Abstract
The mitochondrial genome encodes a very small fraction of the macromolecular components that are required to generate functional mitochondria. Therefore, most components are encoded within the nuclear genome and are imported into mitochondria from the cytosol. Understanding how mitochondria are assembled, function, and dysfunction in diseases requires detailed knowledge of mitochondrial import mechanisms and pathways. The import of nucleus-encoded RNAs is required for mitochondrial biogenesis and function, but unlike pre-protein import, the pathways and cellular machineries of RNA import are poorly defined, especially in mammals. Recent studies have shown that mammalian polynucleotide phosporylase (PNPASE) localizes in the mitochondrial intermembrane space (IMS) to regulate the import of RNA. The identification of PNPASE as the first component of the RNA import pathway, along with a growing list of nucleus-encoded RNAs that are imported and newly developed assay systems for RNA import studies, suggest a unique opportunity is emerging to identify the factors and mechanisms that regulate RNA import into mammalian mitochondria. Here we summarize what is known in this fascinating area of mitochondrial biogenesis, identify areas that require further investigation, and speculate on the impact unraveling RNA import mechanisms and pathways will have for the field going forward.
Keywords: polynucleotide phosphorylase, PNPASE, PNPT1, mitochondria, RNA trafficking, oxidative phosphorylation, reactive oxygen species
1. Introduction
Mitochondrial biogenesis requires the import of nucleus-encoded macromolecules including proteins and RNAs. Compared to the mitochondrial protein import pathways, which have been well characterized [1-4], the pathways importing RNAs into mammalian mitochondria, and the functions of imported RNAs, are just being discovered. Mitochondrial import of nucleus-encoded RNAs including tRNAs , 5S rRNA, RNase P RNA, and MRP RNA is essential for mitochondrial DNA replication, transcription and translation [3, 5-10]. The mitochondrial matrix localization of these small non-coding mammalian RNAs relies on a newly described RNA import regulator, polynucleotide phosphorylase (PNPASE) [11].
PNPASE is a highly conserved 3′-5′ exoribonuclease expressed in organisms that include bacteria, plants, flies, mice, and humans, but is absent in fungi, Trypanosoma, and Archaea [12-16]. PNPASE degrades RNA by phosphorolysis and can also function as a template independent polymerase [17-23]. Prokaryotic and plant PNPASE function in RNA quality control through its RNA polymerase and degradation activities [22-26]. However, the search for a specific function for mammalian PNPASE beyond its overall role in maintaining mitochondrial homeostasis has been confounding. This is because PNPASE was localized in the mitochondrial intermembrane space (IMS), which was believed to be devoid of RNA. The discovery that PNPASE regulates the import of selected nucleus-encoded small RNAs into mitochondria marks a turning point in understanding the function of PNPASE. This exciting discovery still leaves open many unanswered questions, including the mechanism(s) and pathway(s) of PNPASE-regulated RNA import.
2. PNPT1 expression and PNPASE structure
PNPT1, the gene encoding for PNPASE in humans, was first reported in a screen for upregulated genes in both senescent progeroid fibroblasts and terminally differentiated melanoma cells [27]. The PNPT1 gene is ~60kb in length, contains 28 exons, and is located at 2p16.1 (UCSC Genome Browser, Assembly GRCh37/hg19, February, 2009). This genomic region shows deletions and amplifications in human B cell lymphoma and in several genetic disorders [28, 29]. Relatively little is known about the transcriptional or post-transcriptional regulation of PNPT1 expression. So far, the only known inducers of PNPT1 transcription are the type I interferons (IFNs) [13]. Stimulation with IFNα or IFNβ was shown to activate the Janus-activated kinase (JAK)/signal transducer and activators of transcription (STAT) pathway, causing interferon-stimulated gene factor 3 (ISGF3) to bind to an interferon stimulated response element (ISRE) in the PNPT1 promoter, inducing gene transcription [13]. The PNPT1 promoter includes many additional regulatory protein-binding sites, such as a putative site for E2F transcription factor 3 (E2F3), which silences target gene expression during the G1 to S cell cycle phase transition [13]. Since PNPT1 expression was originally identified in senescent and terminally differentiated cells that have left the cell cycle, it would be interesting to know whether E2F3 acts as a PNPT1 repressor and what its role could be in regulating PNPT1 expression in cycling or senescent mammalian cells.
Whether PNPASE protein expression is regulated by type I IFN induction is still unresolved. From our own studies in mice, PNPASE is strongly expressed in all primary tissues examined including brain, heart, lung, liver, intestine, kidney, thymus, and skeletal muscle (data not shown). In one set of human cell line studies, PNPASE was not detectable without IFNβ induction [30, 31]. By contrast, other studies showed abundant PNPASE expression in human cell lines under basal conditions [32-34] with IFNβ exposure failing to induce further expression in some of the lines examined [32, 33]. These differing results may reflect species and cell line specific differences or differences in detection reagents and methods employed. Since PNPT1 knockout is embryonic lethal and hepatocytes or mouse embryonic fibroblasts (MEFs) devoid of PNPASE expression cannot be propagated [11], it might be that all primary mammalian cell types require some level of PNPASE expression. This is sensible to consider, given the role of PNPASE in maintaining mitochondrial homeostasis and its underlying essential functions in RNA import and processing. What role type I IFN induction plays in PNPASE regulation of mitochondrial physiology is currently unknown. However, Type I IFNs are cytokines released by virally-infected cells that generally result in antiviral and growth inhibitory autocrine and paracrine responses [35]. The recent discovery of the mitochondrial antiviral signaling complex (MAVS, CARDIF) located on the mitochondrial outer membrane (OM), and its interaction with processed viral RNA fragments, suggests that juxtaposed PNPASE could have a RNA processing role in cellular antiviral responses, although there is no evidence for this possibility thus far [36].
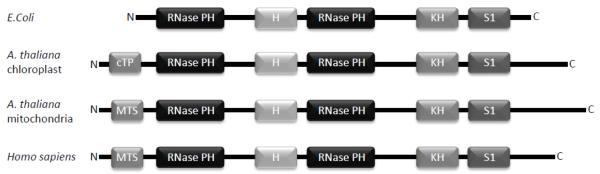
PNPASE proteins from different species share five highly conserved sequence and structural motifs (Fig. 1). Two RNase PH domains, which are homologous to the Escherichia coli tRNA processing RNase PH enzyme, are located at the PNPASE amino-terminus [14]. An alpha-helical domain unique to PNPASE proteins is located between the two RNase PH domains [20, 37]. And KH and S1 RNA binding domains, which are also present in other RNA binding proteins, are located at the PNPASE carboxy-terminus [37-39]. In E. coli, PNPASE can exist on its own, it can be bound to the RhlB RNA helicase, or it can participate in a degradosome complex with RNase E, RhlB, and enolase [15, 21, 40]. Crystallography studies using PNPASE isolated from Streptomyces antibioticus and from E. coli revealed that three PNPASE monomers form a doughnut-shaped structure through interactions of their catalytic RNase PH domains [37, 41]. In such a configuration, the KH and S1 domains of each monomer localize to one face of the doughnut, where they can bind RNA substrates and guide them through the central enzymatic channel in the trimeric complex [37, 41]. A cylindrical structure for PNPASE is also possible, in which two trimers stack into a hexameric configuration with an elongated central enzymatic channel. Plant genomes contain two distinct genes that encode for PNPASE, with one gene containing a chloroplast transit peptide and the other gene having a mitochondrial targeting pre-sequence (MTS) [38, 42]. In contrast with its bacterial counterpart, chloroplast PNPASE is not known to interact with any other proteins and may instead form a homo-hexameric complex [43]. Isolated human PNPASE is ~260-280kDa in native gels, also suggesting a homo-trimeric complex [11, 32, 37, 41]. Interestingly, the archaeal and eukaryotic exosomes, which do not contain PNPASE but also function in RNA turnover and surveillance, have a similar trimeric doughnut-shaped quaternary structure [12, 20].
