis to enter the cells. Both processes are characterized by the formation of relatively large vesicles on the plasma membrane, which are regulated by localized recruitment of the actin cytoskeleton. Phagocytosis is induced by membrane signalling triggered by particle binding to specialized membrane receptors and leading to tight enveloping of the particle by the protruding plasma membrane. Macropinocytosis is usually a cell autonomous process, resulting in massive recruitment of actin beneath the membrane, formation of ruffles and vesicles of variable size filled with extracellular liquid. Bacteria or other particles present in the external milieu can be engulfed with the liquid independently of any specific binding [78-80]. Macropinocytosis can also be induced in non-professional phagocytes by some pathogens to enter the cell. The process has been described for Salmonella, Mycobacteria and Legionella [47, 48, 78, 81, 82].
In macrophages, receptors involved in phagocytosis include the Fc receptor family, the complement receptor (CR3) and lectins [80, 83, 84]. The best known case in macrophages is the signalling pathway linked to the Fcγ receptor. Particle binding leads to receptor clustering and phosphorylation by Src-family kinases, generating docking sites for the Syk kinase, which in turn facilitates binding of docking proteins and PI3K, leading to actin cytoskeleton reorganization [84, 85]. In D. discoideum, the heterotrimeric Gα4βγ protein mediates membrane signals leading to phagocytosis, possibly resulting from receptor clustering. The D. discoideum bona fide phagocytosis receptors are so far unknown [10, 86, 87]. However, adhesion molecules like Phg1, SibA and SadA have been described and it is likely that one or a few of them are adhesion molecules involved in phagocytosis [88-90].
A major role in actin re-organization in the phagocytic and macropinocytic cup is played by membrane phosphoinositides, particularly PI(4,5)P2. This phosphoinositide is the most abundant PI-form of the plasma membrane and recruits several PH-domain containing proteins, among which are the regulators of actin nucleation, such as the Arp2/3 complex, WASP and WAVE, small G proteins of the Rho family and actin binding proteins [91]. Disappearance of PI(4,5)P2 is due to the activity of enzymes such as PI-PLC, PI3K or the PI-5-phosphatase, and is a pre-requisite for actin coat disassembly, vesicle closure and further fusion with vesicles of the endo-lysosomal pathway [92-95]. Both in macrophages and D. discoideum, PI-PLC inhibitors completely inhibit phagocytosis of bacteria, such as E. coli, as well as macropinocytosis, whereas PI3K inactivation interferes with phagocytosis of larger particles or with macropinocytosis [47, 86, 92, 94-98]. Actin assembly during phagocytosis is also regulated by small G proteins of the Rac subfamily, which activate WASP/WAVE family proteins [99, 100]. In D. discoideum there are 18 genes encoding Rac proteins, some of which are involved in phagocytosis or macropinocytosis. Except for RacH, however, which appears to regulate macropinocytosis, but not phagocytosis [101], the results obtained with null mutants and overexpressors for other rac genes underline a high degree of redundancy that explains the absence of phenotypes when a single gene is disrupted [10]. In D. discoideum, macropinocytosis is responsible for the vast majority of pinocytic events [79], and is gain of function due to a few nitrosoguanidine-induced mutations in some axenic strains [1]. The differential requirement for macropinocytosis between wild type natural isolates and axenic strains has recently allowed us to show that L. pneumophila, in contrast to other pathogens, such as M. avium or M. marinum, Neisseria meningitides or S. typhimurium, is taken up exclusively by macropinocytosis [47].
Phagosome Maturation
In less than 5 minutes after engulfment of non-pathogenic bacteria, yeast particles or latex beads, the phagosome or macropinosome fuses with acidic vesicles harbouring the V-H+ ATPase and with vesicles decorated with the Nramp1 protein [102-105]. Fusion with acidic vesicles appears to be regulated, both in D. discoideum and macrophages, by PI(3)P, a PI-form generated mainly via class III PI3K [106-108]. PI3K modulates recruitment of the small G proteins Rab5 and Rab7 to phagosomes, and PI3K inhibitors block phago-lysosome biogenesis [109]. In macrophages, Rab5 is rapidly recruited to newly formed phagosomes and is necessary for the subsequent enrollment of Rab7 either from a soluble pool or by fusion with Rab7-containing endosomes. Acquisition of Rab7 favours recruitment of motor proteins, transport of phagosomes toward the MTOC and fusion with late endosomes and lysosomes [109-111]. Rab7 regulates phagosome fusion with lysosomes, but not with acidic vesicles, not only in macrophages but also in D. discoideum [102, 112], where RabD (homolog of Rab14) appears to stimulate vesicle homotypic fusion, leading to formation of large vesicles containing several bacteria [113]. Studies using invasive and non-invasive Salmonella enterica serovar Typhimurium have shown that several other Rab proteins, in addition to Rab5, 7 or 14, associate selectively with wild type or mutant S. typhimurium, some of which are necessary for phagosome maturation [114]. It appears that phago-lysosome biogenesis is a process involving several small Rab GTPases and cannot be explained only by the single transition between Rab5 and Rab7.
The participation of small GTPases in phagosome maturation is also supported by a recent proteome analysis of L. pneumophila vacuoles purified by magnetic immunoseparation and density gradient centrifugation. Mass spectrometric analysis of purified LCVs revealed 566 host cell proteins, among them known LCV components such as the small GTPases Arf1, Rab1 and Rab7 and novel components such as Rab8, an endosomal regulator of the late secretory pathway, and the endosomal GTPase Rab14. The authors conclude that LCVs also communicate with the late secretory and endosomal pathways [115]. In a parallel study Shevchuk et al. identified in classically purified LCVs 157 host proteins which belong to different functional categories among them a number of cytoskeletal proteins, subunits of the vacuolar ATPase, proteins involved in the stress response and of the proteasome system but no small GTPases, as described above [116].
In order to survive and to establish a replicative niche, pathogens must interfere with the maturation process. They do so by either i) slowing down or stalling maturation, ii) changing the route of the phagosome or iii) escaping from it into the cytosol [17]. Another survival strategy is adaptation to the bactericidal, acidic lysosomal compartment, which is the case for Coxiella burnetii, the agent of Q fever [117, 118].
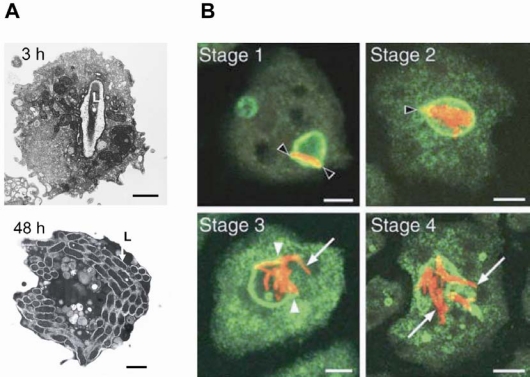
We will first consider results with M. tuberculosis and M. marinum. After uptake by macrophages or by D. discoideum, the pathogen prevents the maturation of the phagosome and replicates inside a compartment that resembles an early endosome [119]. The arrested mycobacterium containing vacuole (MCV) is characterized by the presence of early endosomal markers, the lack of late endosomal or lysosomal markers like the V-ATPase and diminished PI(3)P levels (for review see [120]). Hagedorn and Soldati [121] divided the proliferation of M. marinum in D. discoideum in three distinct phases i) an initial lag phase until 12 hpi, ii) a major proliferation phase from 12 -37 hpi and iii) a plateau or decrease in the cfu after 37 hpi. They further could divide the proliferation phase into four stages (Fig. 1). In the early stage 1, a single mycobacterium resides in a vacuole enriched in vacuolin. The second stage is defined by the proliferation of the bacteria. At the late stages 3 and 4, the vacuolin-positive membrane is ruptured and bacteria are released into the cytosol [121]. After that, M. marinum and M. tuberculosis, but not M. avium, can spread to neighbouring cells via a non-lytic mechanism that requires the host cytoskeleton and an intact mycobacterial ESX-1 secretion system [122].
Fig. (1). Infection of D. discoideum with different pathogens.
A) Transmission electron micrographs of L. pneumophila PhilI JR32 infected D. discoideum cells 3 and 48 hours post infection. 3 h after infection the host cell contains mostly one L. pneumophila (L) within the phagosome. After 48 h the D. discoideum cell is almost entirely filled with L. pneumophila. Scale bars, 2 µm. (Reproduced from figure 1 of [149], modified). B) Immunofluorescence micrographs of phase 2 of M. marinum infection of D. discoideum. Four sequential stages can be distinguished in the establishment and rupture of the vacuolin-positive vacuole. At the early stage 1, a single mycobacterium deformed a vacuole already enriched in vacuolin (black arrowheads). The second stage is defined by the proliferation of the bacteria inside the vacuole which leads to more deformation of the membrane (black arrowhead). At the late stages 3 and 4, the vacuolin-positive membrane was ruptured (arrowheads mark the edges of the membrane sheets generated during niche rupture) and bacteria were released into the cytosol (arrows). M. marinum is labelled in red and vacuolin in green. The scale bar represents 5 µm (Reproduced from figure 3 of [121], modified).
