Abstract
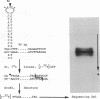
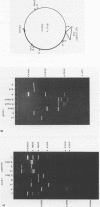
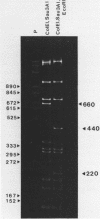
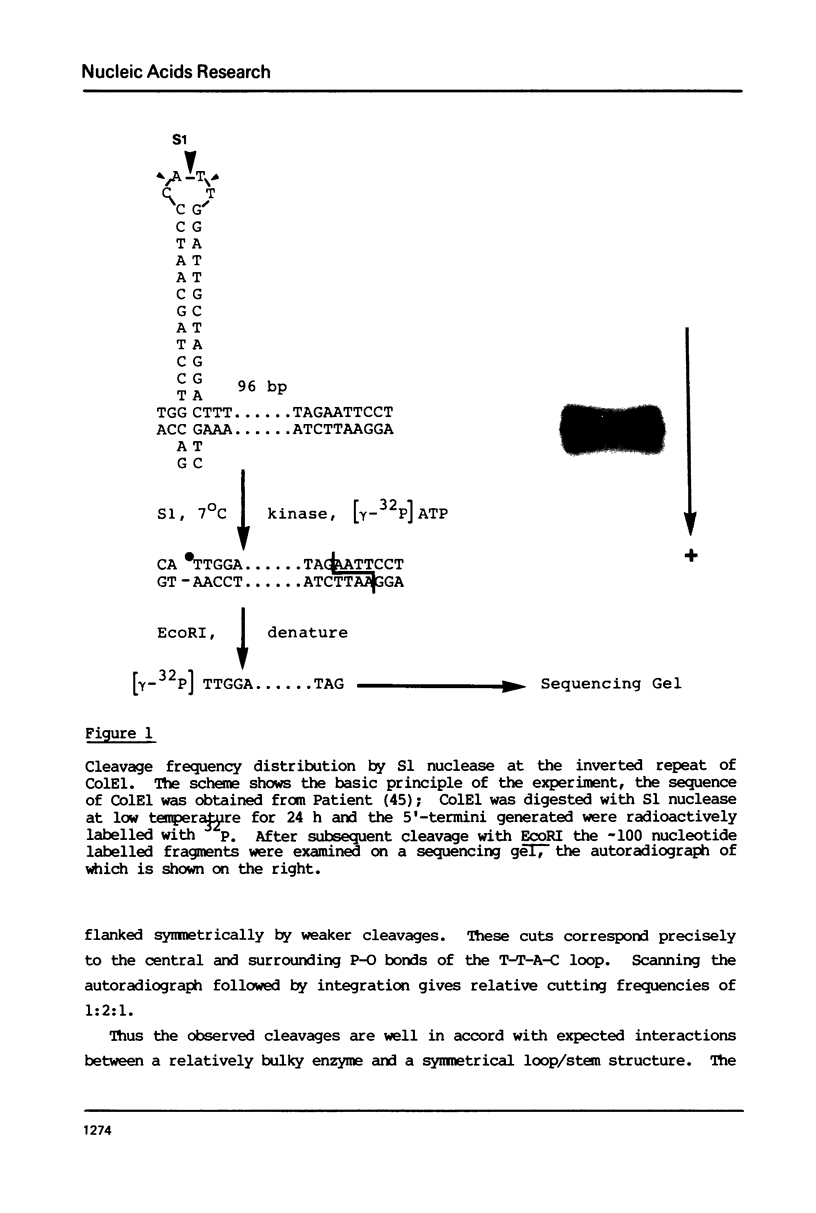
Short inverted repeat sequences adopt hairpin stem-loop type structures in supercoiled closed circular DNA molecules, demonstrated by S1 nuclease cleavage. Fine mapping of cleavage frequencies is in good agreement with expected cleavage patterns based upon the interaction between an unpaired loop and a sterically bulky enzyme molecule. Whilst the topological properties of underwound DNA circles depend ultimately upon reduced linkage, necessarily a global molecular property, hairpin loop formation is an essentially local property. Thus molecular size is unimportant for the S1 hypersensitivity of the Co1E1 inverted repeat. Furthermore, a 440 bp Sau3AI, EcoRI fragment of Co1E1 which contains the inverted repeat has been cloned into pBR322 whereupon it exhibits S1 cleavage similar to Co1E1 in the supercoiled recombinant molecule. The effect is therefore both local and transmissible. Direct competition, between inverted repeats in the recombinant, coupled with close examination of flanking sequences, enables some simple 'rules' for base pairing in hairpin loops to be formulated. Whilst limited G-T and A-C base pairing appears not to be destabilising, A-G, T-C or loop outs are highly destabilising.