Fig. 1.
PNPASE structural domains are conserved between species. PNPASEs contain two RNase PH domains that are homologous to the bacterial tRNA processing enzyme, RNase PH, and these domains catalyze RNA degradation. The alpha helical domain (H) is unique to PNPASE, and the KH and S1 domain at the C terminus bind RNA. In contrast to bacteria and other mammals, plants express two forms of PNPASE, one with a chloroplast transit peptide (cTP) and the other with a mitochondrial targeting sequence (MTS). (Adapted and reprinted with permission from Chen, et al., Trends in cell biology 17 (2007) 600-608)
3. Subcellular localization of PNPASE
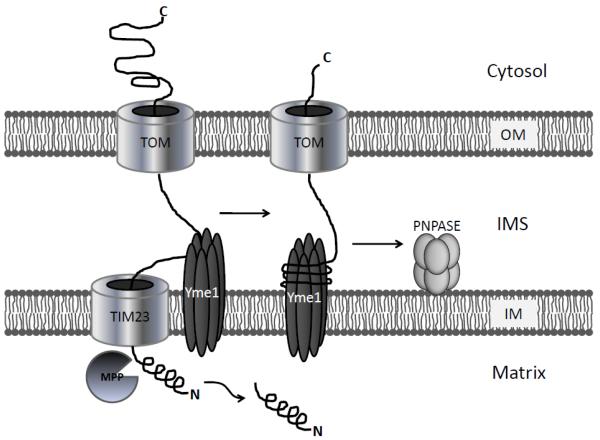
Mammalian PNPASE has an amino-terminal MTS and has been localized to mitochondria in both immunofluorescence and cell-fractionation studies [32, 44, 45]. Since PNPASE processes RNA it was anticipated to reside in the mitochondrial matrix where mtRNA transcription, processing, and translation occur. However, sub-fractionation and protease-protection assays using isolated mouse liver mitochondria, or yeast mitochondria expressing exogenous human PNPASE, repeatedly showed an unexpected intermembrane space (IMS) localization [11, 32]. This result was very surprising because RNAs are not known to reside in the mitochondrial IMS. Carbonate extraction studies further showed that PNPASE is a peripheral inner membrane (IM)-bound protein facing the IMS [32]. To reach its IMS location, PNPASE traffics through the translocase of the outer mitochondrial membrane (TOM) as a precursor, followed by engagement of the translocase of the inner mitochondrial membrane (TIM)-23 complex. These trafficking steps require an intact ΔΨ and are followed by the amino-terminus of PNPASE extending through the TIM23 complex so that the matrix processing peptidase (MPP), consisting of a Mas1/Mas2 heterodimer, can cleave away the 37-residue MTS. Then, the IM-bound i-AAA (ATPases associated with several diverse cellular activities) protease, Yme1, mediates the release of the mature amino-terminal portion of PNPASE into the IMS and functions as a translocation motor to pull the carboxy-terminal portion of PNPASE through the TOM complex into the IMS [44] (Fig. 2). IMS-imported and processed PNPASE then assembles into a functional homo-oligomeric complex, consisting of a trimer or a dimer of trimers [44].
Fig. 2.
Mammalian PNPASE is imported into the mitochondrial IMS via a Yme1-dependent mechanism. After passing through the TOM complex at the OM, the PNPASE amino-terminus extends through the TIM23 complex at the IM into the matrix, where the 37 amino-acid MTS is removed by the matrix-processing peptidase (MPP), Mas1/Mas2. The mature amino-terminus is then released into the IMS with the help of IM-localized Yme1 iAAA protease, which also helps to reel the remaining portion of PNPASE into the IMS from the cytosol. Mature, processed PNPASE then assembles into a homo-oligomeric complex, consisting of a trimer or a dimer of trimers attached to the IM facing the IMS.
The IMS localization of mammalian PNPASE is highly reproducible. However, whether all PNPASE localizes exclusively in the IMS of mammalian mitochondria is controversial. Over-expressed PNPASE with a carboxy-terminal HA-tag was detected in the cytosol of the HO-1 melanoma cell line [31]. PNPASE has also been reported as an interacting partner for TCL1, a cytosol-localized nonenzymatic oncoprotein that promotes B- and T-cell malignancies, and as a weak binding partner for hSUV3, a mitochondrial matrix RNA helicase [46, 47]. These extra-IMS locations, however, could be cell-type specific, condition-specific, or experimental artifact. For example, terminal differentiation, cellular senescence, or apoptosis could alter the distribution of PNPASE in a cell. Over-expression studies can also lead to the accumulation of proteins in locations that are normally devoid of them. Finally, the solubilization step used to identify interacting proteins can bring together proteins that normally reside in different compartments of a cell.
4. The maintenance of mitochondrial homeostasis by PNPASE
The effect of reduced PNPASE expression on mitochondrial structure and function has been well studied using shRNA approaches. In a variety of mammalian cell types PNPASE expression above a critical threshold is required for mitochondrial homeostasis. Conversely, a reduction in PNPASE expression that fails to reach a critical knockdown threshold results in no observable changes in mitochondrial morphology or function [11, 32, 48]. PNPASE reduction to ~20-30% or less of the wild-type expression level caused filamentous mitochondrial networks to fragment with a drop in ΔΨ and a 2-fold or greater reduction in the activities of linked respiratory chain complexes I and III, II and III, or individual complexes IV and V [32]. These data indicate that PNPASE expression is required for efficient oxidative phosphorylation. PNPASE deficiency also leads to cellular changes secondary to mitochondrial dysfunction that include lactate accumulation, reduction in steady state ATP levels, and reduced cell proliferation [32].
Pnpt1 knockout mice have been generated. A whole-animal Pnpt1 knockout was embryonic lethal, indicating an essential role for PNPASE in early mammalian development [11]. By contrast, a liver-specific conditional Pnpt1 knockout (HepKO, for hepatocyte knockout) was viable. The HepKO mouse was generated using albumin-CRE recombinase to excise loxP recombination sites that were inserted flanking exon 2 of Pnpt1. A 2 to 4-fold PNPASE reduction in HepKO liver cells was achieved at ~6 to 8 weeks of age followed by a rebound in PNPASE expression thereafter, likely due to incomplete Pnpt1 knockout from CRE recombinase-mediated excision and liver regeneration from hepatocytes still expressing Pnpt1 [11]. Transmission electron microscopy of HepKO liver cells showed disordered, circular, and smooth IM cristae in contrast to the ordered, linear, stacked cristae with convolutions exhibited by wild-type control liver mitochondria. Oxygen consumption studies with a Clark-type electrode also revealed a 1.5 to 2-fold decrease in the activity of respiratory complex IV and complexes II+III+IV. These ultrastructure and functional results validate a role for PNPASE in maintaining mitochondrial homesostasis in a physiological, in vivo setting.
In addition to PNPASE loss-of-function studies, gain-of-function studies, despite the risk for aberrant PNPASE localization, also seem to support a role for PNPASE in mitochondrial homeostasis. PNPASE over-expression results in increased reactive oxygen species (ROS) accumulation over time, which subsequently leads to NF-κB activation and an increase in NF-κB-regulated pro-inflammatory cytokines, including IL-6, IL-8, RANTES, and MMP-3 [49, 50]. The mechanism of ROS accumulation with PNPASE over-expression is unclear, although most ROS is produced within mitochondria as a result of respiratory chain activities [51-53]. PNPASE reduction inhibits respiratory chain production, respiratory complex activities, and oxygen consumption [11, 32]. Excess PNPASE could have the opposite effect, with increased respiratory activity and mitochondrial ROS production, although this idea requires experimental validation.