In contrast, L. pneumophila changes the route of the Legionella containing vacuole (LCV) and is found in a compartment that is different from that of a non-pathogen. The LCV first associates with mitochondria and with vesicles derived from the ER. It then binds ribosomes and becomes similar to rough ER (for review see [24]). The LCV is characterised by the ER resident protein calnexin, the v-SNARE Sec22b and the small GTPases Arf1 and Rab1 [123, 124]. Finally the calnexin-positive LCVs undergo a transition from tight to spacious vacuoles a few hours post-infection [50]. In D. discoideum the LCV recruits quite rapidly Nramp1, but not the V-H+ ATPase nor vacuolin. Only late in infection are the V-H+ ATPase or the post-lysosomal marker vacuolin found in large vacuoles containing replicating bacteria [47]. Whether L. pneumophila uses the post-lysosomal pathway for exiting the cell, as shown for mycobacteria, is unclear; extensive cell lysis occurs 48 hours post-infection, which suggesta that the bacteria leave the cells by lysing them.
It turned out that the PI metabolism is critically involved in these processes as L. pneumophila secretes effector proteins via the Icm/Dot type 4 secretion system that bind to PI(4)P on the LCV [125-127]. Furthermore, bacterial replication was more efficient in D. discoideum cells lacking the inositol polyphosphate 5-phosphatase, Dd5P4, a homologue of human OCRL1 (Oculocerebrorenal syndrome of Lowe), implicated in retrograde endosome to Golgi trafficking [30], and in D. discoideum mutants of phosphatidylinositol-3 kinases (PI3Ks) and PTEN [47, 125]. Interestingly, inactivating PI3K has no effect on calnexin or Nramp1 recruitment to LCV, whereas fusion with acidic vesicles is further blocked, suggesting that L. pneumophila may hinder V-H+ ATPase recruitment by altering the phosphoinositide composition of the LCV, thus favouring formation of a replication vacuole [47].
In D. discoideum Rab14 induces phagosome homotypic fusion, leading to formation of large vesicles. In macrophages, Rab14 silencing or expression of Rab14 dominant-negative mutants lead to phagolysosomal maturation of phagosomes containing live mycobacteria, whereas overexpression of Rab14 or of a constitutively active Rab14 mutant blocks maturation of phagosomes containing dead bacteria [128]. Similarly, Rab22 that is transiently expressed on latex beads containing phagosomes was, instead, retained on M. tuberculosis-containing phagosomes [129]. Therefore the presence of Rab14 or Rab22 in macrophages seems to be important to inhibit or delay phago-lysosomal biogenesis.
Ion acquisition is important for intracellular survival of pathogenic bacteria. Mg2+, Mn2+, K+ and Zn2+ have been implicated in S. typhimurium virulence [130-133], whereas Fe2+ is an essential metal for all cells, and it is known that Salmonella, Legionella and Mycobacteria accumulate large amounts of iron [134-136]. In response to iron deprivation, these bacteria express siderophores to recruit iron. Iron availability in the phagosome is limited by the activity of Nramp1, a divalent metal transporter that depletes the phagosome of iron by a mechanism dependent on the proton gradient [105, 137]. Mutations in Nramp1 have been linked to innate susceptibility to mycobacterial diseases and S. typhimurium infection [137-140]. Inactivation of the gene in D. discoideum leads to increased intracellular growth of L. pneumophila and M. avium, whereas its overexpression completely inhibits L. pneumophila growth [105]. L. pneumophila hinders recruitment of the V-H+ ATPase in the Legionella-containing vacuole, without interfering with Nramp1 recruitment. Since a proton gradient is required for Nramp1-dependent depletion of iron, the absence of the vacuolar ATPase generates a milieu in which Nramp1 does not function properly [47].
Macroautophagy
Bacterial pathogens manipulate host cell processes to avoid phago-lysosomal fusion and to establish a replicative niche [24]. The host, on the other hand, initiates elaborate defense processes, of which one appears to be macroautophagy (hereafter autophagy) [141]. Autophagy is an ancient cellular pathway that is conserved from yeast to humans and has presumably evolved to enable cells to survive periods of starvation. More than 30 autophagy (ATG) genes have been identified, mainly in yeast, of which 18 constitute the core machinery for starvation induced autophagy. Cytosolic material is captured into double membrane-bound vesicles that mature into autophagosomes and then, after fusion with lysosomes, become autophagolysosomes. There, the cargo is degraded and then recycled for further use [142]. Autophagy contributes to many physiological and pathological processes, including cell differentiation and development, programmed cell death, cancer and neurodegenerative disorders. There is accumulating evidence that autophagy is also a general and important defense mechanism in the complex interactions between host and pathogen [143]. Some pathogens e.g. M. tuberculosis are targeted for degradation through autophagy [144]. In a recent genome-wide analysis of the host intracellular network that regulates survival of M. tuberculosis it was found that host factors predominantly function through the regulation of autophagy [145]. Other pathogens have developed means to evade autophagy, e.g. Shigella flexneri or even to utilize the autophagosome for replication e.g. Staphylococccus aureus [141, 146, 147].
In D. discoideum the role of autophagy in infection was so far investigated with L. pneumophila and S. typhimurium in several autophagy mutants. Otto et al. reported for atg1, 5, 6, 7 and 8 knock-out mutants that autophagy is dispensable for intracellular L. pneumophila replication. However, the authors did not examine if autophagy might be important in restricting the intracellular replication [148]. A microarray study of the time course of Legionella infection revealed differential regulation of three core autophagy genes that are required in the early phase of autophagosome formation [149]. Interestingly, ATG9 was up-regulated while ATG8 and 16 were down-regulated, suggesting that host and pathogen target different pivotal autophagy genes during infection. It is tempting to speculate that the host tries to up-regulate autophagy via ATG9 while the pathogen counteracts via down-regulation of ATG8 and 16. A knock-out mutant of the ATG9 gene showed a strong phagocytosis defect that was particularly apparent when cells were infected with L. pneumophila. However, those Legionellae that entered the host could multiply better in mutant than in wild-type cells. This was due to a less efficient clearance in the early phase and a more efficient replication in the late phase of infection [150]. In an elegant recent study two model organisms, Caenorhabditis elegans and D. discoideum, were used to examine the effects of autophagy gene inactivation on infection with S. typhimurium. In both organisms, the inactivation of autophagy genes increased the intracellular replication of S. typhimurium [77]. In support of a role of autophagy in infection with L. pneumophila it is also worth mentioning that the LCV is associated with markers of autophagy, such as ATG7 and 8. If the LCV is a modified autophagosome, then autophagy must be arrested for the bacteria to maintain intracellular replication [151]. Thus, consistent with studies using macrophages and other models the data from D. discoideum support a protective role of autophagy during pathogen infection, raising the possibility that cellular defense against pathogens could be induced by drugs that stimulate autophagy.
D. DISCOIDEUM AS EXPERIMENTAL SYSTEM FOR DRUG TESTING
As mentioned in the previous sections, D. discoideum cells share with higher eukaryotes several cellular processes and underlying homologous genes [9]. In addition, the cells are not encased in a rigid cell wall and the plasma membrane is thus directly exposed to the extracellular milieu. The composition of the plasma membrane is not basically different from that of higher eukaryotes, except that cholesterol is substituted with ergosterol and that, among the carbohydrate residues of proteins or glycolipids, sialic acid is not found [152]. It is therefore not surprising that pharmacological approaches have been regularly used with D. discoideum cells and that many drugs affecting mammalian cells have proven to be effective also in D. discoideum, though in some cases higher concentrations are required.
PLC and PI3K inhibitors, such as U73122, wortmannin, or LY294002, have been used to characterize phagocytosis and macropinocytosis [86, 97] as well as chemotaxis [153-155]. Actin assembly can be inhibited by cytochalasin or latrunculin A, thus inhibiting spontaneous and chemotactic cell motility as well as phagocytosis and macropinocytosis [156-159]. PLA2 inhibitors do not affect phagocytosis [86, 97], but they have been shown to inhibit calcium signalling [160] and, when used in combination with PI3K inhibitors, chemotaxis [155, 161]. Intracellular and extracellular calcium chelators, such as BAPTA-AM or EDTA and EGTA, have been used to study, among others, cell-cell adhesion and phagocytosis [86, 162-164]. Tyrosine kinase and phosphatase inhibitors helped in showing actin phosphorylation changes [165-168]. Valproic acid or cisplatin have been used to study lithium signalling and effects on gene expression, growth and development [169-172].