Full text
PDF



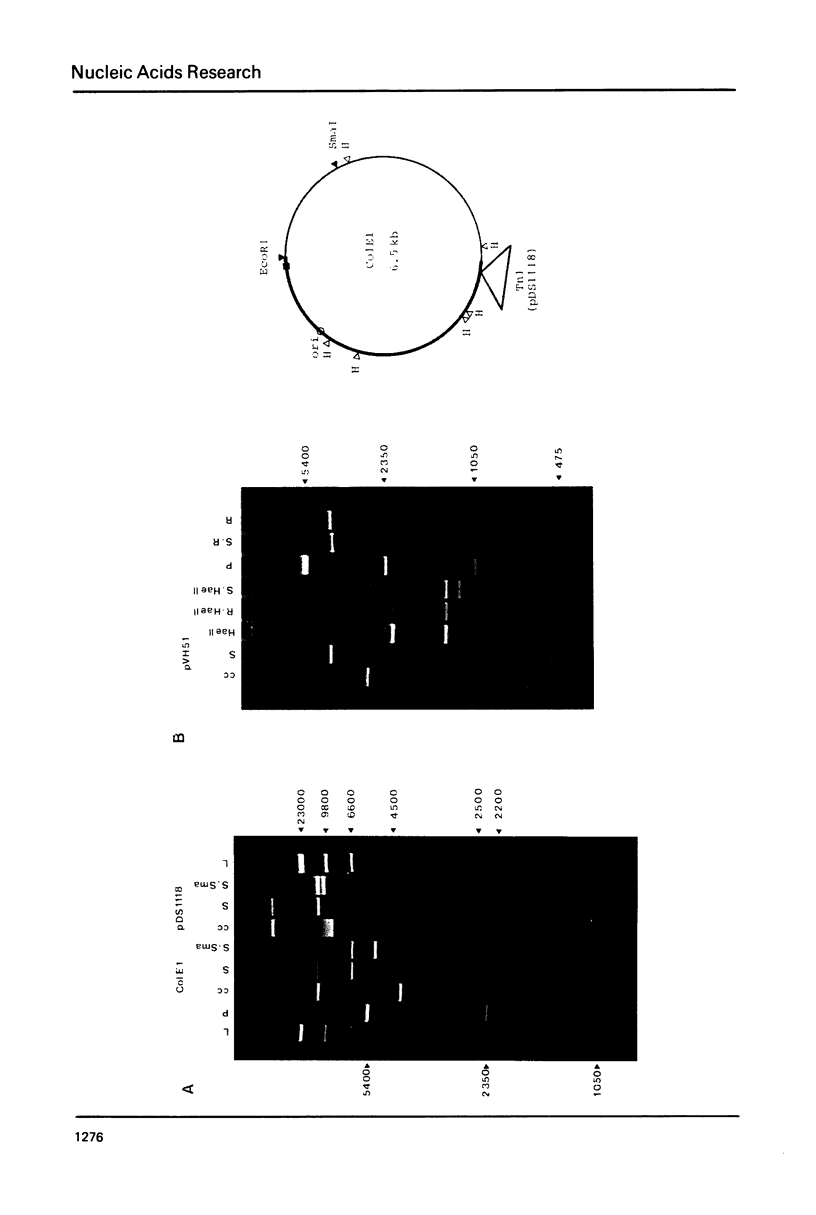


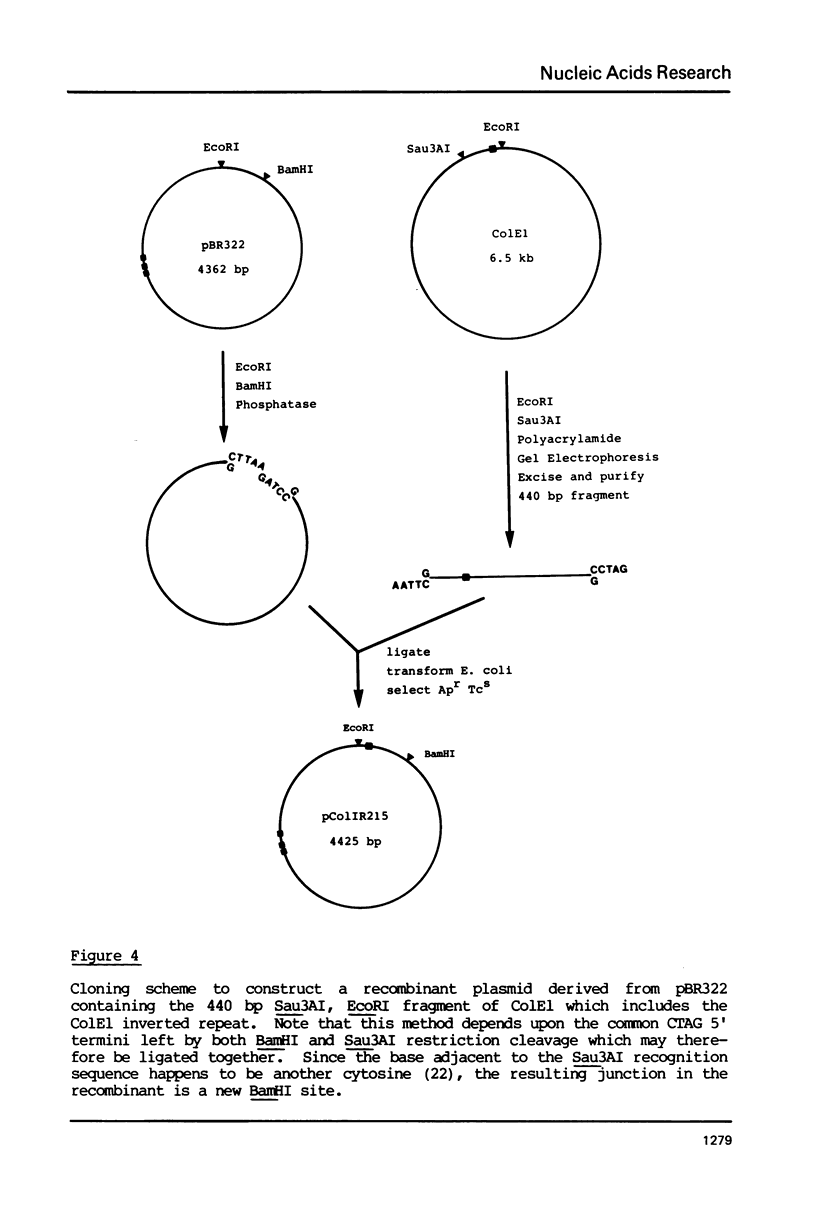
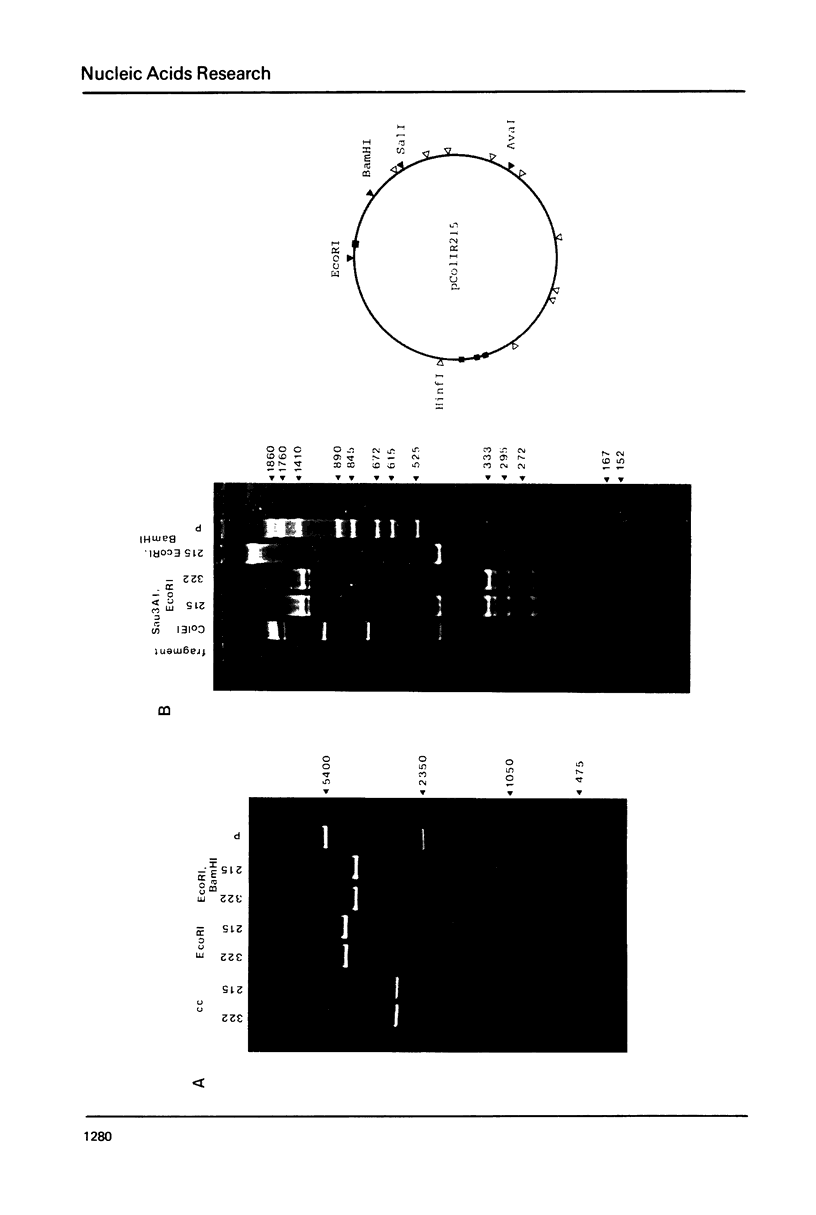









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Chandrasekaran R., Birdsall D. L., Leslie A. G., Ratliff R. L. Left-handed DNA helices. Nature. 1980 Feb 21;283(5749):743–745. doi: 10.1038/283743a0. [DOI] [PubMed] [Google Scholar]
- Bauer W. R. Structure and reactions of closed duplex DNA. Annu Rev Biophys Bioeng. 1978;7:287–313. doi: 10.1146/annurev.bb.07.060178.001443. [DOI] [PubMed] [Google Scholar]
- Betlach M., Hershfield V., Chow L., Brown W., Goodman H., Boyer H. W. A restriction endonuclease analysis of the bacterial plasmid controlling the ecoRI restriction and modification of DNA. Fed Proc. 1976 Jul;35(9):2037–2043. [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Betlach M. C., Heyneker H. L., Shine J., Rodriguez R. L., Boyer H. W. Origin of replication of pBR345 plasmid DNA. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5265–5269. doi: 10.1073/pnas.74.12.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchan P., Wang J. C., Echols H. Effect of circularity and superhelicity on transcription from bacteriophagelambda DNA. Proc Natl Acad Sci U S A. 1973 Nov;70(11):3077–3081. doi: 10.1073/pnas.70.11.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan G., Sherratt D. The transposon Tn1 as a probe for studying ColE1 structure and function. Mol Gen Genet. 1977 Mar 7;151(2):151–160. doi: 10.1007/BF00338689. [DOI] [PubMed] [Google Scholar]