5. PNPASE, mtRNA processing, and RNA import into mitochondria
A reduction in respiration and the synthesis of mitochondrion-encoded proteins of the respiratory chain in HepKO liver cells compared to wild-type controls led to studies of PNPASE in mtRNA transcription and translation. HepKO liver cells showed a surprising reduction in mature mtRNA transcripts and encoded proteins, directly related to deficient mtRNA processing [11]. All of the mammalian mtRNA processing molecules are encoded in the nuclear genome and synthesized either in the nucleus (RNAs) or cytosol (proteins), followed by import into mitochondria [10, 53-56]. PNPASE deficiency had no significant effect on the mitochondrial import of nucleus-encoded proteins [11]. By contrast, HepKO liver cells showed a significant reduction in the RNA component of the RNase P mtRNA processing complex. The reduction in RNase P RNA was not from increased degradation, indicating instead a defect in RNase P RNA import with PNPASE deficiency [11]. Studies using a variety of in vitro and in vivo experimental systems have now demonstrated a direct role for PNPASE in regulating the import of multiple non-coding RNAs into mitochondria. The RNA import function of PNPASE is separable from its RNA-processing function, as an engineered point mutation that cripples its RNA-processing activity has no effect on RNA import into mitochondria [11].
The molecular mechanism by which PNPASE distinguishes RNAs for processing versus those for import remains to be clarified. PNPASE recognizes an RNA stem-loop structure in some and perhaps all of the RNAs it helps to import [11]. It is not known whether mammalian PNPASE binds RNA stem-loop structures with its KH/S1 RNA binding domain in distinct ways to either trigger RNA import or processing. RNA structural elements regulate PNPASE functions in chloroplasts and prokaryotes and a stem-loop structure protects RNAs from degradation by chloroplast PNPASE [23, 57, 58]. It would be of great interest to determine whether mammalian PNPASE functions in a similar manner.
5.1 RNAs imported into mitochondria
Most non-coding RNAs that function in mitochondria are encoded within the mtDNA [59]. On one end of the spectrum, the mtDNA of land plants and protists, such as Chlamydomonas, Paramecium, and Tetrahymena lack one or a few tRNAs that are required to translate mtRNA. At the other extreme, the trypanosomatid protozoa lack all of the mitochondrial tRNA genes. And for Saccharomyces cerevisiae and Marchantia polymorpha, the mtDNA encodes a set of tRNAs that is sufficient for translating all mtRNA codons, but nucleus-encoded tRNAs are nevertheless imported for unknown reasons [60, 61]. Mammalian mitochondria do not import tRNAs, except maybe the nucleus-encoded tRNACUGGln and tRNAUUGGln . Additional nucleus-encoded small RNA molecules are also present in mammalian mitochondria [54, 62-65].
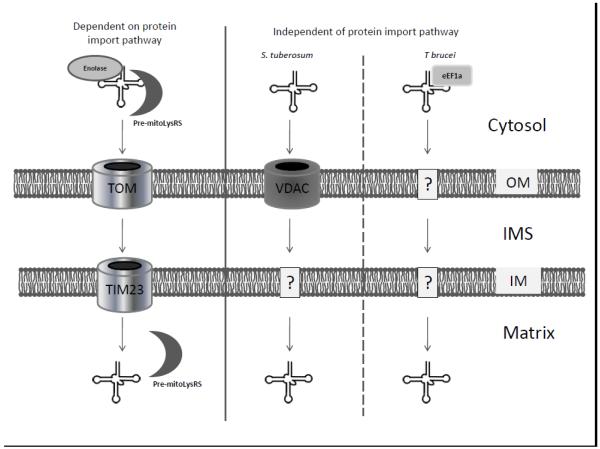
Early studies on tRNA import into mitochondria have shown that it is a selective process, with the selectivity determined by specific tRNA structural or sequence motifs [66]. RNA import is ATP-dependent and involves protease-sensitive receptors at the mitochondrial OM. Thus far, two main general mechanisms for tRNA import into mitochondria have been identified. One mechanism is similar to protein import and is used by yeast to import tRNACUULys (Fig. 3) [67, 68]. This tRNA import mechanism requires an intact ΔΨ and utilizes the protein import pathway. An association with the precursor mitochondrial tRNA synthetase and aminoacylation of the tRNA by the cognate cytosolic synthetase are also required for import. In addition, the metabolic enzyme enolase helps to deliver a tRNA/synthetase complex to the import machinery [69-71]. A second mechanism is independent of the protein import pathway and does not require cytosolic factors even though they may still have an influence when present (Fig. 3) [72]. This additional mechanism has been identified in most organisms that import tRNA into mitochondria including yeast [66, 73], but this import pathway is less well defined. The OM voltage-dependent anion channel (VDAC) has an essential role for tRNA import in plants in vitro, but a homologous protein is dispensable for tRNA import in the protozoan, Trypanosoma brucei (Fig. 3) [74, 75]. For Trypanosoma brucei, elongation factor 1a (eEF1a) mediates mitochondrial tRNA import (Fig. 3) [76].
Fig. 3.
Routes of mitochondrial tRNA import. Two mitochondrial tRNA import mechanisms have been reported. Shown on the far left is tRNA import that utilizes the protein import pathway, including the TOM complex at the OM and the TIM23 complex at the IM. The imported tRNA is delivered to the mitochondrial OM bound to pre-mitochondrial lysyl tRNA synthetase (pre-mitoLysRS) and enolase after aminoacylation by its cognate cytosolic synthetase. Enolase dissociates at the surface of mitochondria and the remainder of the protein-tRNA complex is imported together into the matrix. Thus far, this mechanism has only been described for the import of tRNACUULys into yeast mitochondria. A second mechanism has been reported in organisms ranging from kinetoplastids to human that does not depend on the protein import pathway. Different factors have been identified in this pathway that are organism-specific, including the VDAC channel in the OM for plants and the translation elongation factor, eEF1a, for T. brucei.
RNase MRP is a site-specific endoribonuclease. It was originally isolated from mouse mitochondria and in vitro shows activity in processing mtRNA transcripts to form primers complementary to the mtDNA origin of replication [65]. RNase MRP has a nucleus-encoded RNA component that contains a decamer sequence complementary to a conserved region of mtRNA substrates [65]. Most RNase MRP, however, resides in the nucleus [77, 78]. In Saccharomyces cerevisiae, nuclear RNase MRP processes rRNA precursors to generate the mature 5.8S rRNA [79-82]. During mitosis in S. cerevisiae, nuclear RNase MRP is transiently relocated to temporal asymmetric MRP (TAM) bodies in the cytosol, where it cleaves the mRNA for a B-type cyclin, Clb2, to promote the end of mitosis [82, 83]. The S. cerevisiae mitochondrial and nuclear RNase MRP have identical RNA components but contain distinct protein components and show differing enzymatic activities [84].