In infection studies, drugs have been used to characterize L. pneumophila uptake and replication [173]. Pharmacological or genetic inhibition of PI3K stimulates L. pneumophila infection [47, 125]. Addition of the PI3K inhibitor LY294002 at different time points during infection has recently been used to identify a short period immediately after bacterial uptake, which is sensitive to addition of the drug, stimulating intracellular replication of the bacteria [47]. Similarly, it has been shown pharmacologically that PLC and actin assembly are required for L. pneumophila uptake, but do not seem to play a role for establishment of the replicative niche [47].
These scattered and largely incomplete examples emphasize that D. discoideum cells can be conveniently used for drug testing [174, 175], in combination with the variety of assays that have been developed to study phagocytosis, infection as well as cell motility, chemotaxis, cell-substratum and cell-cell adhesion, signalling, growth, cell differentiation or development [164].
CONCLUSIONS
Investigations with model organisms have significantly contributed to our understanding of host-pathogen interactions and have lead to the discovery of many host genes that are either involved in the defence response or required for the pathogen to establish its replicative niche. D. discoideum is particularly suited for infection studies, because it is a professional phagocyte, its genome is completely sequenced and excellent genetic, biochemical and cell biological tools are available [2, 8, 10, 11]. Mainly, D. discoideum was used to study the host response upon infection with different pathogens in particular L. pneumophila, M. marinum and M. avium and S. typhimurium. This led to the discovery of a variety of bacterial and host cell factors, among them many genes encoding cytoskeletal and signaling proteins that are important in the infection process. A further advantage of D. discoideum is that it can be easily used for drug testing and as screening host for wild-type or mutagenised pathogenic and non-pathogenic bacteria [27, 28]. Furthermore, untargeted mutational screens to find crucial host factors can be carried out [26]. In summary, the properties of D. discoideum in combination with the impressive armoury of tools that is available will help to further dissect host pathogen crosstalk in the years to come.
ACKNOWLEDGEMENTS
LE acknowledges support by the DFG (SFB670), the DAAD (Vigoni program) and Köln Fortune. SB is supported by the Piemonte Region (RSF projects ’08-’10) and the Vigoni program.
REFERENCES
- 1.Kessin RH. Dictyostelium - evolution, cell biology, and the development of multicellularity. Cambridge UK: Cambridge Univ Press; 2001. [Google Scholar]
- 2.Eichinger L. Revamp a model-status and prospects of the Dictyostelium genome project. Curr Genet. 2003;44(2):59–72. doi: 10.1007/s00294-003-0416-1. [DOI] [PubMed] [Google Scholar]
- 3.King JS, Insall RH. Chemotaxis: finding the way forward with Dictyostelium. Trends Cell Biol. 2009;19(10):523–30. doi: 10.1016/j.tcb.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Weijer CJ. Collective cell migration in development. J Cell Sci. 2009;122(Pt 18):3215–23. doi: 10.1242/jcs.036517. [DOI] [PubMed] [Google Scholar]
- 5.Robinson DN, Spudich JA. Mechanics and regulation of cytokinesis. Curr Opin Cell Biol. 2004;16(2):182–8. doi: 10.1016/j.ceb.2004.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chisholm RL, Firtel RA. Insights into morphogenesis from a simple developmental system. Nat Rev Mol Cell Biol. 2004;5(7):531–41. doi: 10.1038/nrm1427. [DOI] [PubMed] [Google Scholar]
- 7.Glöckner G, Eichinger L, Szafranski K, et al. Sequence and analysis of chromosome 2 of Dictyostelium discoideum. Nature. 2002;418(6893):79–85. doi: 10.1038/nature00847. [DOI] [PubMed] [Google Scholar]
- 8.Eichinger L, Pachebat JA, Glockner G, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435(7038):43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams JG, Noegel AA, Eichinger L. Manifestations of multicellularity: Dictyostelium reports in. Trends Genet. 2005;21(7):392–8. doi: 10.1016/j.tig.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Bozzaro S, Bucci C, Steinert M. Phagocytosis and host-pathogen interactions in Dictyostelium with a look at macrophages. Int Rev Cell Mol Biol. 2008;271:253–300. doi: 10.1016/S1937-6448(08)01206-9. [DOI] [PubMed] [Google Scholar]
- 11.Cosson P, Soldati T. Eat, kill or die: when amoeba meets bacteria. Curr Opin Microbiol. 2008;11(3):271–6. doi: 10.1016/j.mib.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Clarke M. Recent insights into host-pathogen interactions from Dictyostelium. Cell Microbiol. 2010;12(3):283–91. doi: 10.1111/j.1462-5822.2009.01413.x. [DOI] [PubMed] [Google Scholar]
- 13.Greub G, Raoult D. Microorganisms resistant to free-living amoebae. Clin Microbiol Rev. 2004;17(2):413–33. doi: 10.1128/CMR.17.2.413-433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steenbergen JN, Shuman HA, Casadevall A. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc Natl Acad Sci USA. 2001;98(26):15245–50. doi: 10.1073/pnas.261418798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goy G, Thomas V, Rimann K, Jaton K, Prod'hom G, Greub G. The Neff strain of Acanthamoeba castellanii, a tool for testing the virulence of Mycobacterium kansasii. Res Microbiol. 2007;158(4):393–7. doi: 10.1016/j.resmic.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Duhon D, Cardelli J. The regulation of phagosome maturation in Dictyostelium. J Muscle Res Cell Motil. 2002;23:803–8. doi: 10.1023/a:1024435913949. [DOI] [PubMed] [Google Scholar]
- 17.Haas A. The phagosome: compartment with a license to kill. Traffic. 2007;8(4):311–30. doi: 10.1111/j.1600-0854.2006.00531.x. [DOI] [PubMed] [Google Scholar]
- 18.Depraitere C, Darmon M. [Growth of "Dictyostelium discoideum" on different species of bacteria (author's transl)] Ann Microbiol (Paris) 1978;129 B(3):451–61. [PubMed] [Google Scholar]
- 19.Solomon JM, Rupper A, Cardelli JA, Isberg RR. Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect Immun. 2000;68(5):2939–47. doi: 10.1128/iai.68.5.2939-2947.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hägele S, Kohler R, Merkert H, Schleicher M, Hacker J, Steinert M. Dictyostelium discoideum: a new host model system for intracellular pathogens of the genus Legionella. Cell Microbiol. 2000;2(2):165–71. doi: 10.1046/j.1462-5822.2000.00044.x. [DOI] [PubMed] [Google Scholar]
- 21.Dorer MS, Isberg RR. Non-vertebrate hosts in the analysis of host-pathogen interactions. Microbes Infect. 2006;8:1637–46. doi: 10.1016/j.micinf.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 22.Jin T, Xu X, Fang J, et al. How human leukocytes track down and destroy pathogens: lessons learned from the model organism Dictyostelium discoideum. Immunol Res. 2009;43(1-3):118–27. doi: 10.1007/s12026-008-8056-7. [DOI] [PubMed] [Google Scholar]
- 23.Hilbi H, Weber SS, Ragaz C, Nyfeler Y, Urwyler S. Environmental predators as models for bacterial pathogenesis. Environ Microbiol. 2007;9(3):563–75. doi: 10.1111/j.1462-2920.2007.01238.x. [DOI] [PubMed] [Google Scholar]
- 24.Isberg RR, O'Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nature Rev Microbiol. 2009;7(1):12–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuspa A, Loomis WF. Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc Natl Acad Sci USA. 1992;89(18):8803–7. doi: 10.1073/pnas.89.18.8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z, Dugan AS, Bloomfield G, et al. The amoebal MAP kinase response to Legionella pneumophila is regulated by DupA. Cell Host Microbe. 2009;6(3):253–67. doi: 10.1016/j.chom.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cosson P, Zulianello L, Join-Lambert O, et al. Pseudomonas aeruginosa virulence analyzed in a Dictyostelium discoideum host system. J Bacteriol. 2002;184(11):3027–33. doi: 10.1128/JB.184.11.3027-3033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Froquet R, Lelong E, Marchetti A, Cosson P. Dictyostelium discoideum: a model host to measure bacterial virulence. Nature Protoc. 2009;4(1):25–30. doi: 10.1038/nprot.2008.212. [DOI] [PubMed] [Google Scholar]