- Drew H., Takano T., Tanaka S., Itakura K., Dickerson R. E. High-salt d(CpGpCpG), a left-handed Z' DNA double helix. Nature. 1980 Aug 7;286(5773):567–573. doi: 10.1038/286567a0. [DOI] [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Itoh T., Tomizawa J. I. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershfield V., Boyer H. W., Chow L., Helinski D. R. Characterization of a mini-ColC1 plasmid. J Bacteriol. 1976 Apr;126(1):447–453. doi: 10.1128/jb.126.1.447-453.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossenlopp P., Oudet P., Chambon P. Animal DNA-dependent RNA polymerases. Studies on the binding of mammalian RNA polymerases AI and B to Simian virus 40 DNA. Eur J Biochem. 1974 Jan 16;41(2):397–411. doi: 10.1111/j.1432-1033.1974.tb03281.x. [DOI] [PubMed] [Google Scholar]
- Lescure B., Chestier A., Yaniv M. Transcription of polyoma virus DNA in vitro. II. Transcription of superhelical and linear polyoma DNA by RNA polymerase II. J Mol Biol. 1978 Sep 5;124(1):73–85. doi: 10.1016/0022-2836(78)90148-1. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Houghton M. The interaction of RNA polymerase II from wheat with supercoiled and linear plasmid templates. Nucleic Acids Res. 1979 Feb;6(2):507–523. doi: 10.1093/nar/6.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim V. I., Mazanov A. L. Tertiary structure for palindromic regions of DNA. FEBS Lett. 1978 Apr 1;88(1):118–123. doi: 10.1016/0014-5793(78)80621-8. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Max E. E., Seidman J. G., Leder P. Sequences of five potential recombination sites encoded close to an immunoglobulin kappa constant region gene. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3450–3454. doi: 10.1073/pnas.76.7.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGavin S. Models of specifically paired like (homologous) nucleic acid structures. J Mol Biol. 1971 Jan 28;55(2):293–298. doi: 10.1016/0022-2836(71)90201-4. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Nash H. A. Restriction assay for integrative recombination of bacteriophage lambda DNA in vitro: requirement for closed circular DNA substrate. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3524–3528. doi: 10.1073/pnas.73.10.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka Y., Leder P. The complete sequence of a chromosomal mouse alpha--globin gene reveals elements conserved throughout vertebrate evolution. Cell. 1979 Nov;18(3):875–882. doi: 10.1016/0092-8674(79)90139-9. [DOI] [PubMed] [Google Scholar]
- Oka A., Nomura N., Morita M., Sugisaki H., Sugimoto K., Takanami M. Nucleotide sequence of small ColE1 derivatives: structure of the regions essential for autonomous replication and colicin E1 immunity. Mol Gen Genet. 1979 May 4;172(2):151–159. doi: 10.1007/BF00268276. [DOI] [PubMed] [Google Scholar]
- Patient R. K. Characterization of in vitro transcription initiation and termination sites in Col E1 DNA. Nucleic Acids Res. 1979 Jun 25;6(8):2647–2665. doi: 10.1093/nar/6.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Friedmann T., Air G. M., Barrell B. G., Brown N. L., Fiddes J. C., Hutchison C. A., 3rd, Slocombe P. M., Smith M. The nucleotide sequence of bacteriophage phiX174. J Mol Biol. 1978 Oct 25;125(2):225–246. doi: 10.1016/0022-2836(78)90346-7. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Uhlenbeck O. C., Levine M. D. Estimation of secondary structure in ribonucleic acids. Nature. 1971 Apr 9;230(5293):362–367. doi: 10.1038/230362a0. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. I., Ohmori H., Bird R. E. Origin of replication of colicin E1 plasmid DNA. Proc Natl Acad Sci U S A. 1977 May;74(5):1865–1869. doi: 10.1073/pnas.74.5.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg A. J., Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980 Jan 10;283(5743):216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]
- Vogt V. M. Purification and further properties of single-strand-specific nuclease from Aspergillus oryzae. Eur J Biochem. 1973 Feb 15;33(1):192–200. doi: 10.1111/j.1432-1033.1973.tb02669.x. [DOI] [PubMed] [Google Scholar]
- WATSON J. D., CRICK F. H. The structure of DNA. Cold Spring Harb Symp Quant Biol. 1953;18:123–131. doi: 10.1101/sqb.1953.018.01.020. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wilson J. H. Nick-free formation of reciprocal heteroduplexes: a simple solution to the topological problem. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3641–3645. doi: 10.1073/pnas.76.8.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing R., Drew H., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Crystal structure analysis of a complete turn of B-DNA. Nature. 1980 Oct 23;287(5784):755–758. doi: 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]
- Yang H. L., Heller K., Gellert M., Zubay G. Differential sensitivity of gene expression in vitro to inhibitors of DNA gyrase. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3304–3308. doi: 10.1073/pnas.76.7.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziff E. B., Evans R. M. Coincidence of the promoter and capped 5' terminus of RNA from the adenovirus 2 major late transcription unit. Cell. 1978 Dec;15(4):1463–1475. doi: 10.1016/0092-8674(78)90070-3. [DOI] [PubMed] [Google Scholar]