Another nucleus-encoded RNA that is imported into mammalian mitochondria is the RNA component of RNase P [10, 54]. RNase P is an endoribonuclease that processes the 5′ end of mitochondrial tRNAs [10]. The mammalian mitochondrial genome consists of a circular chromosome with tightly packed genes. Large polycistronic RNA transcripts are generated from the heavy and light strand promoters. tRNAs often separate the coding regions for electron transport chain complex subunits and, unlike nucleus-encoded tRNA precursors, do not have individual 3′ or 5′ pre-sequences [53, 85]. In most arrangements, multiple tRNAs are grouped together without an intervening pre-sequence in the polycistronic transcripts. The processing of polycistronic transcripts to individual mtRNAs and mature tRNAs requires RNase P enzymatic activities. Earlier studies showed that the RNA component of RNase P was imported into mammalian mitochondrial and was identical to the RNA component of nuclear RNase P [10, 54]. By contrast, the RNA component of mitochondrial RNase P in S. cerevisiae is encoded within its mtDNA [86]. Recently, a functional protein-only mitochondrial RNase P enzyme was isolated from human mitochondria. This alternative mitochondrial RNase P has three protein subunits and processes single tRNA precursors with artificial 5′ pre-sequences in vitro [56]. It remains to be determined whether the protein only mitochondrial RNase P can efficiently process physiological mitochondrial tRNA substrates, especially those that are sequentially linked in polycistronic transcripts with no intervening pre-sequences. Interestingly, a tRNA processing assay using mitoplast lysate with or without nuclease pre-treatment to eliminate RNA showed that an RNA component or components are required for efficient processing of abutting tRNA precursors, further confirming the existence of a RNA-containing mitochondrial RNase P complex [11]. Most likely, protein-only and RNA-containing RNase P complexes coexist in mammalian mitochondria.
The nucleus-encoded 5S rRNA was localized within mammalian mitochondria [64, 87]. The 5S rRNA is the most abundant imported mammalian RNA and an essential ribosome component in almost all living organisms, providing regulatory interactions among most functional sites within the translating ribosomal machinery [88]. Although 5S rRNA has been detected in most non-mammalian organisms, the exact function of 5S rRNA inside mitochondria is still unresolved [89]. Recent studies suggest that 5S rRNA is a component of the mitochondrial ribosome, but based on prior structural analysis of the mitochondrial ribosome its role could be very different from its cytosolic counterpart [7, 89, 90].
There are also recent reports suggesting that specific microRNAs (miRNAs) localize inside mitochondria [63]. miRNAs are a class of small (~22 ribonucleotide) non-coding RNAs that regulate gene expression post-transcriptionally by binding to imperfectly or perfectly matched complementary sequences at the 3′ end of target mRNAs, usually resulting in translational repression or transcript degradation and gene silencing [91, 92]. They are present in almost all eukaryotes except fungi, algae, and marine plants, and are all nucleus-encoded [93]. The human nuclear genome is estimated to encode for over 1,000 miRNAs. Fifteen miRNAs were identified in rat liver mitochondria and five of these were confirmed using specific probes [63]. Based on target analysis using sequence algorithms, such as Miranda and TargetScan, it appears that the miRNAs neither target mtRNA transcripts nor complement the nuclear RNAs that encode imported mitochondrial proteins, but instead seem to be involved in the regulation of genes associated with apoptosis, cell proliferation, and differentiation [63, 94]. Hence, it was proposed that mitochondria might serve as reservoirs for specific miRNAs that modulate these cellular processes. Most recently, miRNAs along with Argonaute 2, a component of the RNA-induced silencing complex (RISC), were co-immunoprecipitated with the COX3 mtRNA transcript, suggesting that miRNA-mediated translational regulation could also occur in mammalian mitochondria [95]. Whether miRNAs localize in mitochondria is still under debate. A recent study showed that the enrichment of some miRNAs in mitochondria disappeared once mitoplast procedures were performed [96], suggesting that these miRNAs could be low level contaminants, or more interestingly, could be associated with the outer membrane or localized inside the IMS.
A virally-encoded RNA was also suggested to be translocated into mammalian mitochondria. The 2.7-kb cytomegalovirus (CMV)-encoded β2.7 RNA targets the Grim-19 (NDUFA13) protein of complex I in the electron transport chain [94]. CMV β2.7 RNA associates with Grim-19 to maintain mitochondrial function in support of viral replication. Interestingly, the mitochondrial OM protein MAVS is activated by viral RNA binding and functions to protect cells from infection by inducing a type I IFN anti-viral response [36, 97]. At present, whether viral RNAs are localized inside mitochondria has not been firmly established. Candidate viral RNAs could associate with the cytosolic precursor of proteins rather than the mature mitochondrion-localized proteins. If mitochondrial localization indeed occurs, nothing is known about the mechanism(s) of viral RNA import or the mechanism(s) for providing processed viral RNA fragments to MAVS. An interesting candidate for mechanistic studies is PNPASE, as it processes RNA, is localized in the IMS, and its expression may be regulated in concert with both Grim-19 and MAVS, as all 3 genes appear to be induced by type 1 interferons [13, 32].
5.2 tRNA import into yeast and mammalian mitochondria
As already mentioned, nucleus-encoded tRNACUULys is imported into isolated yeast mitochondria in vitro [67, 68] with tRNA synthetase binding, aminoacylation [69, 70] and help from cytosolic enolase [71]. This import utilizes the TOM and TIM23 translocons and requires ATP and an intact ΔΨ, similar to protein translocation [98]. The yeast tRNA import pathway appears conserved in mammalian systems because isolated mitochondria from HeLa or HepG2 cells can be substituted for yeast mitochondria [68], and yeast cytosolic import factors remain essential for the process. However, studies addressing whether human cytosolic import factors were able to replace yeast cytosolic factors yielded conflicting results [68]. It is not known whether nucleus-encoded mammalian tRNACUULys can be imported into yeast or mammalian mitochondria under the same conditions. The in vitro transcribed mitochondrion-encoded tRNALys, however, was imported into isolated human mitochondria. In this case, human cytosolic factors were essential for import and could not be replaced by yeast cytosolic factors [64]. In vivo, yeast tRNALys derivatives can also be imported into mammalian mitochondria and rescue defects caused by mitochondrial tRNALys mutations [99].
Yeast tRNAGln is also imported into isolated yeast mitochondria without requiring added cytosolic factors [100]. Recently, mammalian mitochondria were shown to import nucleus-encoded mammalian tRNACUGGln and tRNAUUGGln [62]. Both tRNACUGGln and tRNAUUGGln were also identified in mitochondria isolated from human and rat liver using sub-cellular fractionation and RT-PCR [62], providing in vivo relevance. These tRNAs were also imported into isolated mammalian mitochondria independent of added cytosolic factors. Import required ATP but not an intact ΔΨ [62].
5.3 RNA import mechanisms
In vitro and in vivo assay systems were developed to study the function of PNPASE in mitochondrial RNA import because the RNA component of RNase P was markedly decreased in PNPASE deficient cells [11]. Systems to test import included (1) liver mitochondria isolated from HepKO mice, (2) MEFs isolated from Pnpt1 loxP-flanked mice infected with a CMV-CRE recombinase-expressing retrovirus to reduce PNPASE expression, and (3) several mammalian cell lines containing shRNAs targeting Pnpt1 transcripts [11]. In these assay systems, the import of RNase P RNA into mitochondria correlated with PNPASE abundance. Additional nucleus-encoded RNAs, such as MRP RNA and 5S rRNA, that also localize to mitochondria also showed PNPASE-dependent import in these assay systems. Yeast mitochondria were also tested since the RNA component of RNase P is encoded within the yeast mitochondrial genome. Heterologous expression of PNPASE supported the import of RNase P RNA into yeast mitochondria, suggesting general mechanisms and/or features of the RNA import pathway are conserved in yeast, rodents, and humans [11].