- 29.Steinert M, Heuner K. Dictyostelium as host model for pathogenesis. Cell Microbiol. 2005;7(3):307–14. doi: 10.1111/j.1462-5822.2005.00493.x. [DOI] [PubMed] [Google Scholar]
- 30.Weber SS, Ragaz C, Hilbi H. The inositol polyphosphate 5-phosphatase OCRL1 restricts intracellular growth of Legionella, localizes to the replicative vacuole and binds to the bacterial effector LpnE. Cell Microbiol. 2009;11(3):442–60. doi: 10.1111/j.1462-5822.2008.01266.x. [DOI] [PubMed] [Google Scholar]
- 31.Fraser DW, Tsai TR, Orenstein W, et al. Legionnaires' disease: description of an epidemic of pneumonia. N Engl J Med. 1977;297(22):1189–97. doi: 10.1056/NEJM197712012972201. [DOI] [PubMed] [Google Scholar]
- 32.Cordevant C, Tang JS, Cleland D, Lange M. Characterization of members of the Legionellaceae family by automated ribotyping. J Clin Microbiol. 2003;41(1):34–43. doi: 10.1128/JCM.41.1.34-43.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fields BS, Benson RF, Besser RE. Legionella and Legionnaires' disease: 25 years of investigation. Clin Microbiol Rev. 2002;15(3):506–26. doi: 10.1128/CMR.15.3.506-526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwaik YA. Fatal attraction of mammalian cells to Legionella pneumophila. Mol Microbiol. 1998;30(4):689–95. doi: 10.1046/j.1365-2958.1998.01092.x. [DOI] [PubMed] [Google Scholar]
- 35.Lamoth F, Greub G. Amoebal pathogens as emerging causal agents of pneumonia. FEMS Microbiol Rev. 2010;34(3):260–80. doi: 10.1111/j.1574-6976.2009.00207.x. [DOI] [PubMed] [Google Scholar]
- 36.Horwitz MA. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J Exp Med. 1983;158(6):2108–26. doi: 10.1084/jem.158.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horwitz MA, Maxfield FR. Legionella pneumophila inhibits acidification of its phagosome in human monocytes. J Cell Biol. 1984;99(6):1936–43. doi: 10.1083/jcb.99.6.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wintermeyer E, Ludwig B, Steinert M, Schmidt B, Fischer G, Hacker J. Influence of site specifically altered Mip proteins on intracellular survival of Legionella pneumophila in eukaryotic cells. Infect Immun. 1995;63(12):4576–83. doi: 10.1128/iai.63.12.4576-4583.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Francione L, Smith PK, Accari SL, et al. Legionella pneumophila multiplication is enhanced by chronic AMPK signalling in mitochondrially diseased Dictyostelium cells. Dis Model Mech. 2009;2(9-10):479–89. doi: 10.1242/dmm.003319. [DOI] [PubMed] [Google Scholar]
- 40.Swanson MS, Hammer BK. Legionella pneumophila pathogesesis: a fateful journey from amoebae to macrophages. Annu Rev Microbiol. 2000;54:567–613. doi: 10.1146/annurev.micro.54.1.567. [DOI] [PubMed] [Google Scholar]
- 41.Gao LY, Abu Kwaik Y. Activation of caspase 3 during Legionella pneumophila-induced apoptosis. Infect Immun. 1999;67(9):4886–94. doi: 10.1128/iai.67.9.4886-4894.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang C, Kuspa A. Transcriptional down-regulation and rRNA cleavage in Dictyostelium discoideum mitochondria during Legionella pneumophila infection. PLoS One. 2009;4(5):e5706. doi: 10.1371/journal.pone.0005706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;79(5352):73–6. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 44.Cazalet C, Rusniok C, Bruggemann H, et al. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet. 2004;36(11):1165–73. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- 45.Lau HY, Ashbolt NJ. The role of biofilms and protozoa in Legionella pathogenesis: implications for drinking water. J Appl Microbiol. 2009;107(2):368–78. doi: 10.1111/j.1365-2672.2009.04208.x. [DOI] [PubMed] [Google Scholar]
- 46.Steinert M, Leippe M, Roeder T. Surrogate hosts: protozoa and invertebrates as models for studying pathogen-host interactions. Int J Med Microbiol. 2003;293(5):321–32. doi: 10.1078/1438-4221-00275. [DOI] [PubMed] [Google Scholar]
- 47.Peracino B, Balest A, Bozzaro S. Differential regulation by phosphoinositides of bacterial phagocytosis and Nramp1 (Slc11a1)-induced resistance to Legionella infection in Dictyostelium. J Cell Sci. 2010;123(23):4039–51. doi: 10.1242/jcs.072124. [DOI] [PubMed] [Google Scholar]
- 48.Watarai M, Derre I, Kirby J, Growney JD, Dietrich WF, Isberg RR. Legionella pneumophila is internalized by a macropinocytotic uptake pathway controlled by the Dot/Icm system and the mouse Lgn1 locus. J Exp Med. 2001;194(8):1081–96. doi: 10.1084/jem.194.8.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Solomon JM, Isberg RR. Growth of Legionella pneumophila in Dictyostelium discoideum: a novel system for genetic analysis of host-pathogen interactions. Trends Microbiol. 2000;8(10):478–80. doi: 10.1016/s0966-842x(00)01852-7. [DOI] [PubMed] [Google Scholar]
- 50.Lu H, Clarke M. Dynamic properties of Legionella-containing phagosomes in Dictyostelium amoebae. Cell Microbiol. 2005;7:995–1007. doi: 10.1111/j.1462-5822.2005.00528.x. [DOI] [PubMed] [Google Scholar]
- 51.Rook GA, Hamelmann E, Brunet LR. Mycobacteria and allergies. Immunobiology. 2007;212(6):461–73. doi: 10.1016/j.imbio.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 52.Dye C, Scheele S, Dolin P, Pathania V, Raviglione MC. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA. 1999;282(7):677–86. doi: 10.1001/jama.282.7.677. [DOI] [PubMed] [Google Scholar]
- 53.DiGiuseppe Champion PA, Champion MM, Manzanillo P, Cox JS. ESX-1 secreted virulence factors are recognized by multiple cytosolic AAA ATPases in pathogenic mycobacteria. Mol Microbiol. 2009;73(5):950–62. doi: 10.1111/j.1365-2958.2009.06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cole ST, Brosch R, Parkhill J, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393(6685):537–44. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 55.Ramakrishnan L, Federspiel NA, Falkow S. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science. 2000;288(5470):1436–9. doi: 10.1126/science.288.5470.1436. [DOI] [PubMed] [Google Scholar]
- 56.Chaitra MG, Shaila MS, Nayak R. Detection of interferon gamma-secreting CD8+ T lymphocytes in humans specific for three PE/PPE proteins of Mycobacterium tuberculosis. Microbes Infect. 2008;10(8):858–67. doi: 10.1016/j.micinf.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 57.Mehta PK, Pandey AK, Subbian S, et al. Identification of Mycobacterium marinum macrophage infection mutants. Microb Pathog. 2006;40(4):139–51. doi: 10.1016/j.micpath.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 58.Gluckman SJ. Mycobacterium marinum. Clin Dermatol. 1995;13(3):273–6. doi: 10.1016/0738-081x(95)00023-9. [DOI] [PubMed] [Google Scholar]
- 59.Tobin DM, Ramakrishnan L. Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cell Microbiol. 2008;10(5):1027–39. doi: 10.1111/j.1462-5822.2008.01133.x. [DOI] [PubMed] [Google Scholar]
- 60.Shiloh MU, DiGiuseppe Champion PA. To catch a killer. What can mycobacterial models teach us about Mycobacterium tuberculosis pathogenesis? Curr Opin Microbiol. 2010;13(1):86–92. doi: 10.1016/j.mib.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pozos TC, Ramakrishnan L. New models for the study of Mycobacterium-host interactions. Curr Opin Immunol. 2004;16(4):499–505. doi: 10.1016/j.coi.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 62.Zhang S, Kingsley RA, Santos RL, et al. Molecular pathogenesis of Salmonella enterica serotype typhimurium-induced diarrhea. Infect Immun. 2003;71(1):1–12. doi: 10.1128/IAI.71.1.1-12.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou D, Galan J. Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect. 2001;3(14-15):1293–8. doi: 10.1016/s1286-4579(01)01489-7. [DOI] [PubMed] [Google Scholar]