A physical interaction between RNase P RNA with PNPASE adds support for a direct PNPASE import mechanism and suggests that PNPASE may function as a RNA import receptor in the IMS. RNase P RNA has mitochondrial targeting features. A systematic dissection identified a 20-ribonucleotide sequence that was essential for in vitro import of human RNase P RNA [11]. When appended to a non-mitochondrial RNA, this core sequence targeted the non-mitochondrial RNA to the mitochondrion and PNPASE bound directly to the fusion transcript. Interestingly, the identified import sequence is predicted to form a stem-loop structure. A ~20 ribonucleotide non-identical sequence was identified in MRP RNA that was predicted to form a stem-loop structure. This MRP RNA sequence could also direct non-mitochondrial RNA import when placed into chimeric fusion transcripts. Thus, a common theme for imported RNAs may be the formation of a stem-loop that facilitates PNPASE binding without activating its RNA processing functions. Currently, there are no reports on the mechanism(s) for miRNA import into mitochondria.
Components of the import pathway for 5S rRNA into mammalian mitochondria have been described. Basic requirements include ATP, an intact ΔΨ, the protein translocases, and cytosolic factors [62]. The cytosolic factors include the cytosolic precursor of the mitochondrial ribosomal protein L-18 (MRP-L18) [7] and the mitochondrial thiosulfate sulfurtransferase, rhodanese [101]. MRP-L18 first binds to the 5S rRNA, inducing a conformational change. Misfolded rhodanese then binds to the 5S rRNA and the combination acts as reciprocal chaperones to facilitate import [101]. Inhibition of rhodanese expression decreased 5S rRNA import and mitochondrial translation in vivo [101]. Two distinct structural elements of 5S rRNA are required for its mitochondrial import. One is in the proximal part of helix 1 containing a conserved uncompensated G:U pair, and the second is associated with the loop E-helix IV region with several non-canonical structural features [87, 102]. Whereas PNPase-dependent import did not seem to require cytosolic proteins, the import efficiency seems more robust with cytosolic factors, suggesting that multiple import pathways may be used for 5S rRNA import.
5.4 The mechanism of PNPASE-dependent RNA import into mitochondria
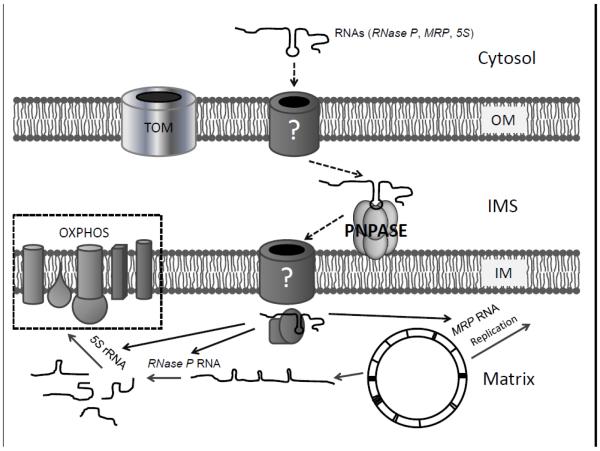
For the import of nucleus-encoded mammalian tRNACUGGln and tRNAUUGGln, ATP is required, but unlike the import of yeast tRNALys and 5S rRNA, import into mammalian mitochondria does not require an intact ΔΨ or cytosolic factors [62]. Interestingly, PNPASE enhances RNA import in yeast, which does not have a PNPASE homolog, indicating that PNPASE can augment a distinct RNA import mechanism directly or independently [11]. An intact ΔΨ is required for PNPASE-dependent import of RNase P RNA and no cytosolic factors are required (Fig. 4) [11]. In mammalian cells PNPASE expression is required for cell survival, so how essential PNPASE is for RNA import remains unclear. Also, it is not known whether there are PNPASE dependent and independent mitochondrial RNA import mechanisms in mammals, as may be the case in PNPASE-modified yeast. Identification of additional RNA import pathway components, including OM and IM channels, is essential to dissect mechanisms of RNA import into mammalian mitochondria.
Fig. 4.
PNPASE regulates the import of nucleus-encoded small RNAs including RNase P RNA, MRP RNA, and 5S rRNA into the mitochondrial matrix. PNPASE promotes the import of RNAs from the cytosol into the matrix by binding to specific stem-loop motifs in the imported RNAs. Imported non-coding RNAs function in mitochondrial replication, transcription, and translation. The mitochondrial translation products are components of the oxidative phosphorylation (OXPHOS) complexes I-IV.
Although PNPASE RNA degradation and mitochondrial RNA import activities are separable [11], it is not known whether PNPASE has a gatekeeper function for RNA import, degrading misplaced RNAs in the IMS. A stem-loop structure protects RNA from degradation by PNPASE in chloroplasts and stem-loop sequences in human RNase P and MRP RNAs could have a similar role, protecting these RNAs from PNPASE degradation during import [23, 57]. This speculated gatekeeper function could be a differential property of mammalian PNPASE in comparison to its bacterial and chloroplast counterparts. Recombinant mammalian PNPASE does not preferentially bind poly(A) containing RNAs, consistent with data that PNPASE does not metabolize poly(A) mRNA tails [103]. Human PNPASE also has a lower poly(A) polymerase activity than its E. coli counterpart. Poly(A) polymerization activity depends on the concentration of ADP, which is significantly lower in the IMS compared to the matrix [103]. It would be interesting to know whether PNPASE degradation activity is regulated by pH, as the mitochondrial IMS has a redox buffering system and lower pH than the matrix.
5.5 Mammalian PNPASE and mtRNA processing
In mammalian mitochondria, RNA editing is a critical step in gene expression. Large polycistronic RNA transcripts are generated from the heavy and light strand promoters [85]. RNA cleavage occurs at the 3′ and 5′ ends of the tRNAs that intervene between RNA coding genes, followed by polyadenylation of mtRNAs [104-106]. For several mitochondrial genes with a partial U or UA stop codon, the polyadenylation step completes the stop codon [104]. Whether polyadenylation also has an additional role in enhancing mtRNA stability is still debated. Detection and isolation of truncated, non-abundant, polyadenylated mitochondrial RNAs suggests that polyadenylation could occur with RNA transcripts destined for degradation, whereas another study shows deadenylation sometimes leads to quick decay of certain mutant transcripts [107, 108].
In bacteria, organelles, and some eukaryotes, polyadenylation is catalyzed by one of the poly(A) polymerases (PAPs) or PNPASE [109]. Previously, it was proposed that mammalian PNPASE could act as a mtRNA poly(A) polymerase and also degrade polyadenylated mtRNAs because it exhibits both polymerization and RNA degradation biochemical activities [103]. However, the IMS location of mammalian PNPASE potentially excludes a direct role in these processes, although an indirect effect on mtRNA processing may be possible [32]. shRNA against PNPT1 altered mtRNA polyadenylation without changing the mtRNA abundance [32, 48, 110]. In other studies, shRNA against PNPT1 affected polyadenylation to different extents for different mtRNAs. For example, 5′ processing of COX1 mtRNA was defective and its poly(A) tail was abolished [110]. By contrast, PNPASE knockdown had no effect on COX3 polyadenylation and even led to longer poly(A) tail extensions for ND5 and ND3 mtRNAs [110]. Whether PNPASE regulates mtRNA polyadenylation by changing mitochondrial ATP concentrations is also unclear. Transient PNPT1 shRNA knockdown caused a decrease in cellular ATP, which could alter mtRNA poly(A) tail lengths [48, 110]. However, stable PNPT1 shRNA knockdown showed inconsistent effects, with ATP levels decreased, slightly increased, or unaffected in different cell clones, all of which showed similar defects in mtRNA polyadenylation [110]. Also, the effect that PNPT1 knockdown and ATP depletion had on mtRNA poly(A) tail lengths appears distinct, with the latter causing a decrease in poly(A) tail lengths in ND3 and ND5 mtRNA transcripts compared to an increase in poly(A) tail lengths by PNPT1 knockdown [110]. Other work showed that the human mitochondrial matrix protein hSUV3 regulates mtRNA stability and the removal of non-coding processing intermediates. PNPASE was reported to interact with hSUV3, but this would place PNPASE in the mitochondrial matrix [47]. Even though it is impossible to rule out a small fraction of PNPASE in the matrix, an equally plausible explanation for this interaction between hSUV3 and PNPASE is that it occurs during the solubilization step in co-immunprecipitation assays. In fact, human mtPAP has been shown to polyadenylate the mature 3′ ends of mtRNAs [48, 111].