- 64.Patel JC, Galan JE. Manipulation of the host actin cytoskeleton by Salmonella--all in the name of entry. Curr Opin Microbiol. 2005;8(1):10–5. doi: 10.1016/j.mib.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 65.Drecktrah D, Knodler LA, Ireland R, Steele-Mortimer O. The mechanism of Salmonella entry determines the vacuolar environment and intracellular gene expression. Traffic. 2006;7(1):39–51. doi: 10.1111/j.1600-0854.2005.00360.x. [DOI] [PubMed] [Google Scholar]
- 66.McGhie EJ, Brawn LC, Hume PJ, Humphreys D, Koronakis V. Salmonella takes control: effector-driven manipulation of the host. Curr Opin Microbiol. 2009;12(1):117–24. doi: 10.1016/j.mib.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Valdez Y, Ferreira RB, Finlay BB. Molecular mechanisms of Salmonella virulence and host resistance. Curr Top Microbiol Immunol. 2009;337:93–127. doi: 10.1007/978-3-642-01846-6_4. [DOI] [PubMed] [Google Scholar]
- 68.Rathman M, Sjaastad MD, Falkow S. Acidification of phagosomes containing Salmonella typhimurium in murine macrophages. Infect Immun. 1996;64(7):2765–73. doi: 10.1128/iai.64.7.2765-2773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Steele-Mortimer O, Meresse S, Gorvel JP, Toh BH, Finlay BB. Biogenesis of Salmonella typhimurium-containing vacuoles in epithelial cells involves interactions with the early endocytic pathway. Cell Microbiol. 1999;1(1):33–49. doi: 10.1046/j.1462-5822.1999.00003.x. [DOI] [PubMed] [Google Scholar]
- 70.Shea JE, Hensel M, Gleeson C, Holden DW. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93(6):2593–7. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cirillo DM, Valdivia RH, Monack DM, Falkow S. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol. 1998;30(1):175–88. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 72.Hensel M, Shea JE, Waterman SR, et al. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol Microbiol. 1998;30(1):163–74. doi: 10.1046/j.1365-2958.1998.01047.x. [DOI] [PubMed] [Google Scholar]
- 73.Ibarra JA, Steele-Mortimer O. Salmonella--the ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell Microbiol. 2009;11(11):1579–86. doi: 10.1111/j.1462-5822.2009.01368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boucrot E, Henry T, Borg JP, Gorvel JP, Meresse S. The intracellular fate of Salmonella depends on the recruitment of kinesin. Science. 2005;308(5725):1174–8. doi: 10.1126/science.1110225. [DOI] [PubMed] [Google Scholar]
- 75.Henry T, Gorvel JP, Meresse S. Molecular motors hijacking by intracellular pathogens. Cell Microbiol. 2006;8(1):23–32. doi: 10.1111/j.1462-5822.2005.00649.x. [DOI] [PubMed] [Google Scholar]
- 76.Skriwan C, Fajardo M, Hagele S, et al. Various bacterial pathogens and symbionts infect the amoeba Dictyostelium discoideum. Int J Med Microbiol. 2002;291(8):615–24. doi: 10.1078/1438-4221-00177. [DOI] [PubMed] [Google Scholar]
- 77.Jia K, Thomas C, Akbar M, et al. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc Natl Acad Sci USA. 2009;106(34):14564–9. doi: 10.1073/pnas.0813319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Amyere M, Mettlen M, Van der Smissen P, et al. Origin, originality, functions, subversions and molecular signalling of macropinocytosis. Int J Med Microbiol. 2002;291:487–94. doi: 10.1078/1438-4221-00157. [DOI] [PubMed] [Google Scholar]
- 79.Maniak M. Conserved features of endocytosis in Dictyostelium. Int Rev Cytol. 2002;221(6):257–87. doi: 10.1016/s0074-7696(02)21014-1. [DOI] [PubMed] [Google Scholar]
- 80.Swanson JA. Shaping cups into phagosomes and macropinosomes. Nat Rev Mol Cell Biol. 2008;9(8):639–49. doi: 10.1038/nrm2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Francis CL, Ryan TA, Jones BD, Smith SJ, Falkow S. Ruffles induced by Salmonella and other stimuli direct macropinocytosis of bacteria. Nature. 1993;364(6438):639–42. doi: 10.1038/364639a0. [DOI] [PubMed] [Google Scholar]
- 82.Alpuche-Aranda CM, Racoosin EL, Swanson JA, Miller SI. Salmonella stimulate macrophage macropinocytosis and persist within spacious phagosomes. J Exp Med. 1994;179(2):601–8. doi: 10.1084/jem.179.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Greenberg S. Signal transduction of phagocytosis. Trends Cell Biol. 1995;5(3):93–9. doi: 10.1016/s0962-8924(00)88957-6. [DOI] [PubMed] [Google Scholar]
- 84.Cox D, Greenberg S. Phagocytic signaling strategies: Fc(gamma)receptor-mediated phagocytosis as a model system. Semin Immunol. 2001;13(6):339–45. doi: 10.1006/smim.2001.0330. [DOI] [PubMed] [Google Scholar]
- 85.Gu H, Botelho RJ, Yu M, Grinstein S, Neel BG. Critical role for scaffolding adapter Gab2 in Fc gamma R-mediated phagocytosis. J Cell Biol. 2003;161(6):1151–61. doi: 10.1083/jcb.200212158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peracino B, Borleis J, Jin T, et al. G protein beta subunit-null mutants are impaired in phagocytosis and chemotaxis due to inappropriate regulation of the actin cytoskeleton. J Cell Biol. 1998;141(7):1529–37. doi: 10.1083/jcb.141.7.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gotthardt D, Blancheteau V, Bosserhoff A, Ruppert T, Delorenzi M, Soldati T. Proteomics fingerprinting of phagosome maturation and evidence for the role of a Galpha during uptake. Mol Cell Proteomics. 2006;5(12):2228–43. doi: 10.1074/mcp.M600113-MCP200. [DOI] [PubMed] [Google Scholar]
- 88.Fey P, Stephens S, Titus MA, Chisholm RL. SadA, a novel adhesion receptor in Dictyostelium. J Cell Biol. 2002;159(6):1109–19. doi: 10.1083/jcb.200206067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cornillon S, Gebbie L, Benghezal M, et al. An adhesion molecule in free-living Dictyostelium amoebae with integrin beta features. EMBO Rep. 2006;7(6):617–21. doi: 10.1038/sj.embor.7400701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cornillon S, Pech E, Benghezal M, et al. Phg1p is a nine-transmembrane protein superfamily member involved in dictyostelium adhesion and phagocytosis. J Biol Chem. 2000;275(44):34287–92. doi: 10.1074/jbc.M006725200. [DOI] [PubMed] [Google Scholar]
- 91.Lemmon MA. Pleckstrin homology (PH) domains and phosphoinositides. Biochem Soc Symp. 2007;(74):81–93. doi: 10.1042/BSS0740081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Botelho RJ, Teruel M, Dierckman R, et al. Localized biphasic changes in phosphatidylinositol-4,5-bisphosphate at sites of phagocytosis. J Cell Biol. 2000;151(7):1353–68. doi: 10.1083/jcb.151.7.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vieira OV, Botelho RJ, Rameh L, et al. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J Cell Biol. 2001;155(1):19–25. doi: 10.1083/jcb.200107069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dormann D, Weijer G, Dowler S, Weijer CJ. In vivo analysis of 3-phosphoinositide dynamics during Dictyostelium in phagocytosis and chemotaxis. J Cell Sci. 2004;117:6497–509. doi: 10.1242/jcs.01579. [DOI] [PubMed] [Google Scholar]
- 95.Scott CC, Dobson W, Botelho RJ, et al. Phosphatidylinositol-4,5-bisphosphate hydrolysis directs actin remodeling during phagocytosis. J Cell Biol. 2005;169(1):139–49. doi: 10.1083/jcb.200412162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhou K, Pandol S, Bokoch G, Traynor-Kaplan AE. Disruption of Dictyostelium PI3K genes reduces [32P]phosphatidylinositol 3,4 bisphosphate and [32P]phosphatidylinositol trisphosphate levels, alters F-actin distribution and impairs pinocytosis. J Cell Sci. 1998;111:283–94. doi: 10.1242/jcs.111.2.283. [DOI] [PubMed] [Google Scholar]
- 97.Seastone DJ, Zhang LY, Buczynski G, et al. The small Mr Ras-like GTPase Rap1 and the phospholipase C pathway act to regulate phagocytosis in Dictyostelium discoideum. Mol Biol Cell. 1999;10:393–406. doi: 10.1091/mbc.10.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cheeseman KL, Ueyama T, Michaud TM, et al. Targeting of protein kinase C-epsilon during Fcgamma receptor-dependent phagocytosis requires the epsilonC1B domain and phospholipase C-gamma1. Mol Biol Cell. 2006;17(2):799–813. doi: 10.1091/mbc.E04-12-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ridley AJ. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006;16(10):522–9. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 100.Pollitt AY, Insall RH. Loss of Dictyostelium HSPC300 causes a scar-like phenotype and loss of SCAR protein. BMC Cell Biol. 2009;10:13. doi: 10.1186/1471-2121-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Somesh BP, Neffgen C, Iijima M, Devreotes P, Rivero F. Dictyostelium RacH regulates endocytic vesicular trafficking and is required for localization of vacuolin. Traffic. 2006;7:1194–212. doi: 10.1111/j.1600-0854.2006.00455.x. [DOI] [PubMed] [Google Scholar]
- 102.Rupper A, Grove B, Cardelli J. Rab7 regulates phagosome maturation in Dictyostelium. J Cell Sci. 2001;114:2449–60. doi: 10.1242/jcs.114.13.2449. [DOI] [PubMed] [Google Scholar]
- 103.Gotthardt D, Warnatz HJ, Henschel O, Bruckert F, Schleicher M, Soldati T. High-resolution dissection of phagosome maturation reveals distinct membrane trafficking phases. Mol Biol Cell. 2002;13:3508–20. doi: 10.1091/mbc.E02-04-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Clarke M, Kohler J, Arana Q, Liu TY, Heuser J, Gerisch G. Dynamics of the vacuolar H+-ATPase in the contractile vacuole complex and the endosomal pathway of Dictyostelium cells. J Cell Sci. 2002;115:2893–905. doi: 10.1242/jcs.115.14.2893. [DOI] [PubMed] [Google Scholar]
- 105.Peracino B, Wagner C, Balest A, et al. Function and mechanism of action of Dictyostelium Nramp1 (Slc11a1) in bacterial infection. Traffic. 2006;7(1):22–38. doi: 10.1111/j.1600-0854.2005.00356.x. [DOI] [PubMed] [Google Scholar]
- 106.Ellson CD, Anderson KE, Morgan G, et al. Phosphatidylinositol 3-phosphate is generated in phagosomal membranes. Curr Biol. 2001;11(20):1631–5. doi: 10.1016/s0960-9822(01)00447-x. [DOI] [PubMed] [Google Scholar]
- 107.Yeung T, Ozdamar B, Paroutis P, Grinstein S. Lipid metabolism and dynamics during phagocytosis. Curr Opin Cell Biol. 2006;18(4):429–37. doi: 10.1016/j.ceb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 108.Clarke M, Maddera L, Engel U, Gerisch G. Retrieval of the vacuolar H-ATPase from phagosomes revealed by live cell imaging. PLoS One. 2010;5(1):e8585. doi: 10.1371/journal.pone.0008585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vieira OV, Bucci C, Harrison RE, et al. Modulation of Rab5 and Rab7 recruitment to phagosomes by phosphatidylinositol 3-kinase. Mol Cell Biol. 2003;23(7):2501–14. doi: 10.1128/MCB.23.7.2501-2514.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Harrison RE, Bucci C, Vieira OV, Schroer TA, Grinstein S. Phagosomes fuse with late endosomes and/or lysosomes by extension of membrane protrusions along microtubules: role of Rab7 and RILP. Mol Cell Biol. 2003;23(18):6494–506. doi: 10.1128/MCB.23.18.6494-6506.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kitano M, Nakaya M, Nakamura T, Nagata S, Matsuda M. Imaging of Rab5 activity identifies essential regulators for phagosome maturation. Nature. 2008;453(7192):241–5. doi: 10.1038/nature06857. [DOI] [PubMed] [Google Scholar]
- 112.Laurent O, Bruckert F, Adessi C, Satre M. In vitro reconstituted Dictyostelium discoideum early endosome fusion is regulated by Rab7 but proceeds in the absence of ATP-Mg2+ from the bulk solution. J Biol Chem. 1998;273:793–9. doi: 10.1074/jbc.273.2.793. [DOI] [PubMed] [Google Scholar]
- 113.Harris E, Cardelli J. RabD, a Dictyostelium Rab14-related GTPase, regulates phagocytosis and homotypic phagosome and lysosome fusion. J Cell Sci. 2002;115:3703–13. doi: 10.1242/jcs.00050. [DOI] [PubMed] [Google Scholar]
- 114.Smith AC, Heo WD, Braun V, et al. A network of Rab GTPases controls phagosome maturation and is modulated by Salmonella enterica serovar Typhimurium. J Cell Biol. 2007;176(3):263–8. doi: 10.1083/jcb.200611056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Urwyler S, Nyfeler Y, Ragaz C, et al. Proteome analysis of Legionella vacuoles purified by magnetic immunoseparation reveals secretory and endosomal GTPases. Traffic. 2009;10(1):76–87. doi: 10.1111/j.1600-0854.2008.00851.x. [DOI] [PubMed] [Google Scholar]
- 116.Shevchuk O, Batzilla C, Hagele S, et al. Proteomic analysis of Legionella-containing phagosomes isolated from Dictyostelium. Int J Med Microbiol. 2009;299(7):489–508. doi: 10.1016/j.ijmm.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 117.Voth DE, Heinzen RA. Lounging in a lysosome: the intracellular lifestyle of Coxiella burnetii. Cell Microbiol. 2007;9(4):829–40. doi: 10.1111/j.1462-5822.2007.00901.x. [DOI] [PubMed] [Google Scholar]
- 118.Voth DE, Heinzen RA. Coxiella type IV secretion and cellular microbiology. Curr Opin Microbiol. 2009;12(1):74–80. doi: 10.1016/j.mib.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Subbian S, Mehta PK, Cirillo SL, Bermudez LE, Cirillo JD. A Mycobacterium marinum mel2 mutant is defective for growth in macrophages that produce reactive oxygen and reactive nitrogen species. Infect Immun. 2007;75(1):127–34. doi: 10.1128/IAI.01000-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Philips JA. Mycobacterial manipulation of vacuolar sorting. Cell Microbiol. 2008;10(12):2408–15. doi: 10.1111/j.1462-5822.2008.01239.x. [DOI] [PubMed] [Google Scholar]
- 121.Hagedorn M, Soldati T. Flotillin and RacH modulate the intracellular immunity of Dictyostelium to Mycobacterium marinum infection. Cell Microbiol. 2007;9(11):2716–33. doi: 10.1111/j.1462-5822.2007.00993.x. [DOI] [PubMed] [Google Scholar]
- 122.Hagedorn M, Rohde KH, Russell DG, Soldati T. Infection by tubercular mycobacteria is spread by nonlytic ejection from their amoeba hosts. Science. 2009;323(5922):1729–33. doi: 10.1126/science.1169381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Derre I, Isberg RR. Legionella pneumophila replication vacuole formation involves rapid recruitment of proteins of the early secretory system. Infect Immun. 2004;72(5):3048–53. doi: 10.1128/IAI.72.5.3048-3053.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kagan JC, Stein MP, Pypaert M, Roy CR. Legionella subvert the functions of Rab1 and Sec22b to create a replicative organelle. J Exp Med. 2004;199(9):1201–11. doi: 10.1084/jem.20031706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Weber SS, Ragaz C, Reus K, Nyfeler Y, Hilbi H. Legionella pneumophila exploits PI(4)P to anchor secreted effector proteins to the replicative vacuole. PLoS Pathog. 2006;2(5):e46. doi: 10.1371/journal.ppat.0020046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ragaz C, Pietsch H, Urwyler S, Tiaden A, Weber SS, Hilbi H. The Legionella pneumophila phosphatidylinositol-4 phosphate-binding type IV substrate SidC recruits endoplasmic reticulum vesicles to a replication-permissive vacuole. Cell Microbiol. 2008;10(12):2416–33. doi: 10.1111/j.1462-5822.2008.01219.x. [DOI] [PubMed] [Google Scholar]
- 127.Schoebel S, Blankenfeldt W, Goody RS, Itzen A. High-affinity binding of phosphatidylinositol 4-phosphate by Legionella pneumophila DrrA. EMBO Rep. 2010;11(8):598–604. doi: 10.1038/embor.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kyei GB, Vergne I, Chua J, et al. Rab14 is critical for maintenance of Mycobacterium tuberculosis phagosome maturation arrest. EMBO J. 2006;25(22):5250–9. doi: 10.1038/sj.emboj.7601407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Roberts EA, Chua J, Kyei GB, Deretic V. Higher order Rab programming in phagolysosome biogenesis. J Cell Biol. 2006;174(7):923–9. doi: 10.1083/jcb.200603026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Parra-Lopez C, Lin R, Aspedon A, Groisman EA. A Salmonella protein that is required for resistance to antimicrobial peptides and transport of potassium. EMBO J. 1994;13(17):3964–72. doi: 10.1002/j.1460-2075.1994.tb06712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Blanc-Potard AB, Groisman EA. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 1997;16(17):5376–85. doi: 10.1093/emboj/16.17.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ammendola S, Pasquali P, Pistoia C, et al. High-affinity Zn2+ uptake system ZnuABC is required for bacterial zinc homeostasis in intracellular environments and contributes to the virulence of Salmonella enterica. Infect Immun. 