How mammalian PNPASE affects mtRNA processing is not resolved. PNPASE regulates the import of nucleus-encoded small RNAs including RNase P and MRP RNAs [11]. RNase P processes mitochondrial tRNAs and PNPASE knockdown results in the accumulation of partially processed polycistronic mtRNA transcripts, indicating a defect in cleaving polycistronic transcripts into individual coding RNAs, rRNAs, and tRNAs [11]. Interference of this early processing step could inhibit later processing steps, such as mature transcript polyadenylation and degradation. The polycistronic transcript cleavage sites are unique and could require different processing mechanisms, so that defective RNA import could cause variable defects in generating mature transcripts [85].
6. Future directions and concluding remarks
Thus far, PNPASE has been identified as an IMS localized protein that directly regulates RNA import into mammalian mitochondria. The remaining components of this and potentially additional mammalian RNA import pathways, however, remain unknown. The in vivo and in vitro import assay systems established for PNPASE studies provide tools for identifying and dissecting additional import components and mechanisms [11]. Importantly, even though S. cerevisiae does not have a PNPASE homolog, heterologous PNPASE expression enhances the import of RNA in vivo and in vitro, suggesting the import pathways in mammalian cells and S. cerevisiae are compatible, which may provide a system where yeast genetic approaches can be employed.
A second area of potential significance is in the identification of additional imported RNA species and the elucidation of their functions inside mitochondria. So far, mammalian mitochondria have been shown to import RNase P RNA, MRP RNA, 5S rRNA, CMV β2.7 viral RNA, several tRNAs, and a few miRNAs. It is likely that additional nucleus-encoded RNAs, and possibly more viral RNAs, will be identified inside mitochondria. Knowledge of the range of mitochondrial RNA import signals will provide clues as to which RNAs are potential candidates for import. New assay systems may also be required to unravel the functions of imported RNAs.
A fundamental question in considering RNA import into mammalian mitochondria also remains to be addressed. Are there distinct pathways, even PNPASE-dependent and independent pathways, or is there one major import pathway with different RNA substrates requiring similar or distinct accessory factors? In fact, a few features seem to converge in all systems studied to date. ATP seems to be universally required for RNA import and import appears to depend on one or more OM factor(s). So far, it is still unknown whether mammalian PNPASE is involved in tRNA import into mitochondria.
The function(s) of PNPASE in the mitochondrial IMS is still not fully understood. One remaining challenge is to understand a role for its biochemically demonstrated RNA degradation activity. PNPASE degradation and import activities are separable. More work is required to determine whether PNPASE RNA degradation activity plays a role in RNA import, such as a gate-keeper for import or a waste-disposal shunt. Also, what role, if any, PNPASE has in responding to infection and the localization of viral RNA in the IMS remains to be elucidated.
In addition, there are possible roles for mammalian PNPASE in cellular locations other than the IMS. For example, PNPASE has been shown to be released from the IMS into the cytosol late, hours after apoptosis induction and caspase activation [32]. Could PNPASE participate in degrading cytosolic RNAs in cells committed to death? Also, PNPASE may participate in antiviral responses; does type I IFN-induced PNPT1 expression produce additional PNPASE and, if so, does all the increased PNPASE traffic to the mitochondrial IMS? With the nearby MAVS complex on the OM, does PNPASE process IMS localized viral RNAs and, if so, what is its role in delivery to MAVS? Finally, new approaches will be required to determine unequivocally whether small amounts of endogenous mammalian PNPASE localize into the mitochondrial matrix, and, if so, what may be the functional consequences.
In addition to potential significance in viral infection and cell death, the RNA import function of mammalian PNPASE may be manipulated to correct mitochondrial dysfunction. mtDNA mutations have been linked to multiple human neuromuscular diseases [112-114]. Aging is also associated with the accumulation of mtDNA mutations [115, 116]. Currently, there is no efficient approach for overcoming mtDNA mutations in these morbid diseases. Existing therapies target symptoms instead of mending primary defects. Mitochondrial gene therapy provides an interesting approach to treating the root cause of these diseases. One practical application of studies on RNA import into mitochondria is to complement mtDNA alterations by introducing and importing corrective RNAs. Mitochondrial RNA import sequences could potentially be used to import RNAs that normally are not imported, such as those that could repair defective or inefficient electron transport chain mtRNAs. Another possible application is to manipulate the mitochondrial transcription or translation profile. For example, anti-sense RNAs could be imported to inhibit the expression of mutated mtRNA transcripts or to generate models of disease. Overall, the accumulating data indicates that RNAs, like proteins and lipids, traffic to the mitochondria and are required for proper mitochondrial function. Mammalian PNPASE is required for the import of some if not all of these small nucleus-encoded RNAs.
Highlights.