2007;75(12):5867–76. doi: 10.1128/IAI.00559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Papp-Wallace KM, Nartea M, Kehres DG, et al. The CorA Mg2+ channel is required for the virulence of Salmonella enterica serovar typhimurium. J Bacteriol. 2008;190(19):6517–23. doi: 10.1128/JB.00772-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gobin J, Horwitz MA. Exochelins of Mycobacterium tuberculosis remove iron from human iron-binding proteins and donate iron to mycobactins in the M. tuberculosis cell wall. J Exp Med. 1996;183(4):1527–32. doi: 10.1084/jem.183.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Robey M, Cianciotto NP. Legionella pneumophila feoAB promotes ferrous iron uptake and intracellular infection. Infect Immun. 2002;70(10):5659–69. doi: 10.1128/IAI.70.10.5659-5669.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.De Voss JJ, Rutter K, Schroeder BG, Su H, Zhu Y, Barry CE 3rd. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc Natl Acad Sci USA. 2000;97(3):1252–7. doi: 10.1073/pnas.97.3.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Courville P, Chaloupka R, Cellier MF. Recent progress in structure-function analyses of Nramp proton-dependent metal-ion transporters. Biochem Cell Biol. 2006;84(6):960–78. doi: 10.1139/o06-193. [DOI] [PubMed] [Google Scholar]
- 138.Bellamy R. Susceptibility to mycobacterial infections: the importance of host genetics. Genes Immun. 2003;4(1):4–11. doi: 10.1038/sj.gene.6363915. [DOI] [PubMed] [Google Scholar]
- 139.Jabado N, Cuellar-Mata P, Grinstein S, Gros P. Iron chelators modulate the fusogenic properties of Salmonella-containing phagosomes. Proc Natl Acad Sci USA. 2003;100(10):6127–32. doi: 10.1073/pnas.0937287100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Malik S, Abel L, Tooker H, et al. Alleles of the NRAMP1 gene are risk factors for pediatric tuberculosis disease. Proc Natl Acad Sci USA. 2005;102(34):12183–8. doi: 10.1073/pnas.0503368102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Huang J, Klionsky DJ. Autophagy and human disease. Cell Cycle. 2007;6(15):1837–49. doi: 10.4161/cc.6.15.4511. [DOI] [PubMed] [Google Scholar]
- 142.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10(7):458–67. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 143.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9(10):1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 144.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119(6):753–66. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 145.Kumar D, Nath L, Kamal MA, et al. Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell. 2010;140(5):731–43. doi: 10.1016/j.cell.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 146.Schnaith A, Kashkar H, Leggio SA, Addicks K, Kronke M, Krut O. Staphylococcus aureus subvert autophagy for induction of caspase-independent host cell death. J Biol Chem. 2007;282(4):2695–706. doi: 10.1074/jbc.M609784200. [DOI] [PubMed] [Google Scholar]
- 147.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005 Feb 4;307(5710):727–31. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 148.Otto GP, Wu MY, Clarke M, et al. Macroautophagy is dispensable for intracellular replication of Legionella pneumophila in Dictyostelium discoideum. Mol Microbiol. 2004;51(1):63–72. doi: 10.1046/j.1365-2958.2003.03826.x. [DOI] [PubMed] [Google Scholar]
- 149.Farbrother P, Wagner C, Na J, et al. Dictyostelium transcriptional host cell response upon infection with Legionella. Cell Microbiol. 2006;8(3):438–56. doi: 10.1111/j.1462-5822.2005.00633.x. [DOI] [PubMed] [Google Scholar]
- 150.Tung SM, Unal C, Ley A, et al. Loss of Dictyostelium ATG9 results in a pleiotropic phenotype affecting growth, development, phagocytosis and clearance and replication of Legionella pneumophila. Cell Microbiol. 2010;12(6):765–80. doi: 10.1111/j.1462-5822.2010.01432.x. [DOI] [PubMed] [Google Scholar]
- 151.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6(4):463–77. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 152.West CM, McMahon D, Molday RS. Identification of glycoproteins, using lectins as probes, in plasma membranes from Dictyostelium discoideum and human erythrocytes. J Biol Chem. 1978;253(5):1716–24. [PubMed] [Google Scholar]
- 153.Schaloske R, Sordano C, Bozzaro S, Malchow D. Stimulation of calcium influx by platelet activating factor in Dictyostelium. J Cell Sci. 1995;108(Pt 4):1597–603. doi: 10.1242/jcs.108.4.1597. [DOI] [PubMed] [Google Scholar]
- 154.Funamoto S, Milan K, Meili R, Firtel RA. Role of phosphatidylinositol 3' kinase and a downstream pleckstrin homology domain-containing protein in controlling chemotaxis in dictyostelium. J Cell Biol. 2001;153(4):795–810. doi: 10.1083/jcb.153.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kortholt A, King JS, Keizer-Gunnink I, Harwood AJ, Van Haastert PJ. Phospholipase C regulation of phosphatidylinositol 3,4,5-trisphosphate-mediated chemotaxis. Mol Biol Cell. 2007;18(12):4772–9. doi: 10.1091/mbc.E07-05-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Brown SS, Spudich JA. Cytochalasin inhibits the rate of elongation of actin filament fragments. J Cell Biol. 1979;83(3):657–62. doi: 10.1083/jcb.83.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Maniak M, Rauchenberger R, Albrecht R, Murphy J, Gerisch G. Coronin involved in phagocytosis: dynamics of particle-induced relocalization visualized by a green fluorescent protein Tag. Cell. 1995;83(6):915–24. doi: 10.1016/0092-8674(95)90207-4. [DOI] [PubMed] [Google Scholar]
- 158.Konzok A, Weber I, Simmeth E, Hacker U, Maniak M, Muller-Taubenberger A. DAip1, a Dictyostelium homologue of the yeast actin-interacting protein 1, is involved in endocytosis, cytokinesis, and motility. J Cell Biol. 1999;146(2):453–64. doi: 10.1083/jcb.146.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Janetopoulos C, Ma L, Devreotes PN, Iglesias PA. Chemoattractant-induced phosphatidylinositol 3,4,5-trisphosphate accumulation is spatially amplified and adapts, independent of the actin cytoskeleton. Proc Natl Acad Sci USA. 2004;101(24):8951–6. doi: 10.1073/pnas.0402152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Schaloske R, Malchow D. Mechanism of cAMP-induced Ca2+ influx in Dictyostelium: role of phospholipase A2. Biochem J. 1997;327(Pt 1):233–8. doi: 10.1042/bj3270233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chen L, Iijima M, Tang M, et al. PLA2 and PI3K/PTEN pathways act in parallel to mediate chemotaxis. Dev Cell. 2007;12(4):603–14. doi: 10.1016/j.devcel.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Beug H, Katz FE, Stein A, Gerisch G. Quantitation of membrane sites in aggregating Dictyostelium cells by use of tritiated univalent antibody. Proc Natl Acad Sci USA. 1973;70:3150–4. doi: 10.1073/pnas.70.11.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Seastone DJ, Lee E, Bush J, Knecht D, Cardelli J. Overexpression of a novel Rho family GTPase, RacC, induces unusual actin-based structures and positively affects phagocytosis in Dictyostelium discoideum. Mol Biol Cell. 1998;9:2891–904. doi: 10.1091/mbc.9.10.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Eichinger L, Rivero F. Methods in Molecular Biology - Dictyostelium discoideum Protocols. Totowa NJ: Humana Press; 2006. [Google Scholar]
- 165.Schweiger A, Mihalache O, Ecke M, Gerisch G. Stage-specific tyrosine phosphorylation of actin in Dictyostelium discoideum cells. J Cell Sci. 1992;102(Pt 3):601–9. doi: 10.1242/jcs.102.3.601. [DOI] [PubMed] [Google Scholar]
- 166.Howard PK, Sefton BM, Firtel RA. Tyrosine phosphorylation of actin in Dictyostelium associated with cell-shape changes. Science. 1993;259(5092):241–4. doi: 10.1126/science.7678470. [DOI] [PubMed] [Google Scholar]
- 167.Jungbluth A, Eckerskorn C, Gerisch G, Lottspeich F, Stocker S, Schweiger A. Stress-induced tyrosine phosphorylation of actin in Dictyostelium cells and localization of the phosphorylation site to tyrosine-53 adjacent to the DNase I binding loop. FEBS Lett. 1995;375(1-2):87–90. doi: 10.1016/0014-5793(95)01165-b. [DOI] [PubMed] [Google Scholar]