Mitochondrial homeostasis is maintained by PNPASE from an intermembrane space location
PNPASE activities in mitochondrial RNA import and RNA processing are separable
Imported RNAs are required for mitochondrial genome replication, transcription, and translation
PNPASE regulation of mitochondrial metabolism and RNA import are thus far inseparable
Acknowledgments
This research is supported by the NIH (GM061721, GM073981, GM081621, CA90571, and CA156674), the Muscular Dystrophy Association (022398), the American Heart Association (0640076N), the California Institute of Regenerative Medicine (CIRM RB1-01397), and the Whitcome pre-doctoral training program of the UCLA Molecular Biology Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- [3].Duchene AM, Pujol C, Marechal-Drouard L. Import of tRNAs and aminoacyl-tRNA synthetases into mitochondria. Current genetics. 2009;55:1–18. doi: 10.1007/s00294-008-0223-9. [DOI] [PubMed] [Google Scholar]
- [4].Sieber F, Duchene AM, Marechal-Drouard L. Mitochondrial RNA import: from diversity of natural mechanisms to potential applications. International review of cell and molecular biology. 2011;287:145–190. doi: 10.1016/B978-0-12-386043-9.00004-9. [DOI] [PubMed] [Google Scholar]
- [5].Tarassov I, Kamenski P, Kolesnikova O, Karicheva O, Martin RP, Krasheninnikov IA, Entelis N. Import of nuclear DNA-encoded RNAs into mitochondria and mitochondrial translation. Cell cycle (Georgetown, Tex. 2007;6:2473–2477. doi: 10.4161/cc.6.20.4783. [DOI] [PubMed] [Google Scholar]
- [6].Kamenski P, Kolesnikova O, Jubenot V, Entelis N, Krasheninnikov IA, Martin RP, Tarassov I. Evidence for an adaptation mechanism of mitochondrial translation via tRNA import from the cytosol. Molecular cell. 2007;26:625–637. doi: 10.1016/j.molcel.2007.04.019. [DOI] [PubMed] [Google Scholar]
- [7].Smirnov A, Entelis N, Martin RP, Tarassov I. Biological significance of 5S rRNA import into human mitochondria: role of ribosomal protein MRP-L18. Genes & development. 2011;25:1289–1305. doi: 10.1101/gad.624711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chang DD, Clayton DA. A novel endoribonuclease cleaves at a priming site of mouse mitochondrial DNA replication. The EMBO journal. 1987;6:409–417. doi: 10.1002/j.1460-2075.1987.tb04770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Robertson HD, Altman S, Smith JD. Purification and properties of a specific Escherichia coli ribonuclease which cleaves a tyrosine transfer ribonucleic acid presursor. The Journal of biological chemistry. 1972;247:5243–5251. [PubMed] [Google Scholar]
- [10].Doersen CJ, Guerrier-Takada C, Altman S, Attardi G. Characterization of an RNase P activity from HeLa cell mitochondria. Comparison with the cytosol RNase P activity. The Journal of biological chemistry. 1985;260:5942–5949. [PubMed] [Google Scholar]
- [11].Wang G, Chen HW, Oktay Y, Zhang J, Allen EL, Smith GM, Fan KC, Hong JS, French SW, McCaffery JM, Lightowlers RN, Morse HC, 3rd, Koehler CM, Teitell MA. PNPASE regulates RNA import into mitochondria. Cell. 2010;142:456–467. doi: 10.1016/j.cell.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Raijmakers R, Egberts WV, van Venrooij WJ, Pruijn GJ. Protein-protein interactions between human exosome components support the assembly of RNase PH-type subunits into a six-membered PNPase-like ring. Journal of molecular biology. 2002;323:653–663. doi: 10.1016/s0022-2836(02)00947-6. [DOI] [PubMed] [Google Scholar]
- [13].Leszczyniecka M, Su ZZ, Kang DC, Sarkar D, Fisher PB. Expression regulation and genomic organization of human polynucleotide phosphorylase, hPNPase(old-35), a Type I interferon inducible early response gene. Gene. 2003;316:143–156. doi: 10.1016/s0378-1119(03)00752-2. [DOI] [PubMed] [Google Scholar]
- [14].Leszczyniecka M, DeSalle R, Kang DC, Fisher PB. The origin of polynucleotide phosphorylase domains. Molecular phylogenetics and evolution. 2004;31:123–130. doi: 10.1016/j.ympev.2003.07.012. [DOI] [PubMed] [Google Scholar]
- [15].Sarkar D, Fisher PB. Polynucleotide phosphorylase: an evolutionary conserved gene with an expanding repertoire of functions. Pharmacology & therapeutics. 2006;112:243–263. doi: 10.1016/j.pharmthera.2006.04.003. [DOI] [PubMed] [Google Scholar]
- [16].Kao CY, Read LK. Opposing effects of polyadenylation on the stability of edited and unedited mitochondrial RNAs in Trypanosoma brucei. Molecular and cellular biology. 2005;25:1634–1644. doi: 10.1128/MCB.25.5.1634-1644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Andrade JM, Pobre V, Silva IJ, Domingues S, Arraiano CM. The role of 3′-5′ exoribonucleases in RNA degradation. Progress in molecular biology and translational science. 2009;85:187–229. doi: 10.1016/S0079-6603(08)00805-2. [DOI] [PubMed] [Google Scholar]
- [18].Littauer UZ, Soreq H. The regulatory function of poly(A) and adjacent 3′ sequences in translated RNA. Progress in nucleic acid research and molecular biology. 1982;27:53–83. doi: 10.1016/s0079-6603(08)60597-8. [DOI] [PubMed] [Google Scholar]
- [19].Grunberg-Manago M. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annual review of genetics. 1999;33:193–227. doi: 10.1146/annurev.genet.33.1.193. [DOI] [PubMed] [Google Scholar]
- [20].Symmons MF, Williams MG, Luisi BF, Jones GH, Carpousis AJ. Running rings around RNA: a superfamily of phosphate-dependent RNases. Trends in biochemical sciences. 2002;27:11–18. doi: 10.1016/s0968-0004(01)01999-5. [DOI] [PubMed] [Google Scholar]
- [21].Lin-Chao S, Chiou NT, Schuster G. The PNPase, exosome and RNA helicases as the building components of evolutionarily-conserved RNA degradation machines. Journal of biomedical science. 2007;14:523–532. doi: 10.1007/s11373-007-9178-y. [DOI] [PubMed] [Google Scholar]
- [22].Mohanty BK, Kushner SR. Polynucleotide phosphorylase functions both as a 3′ right- arrow 5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:11966–11971. doi: 10.1073/pnas.220295997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yehudai-Resheff S, Hirsh M, Schuster G. Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts. Molecular and cellular biology. 2001;21:5408–5416. doi: 10.1128/MCB.21.16.5408-5416.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Holec S, Lange H, Kuhn K, Alioua M, Borner T, Gagliardi D. Relaxed transcription in Arabidopsis mitochondria is counterbalanced by RNA stability control mediated by polyadenylation and polynucleotide phosphorylase. Molecular and cellular biology. 2006;26:2869–2876. doi: 10.1128/MCB.26.7.2869-2876.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rott R, Zipor G, Portnoy V, Liveanu V, Schuster G. RNA polyadenylation and degradation in cyanobacteria are similar to the chloroplast but different from Escherichia coli. The Journal of biological chemistry. 2003;278:15771–15777. doi: 10.1074/jbc.M211571200. [DOI] [PubMed] [Google Scholar]
- [26].Jarrige A, Brechemier-Baey D, Mathy N, Duche O, Portier C. Mutational analysis of polynucleotide phosphorylase from Escherichia coli. Journal of molecular biology. 2002;321:397–409. doi: 10.1016/s0022-2836(02)00645-9. [DOI] [PubMed] [Google Scholar]
- [27].Leszczyniecka M, Kang DC, Sarkar D, Su ZZ, Holmes M, Valerie K, Fisher PB. Identification and cloning of human polynucleotide phosphorylase, hPNPase old-35, in the context of terminal differentiation and cellular senescence. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:16636–16641. doi: 10.1073/pnas.252643699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Fukuhara N, Tagawa H, Kameoka Y, Kasugai Y, Karnan S, Kameoka J, Sasaki T, Morishima Y, Nakamura S, Seto M. Characterization of target genes at the 2p15-16 amplicon in diffuse large B-cell lymphoma. Cancer science. 2006;97:499–504. doi: 10.1111/j.1349-7006.2006.00209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kirschner LS, Taymans SE, Pack S, Pak E, Pike BL, Chandrasekharappa SC, Zhuang Z, Stratakis CA. Genomic mapping of chromosomal region 2p15-p21 (D2S378-D2S391): integration of Genemap’98 within a framework of yeast and bacterial artificial chromosomes. Genomics. 1999;62:21–33. doi: 10.1006/geno.1999.5957. [DOI] [PubMed] [Google Scholar]