- 168.Gamper M, Kim E, Howard PK, Ma H, Hunter T, Firtel RA. Regulation of Dictyostelium protein-tyrosine phosphatase-3 (PTP3) through osmotic shock and stress stimulation and identification of pp130 as a PTP3 substrate. J Biol Chem. 1999;274(17):12129–38. doi: 10.1074/jbc.274.17.12129. [DOI] [PubMed] [Google Scholar]
- 169.Tillner J, Nau H, Winckler T, Dingermann T. Evaluation of the teratogenic potential of valproic acid analogues in transgenic dictyostelium discoideum strains. Toxicol In Vitro. 1998;12(4):463–9. doi: 10.1016/s0887-2333(98)00016-2. [DOI] [PubMed] [Google Scholar]
- 170.Li G, Alexander H, Schneider N, Alexander S. Molecular basis for resistance to the anticancer drug cisplatin in Dictyostelium. Microbiology. 2000;146(Pt 9):2219–27. doi: 10.1099/00221287-146-9-2219. [DOI] [PubMed] [Google Scholar]
- 171.Williams RS, Cheng L, Mudge AW, Harwood AJ. A common mechanism of action for three mood-stabilizing drugs. Nature. 2002;417(6886):292–5. doi: 10.1038/417292a. [DOI] [PubMed] [Google Scholar]
- 172.Van Driessche N, Alexander H, Min J, Kuspa A, Alexander S, Shaulsky G. Global transcriptional responses to cisplatin in Dictyostelium discoideum identify potential drug targets. Proc Natl Acad Sci USA. 2007;104(39):15406–11. doi: 10.1073/pnas.0705996104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Fajardo M, Schleicher M, Noegel A, et al. Calnexin, calreticulin and cytoskeleton-associated proteins modulate uptake and growth of Legionella pneumophila in Dictyostelium discoideum. Microbiology. 2004;150(Pt 9):2825–35. doi: 10.1099/mic.0.27111-0. [DOI] [PubMed] [Google Scholar]
- 174.Dannat K, Tillner J, Winckler T, Weiss M, Eger K, Dingermann T. Effects of medicinal compounds on the differentiation of the eukaryotic microorganism dictyostelium discoideum: can this model be used as a screening test for reproductive toxicity in humans? Pharmazie. 2003;58(3):204–10. [PubMed] [Google Scholar]
- 175.Alexander S, Min J, Alexander H. Dictyostelium discoideum to human cells: pharmacogenetic studies demonstrate a role for sphingolipids in chemoresistance. Biochim Biophys Acta. 2006;1760(3):301–9. doi: 10.1016/j.bbagen.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 176.Solomon JM, Leung GS, Isberg RR. Intracellular replication of Mycobacterium marinum within Dictyostelium discoideum: efficient replication in the absence of host coronin. Inf Immun. 2003;71:3578–86. doi: 10.1128/IAI.71.6.3578-3586.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Pukatzki S, Kessin RH, Mekalanos JJ. The human pathogen Pseudomonas aeruginosa utilizes conserved virulence pathways to infect the social amoeba Dictyostelium discoideum. Proc Natl Acad Sci USA. 2002;99(5):3159–64. doi: 10.1073/pnas.052704399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pukatzki S, Ma AT, Sturtevant D, et al. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA. 2006;103:1528–33. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Benghezal M, Fauvarque MO, Tournebize R, et al. Specific host genes required for the killing of Klebsiella bacteria by phagocytes. Cell Microbiol. 2006;8(1):139–48. doi: 10.1111/j.1462-5822.2005.00607.x. [DOI] [PubMed] [Google Scholar]
- 180.Colucci AM, Peracino B, Tala A, Bozzaro S, Alifano P, Bucci C. Dictyostelium discoideum as a model host for meningococcal pathogenesis. Med Sci Monit. 2008;14(7):BR134–40. [PubMed] [Google Scholar]
- 181.Aubert DF, Flannagan RS, Valvano MA. A novel sensor kinase-response regulator hybrid controls biofilm formation and type VI secretion system activity in Burkholderia cenocepacia. Infect Immun. 2008;76(5):1979–91. doi: 10.1128/IAI.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Shina MC, Unal C, Eichinger L, et al. A Coronin7 homolog with functions in actin-driven processes. J Biol Chem. 2010;285(12):9249–61. doi: 10.1074/jbc.M109.083725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Schreiner T, Mohrs MR, Blau-Wasser R, et al. Loss of the F-actin binding and vesicle-associated protein comitin leads to a phagocytosis defect. Eukaryot Cell. 2002;1(6):906–14. doi: 10.1128/EC.1.6.906-914.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Li Z, Solomon JM, Isberg RR. Dictyostelium discoideum strains lacking the RtoA protein are defective for maturation of the Legionella pneumophila replication vacuole. Cell Microbiol. 2005;7(3):431–42. doi: 10.1111/j.1462-5822.2004.00472.x. [DOI] [PubMed] [Google Scholar]
- 185.Chen G, Zhuchenko O, Kuspa A. Immune-like phagocyte activity in the social amoeba. Science. 2007;317(5838):678–81. doi: 10.1126/science.1143991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chen J, deFelipe KS, Clarke M, et al. Legionella effectors that promote nonlytic release from protozoa. Science. 2004;303:1358–61. doi: 10.1126/science.1094226. [DOI] [PubMed] [Google Scholar]
- 187.Chen J, Reyes M, Clarke M, Shuman HA. Host cell-dependent secretion and translocation of the lepA and lepB effectors of Legionella pneumophila. Cell Microbiol. 2007;9:1660–71. doi: 10.1111/j.1462-5822.2007.00899.x. [DOI] [PubMed] [Google Scholar]
- 188.Laguna RK, Creasey EA, Li Z, Valtz N, Isberg RR. A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. Proc Natl Acad Sci USA. 2006;103:18745–50. doi: 10.1073/pnas.0609012103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.VanRheenen SM, Luo ZQ, O'Connor T, Isberg RR. Members of a Legionella pneumophila family of proteins with ExoU (phospholipase A) active sites are translocated to target cells. Infect Immun. 2006;74:3597–606. doi: 10.1128/IAI.02060-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Tiaden A, Spirig T, Weber SS, et al. The Legionella pneumophila response regulator LqsR promotes host cell interactions as an element of the virulence regulatory network controlled by RpoS and LetA. Cell Microbiol. 2007;9:2903–20. doi: 10.1111/j.1462-5822.2007.01005.x. [DOI] [PubMed] [Google Scholar]
- 191.Tiaden A, Spirig T, Carranza P, et al. Synergistic contribution of the Legionella pneumophila lqs genes to pathogen-host interactions. J Bacteriol. 2008;190(22):7532–47. doi: 10.1128/JB.01002-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Tiaden A, Spirig T, Sahr T, et al. The autoinducer synthase LqsA and putative sensor kinase LqsS regulate phagocyte interactions, extracellular filaments and a genomic island of Legionella pneumophila. Environ Microbiol. 2010;12(5):1243–59. doi: 10.1111/j.1462-2920.2010.02167.x. [DOI] [PubMed] [Google Scholar]
- 193.Liu Y, Luo ZQ. The Legionella pneumophila effector SidJ is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome. Infect Immun. 2007;75(2):592–603. doi: 10.1128/IAI.01278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Liu M, Conover GM, Isberg RR. Legionella pneumophila EnhC is required for efficient replication in tumour necrosis factor alpha-stimulated macrophages. Cell Microbiol. 2008;10(9):1906–23. doi: 10.1111/j.1462-5822.2008.01180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.de Felipe KS, Glover RT, Charpentier X, et al. Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog. 2008;4(8):e1000117. doi: 10.1371/journal.ppat.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hovel-Miner G, Pampou S, Faucher SP, et al. SigmaS controls multiple pathways associated with intracellular multiplication of Legionella pneumophila. J Bacteriol. 2009;191(8):2461–73. doi: 10.1128/JB.01578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Price CT, Al-Khodor S, Al-Quadan T, et al. Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoS Pathog. 2009;5(12):e1000704. doi: 10.1371/journal.ppat.1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Alibaud L, Kohler T, Coudray A, et al. Pseudomonas aeruginosa virulence genes identified in a Dictyostelium host model. Cell Microbiol. 2008;10(3):729–40. doi: 10.1111/j.1462-5822.2007.01080.x. [DOI] [PubMed] [Google Scholar]