- [30].Sarkar D, Park ES, Fisher PB. Defining the mechanism by which IFN-beta dowregulates c-myc expression in human melanoma cells: pivotal role for human polynucleotide phosphorylase (hPNPaseold-35) Cell death and differentiation. 2006;13:1541–1553. doi: 10.1038/sj.cdd.4401829. [DOI] [PubMed] [Google Scholar]
- [31].Sarkar D, Park ES, Emdad L, Randolph A, Valerie K, Fisher PB. Defining the domains of human polynucleotide phosphorylase (hPNPaseOLD-35) mediating cellular senescence. Molecular and cellular biology. 2005;25:7333–7343. doi: 10.1128/MCB.25.16.7333-7343.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chen HW, Rainey RN, Balatoni CE, Dawson DW, Troke JJ, Wasiak S, Hong JS, McBride HM, Koehler CM, Teitell MA, French SW. Mammalian polynucleotide phosphorylase is an intermembrane space RNase that maintains mitochondrial homeostasis. Molecular and cellular biology. 2006;26:8475–8487. doi: 10.1128/MCB.01002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gewartowski K, Tomecki R, Muchowski L, Dmochow Ska A, Dzwonek A, Malecki M, Skurzak H, Ostrowski J, Stepien PP. Up-regulation of human PNPase mRNA by beta-interferon has no effect on protein level in melanoma cell lines. Acta biochimica Polonica. 2006;53:179–188. [PubMed] [Google Scholar]
- [34].Hayakawa H, Sekiguchi M. Human polynucleotide phosphorylase protein in response to oxidative stress. Biochemistry. 2006;45:6749–6755. doi: 10.1021/bi052585l. [DOI] [PubMed] [Google Scholar]
- [35].Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- [36].Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- [37].Symmons MF, Jones GH, Luisi BF. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure. 2000;8:1215–1226. doi: 10.1016/s0969-2126(00)00521-9. [DOI] [PubMed] [Google Scholar]
- [38].Yehudai-Resheff S, Portnoy V, Yogev S, Adir N, Schuster G. Domain analysis of the chloroplast polynucleotide phosphorylase reveals discrete functions in RNA degradation, polyadenylation, and sequence homology with exosome proteins. The Plant cell. 2003;15:2003–2019. doi: 10.1105/tpc.013326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Stickney LM, Hankins JS, Miao X, Mackie GA. Function of the conserved S1 and KH domains in polynucleotide phosphorylase. Journal of bacteriology. 2005;187:7214–7221. doi: 10.1128/JB.187.21.7214-7221.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Liou GG, Chang HY, Lin CS, Lin-Chao S. DEAD box RhlB RNA helicase physically associates with exoribonuclease PNPase to degrade double-stranded RNA independent of the degradosome-assembling region of RNase E. The Journal of biological chemistry. 2002;277:41157–41162. doi: 10.1074/jbc.M206618200. [DOI] [PubMed] [Google Scholar]
- [41].Shi Z, Yang WZ, Lin-Chao S, Chak KF, Yuan HS. Crystal structure of Escherichia coli PNPase: central channel residues are involved in processive RNA degradation. RNA (New York, N.Y. 2008;14:2361–2371. doi: 10.1261/rna.1244308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Walter M, Kilian J, Kudla J. PNPase activity determines the efficiency of mRNA 3′-end processing, the degradation of tRNA and the extent of polyadenylation in chloroplasts. The EMBO journal. 2002;21:6905–6914. doi: 10.1093/emboj/cdf686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Baginsky S, Shteiman-Kotler A, Liveanu V, Yehudai-Resheff S, Bellaoui M, Settlage RE, Shabanowitz J, Hunt DF, Schuster G, Gruissem W. Chloroplast PNPase exists as a homo-multimer enzyme complex that is distinct from the Escherichia coli degradosome. RNA (New York, N.Y. 2001;7:1464–1475. [PMC free article] [PubMed] [Google Scholar]
- [44].Rainey RN, Glavin JD, Chen HW, French SW, Teitell MA, Koehler CM. A new function in translocation for the mitochondrial i-AAA protease Yme1: import of polynucleotide phosphorylase into the intermembrane space. Molecular and cellular biology. 2006;26:8488–8497. doi: 10.1128/MCB.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Piwowarski J, Grzechnik P, Dziembowski A, Dmochowska A, Minczuk M, Stepien PP. Human polynucleotide phosphorylase, hPNPase, is localized in mitochondria. Journal of molecular biology. 2003;329:853–857. doi: 10.1016/s0022-2836(03)00528-x. [DOI] [PubMed] [Google Scholar]
- [46].French SW, Dawson DW, Chen HW, Rainey RN, Sievers SA, Balatoni CE, Wong L, Troke JJ, Nguyen MT, Koehler CM, Teitell MA. The TCL1 oncoprotein binds the RNase PH domains of the PNPase exoribonuclease without affecting its RNA degrading activity. Cancer letters. 2007;248:198–210. doi: 10.1016/j.canlet.2006.07.006. [DOI] [PubMed] [Google Scholar]
- [47].Wang DD, Shu Z, Lieser SA, Chen PL, Lee WH. Human mitochondrial SUV3 and polynucleotide phosphorylase form a 330-kDa heteropentamer to cooperatively degrade double-stranded RNA with a 3′-to-5′ directionality. The Journal of biological chemistry. 2009;284:20812–20821. doi: 10.1074/jbc.M109.009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Nagaike T, Suzuki T, Katoh T, Ueda T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. The Journal of biological chemistry. 2005;280:19721–19727. doi: 10.1074/jbc.M500804200. [DOI] [PubMed] [Google Scholar]
- [49].Sarkar D, Lebedeva IV, Emdad L, Kang DC, Baldwin AS, Jr., Fisher PB. Human polynucleotide phosphorylase (hPNPaseold-35): a potential link between aging and inflammation. Cancer research. 2004;64:7473–7478. doi: 10.1158/0008-5472.CAN-04-1772. [DOI] [PubMed] [Google Scholar]
- [50].Sarkar D, Fisher PB. Molecular mechanisms of aging-associated inflammation. Cancer letters. 2006;236:13–23. doi: 10.1016/j.canlet.2005.04.009. [DOI] [PubMed] [Google Scholar]
- [51].Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- [52].Adam-Vizi V, Chinopoulos C. Bioenergetics and the formation of mitochondrial reactive oxygen species. Trends in pharmacological sciences. 2006;27:639–645. doi: 10.1016/j.tips.2006.10.005. [DOI] [PubMed] [Google Scholar]
- [53].Bonawitz ND, Rodeheffer MS, Shadel GS. Defective mitochondrial gene expression results in reactive oxygen species-mediated inhibition of respiration and reduction of yeast life span. Molecular and cellular biology. 2006;26:4818–4829. doi: 10.1128/MCB.02360-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Puranam RS, Attardi G. The RNase P associated with HeLa cell mitochondria contains an essential RNA component identical in sequence to that of the nuclear RNase P. Molecular and cellular biology. 2001;21:548–561. doi: 10.1128/MCB.21.2.548-561.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Esakova O, Krasilnikov AS. Of proteins and RNA: the RNase P/MRP family. RNA (New York, N.Y. 2010;16:1725–1747. doi: 10.1261/rna.2214510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- [57].Lisitsky I, Klaff P, Schuster G. Addition of destabilizing poly (A)-rich sequences to endonuclease cleavage sites during the degradation of chloroplast mRNA. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:13398–13403. doi: 10.1073/pnas.93.23.13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Yehudai-Resheff S, Schuster G. Characterization of the E.coli poly(A) polymerase: nucleotide specificity, RNA-binding affinities and RNA structure dependence. Nucleic acids research. 2000;28:1139–1144. doi: 10.1093/nar/28.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Entelis NS, Kolesnikova OA, Martin RP, Tarassov IA. RNA delivery into mitochondria. Advanced drug delivery reviews. 2001;49:199–215. doi: 10.1016/s0169-409x(01)00135-1. [DOI] [PubMed] [Google Scholar]
- [60].Martin RP, Schneller JM, Stahl AJ, Dirheimer G. Import of nuclear deoxyribonucleic acid coded lysine-accepting transfer ribonucleic acid (anticodon C-U-U) into yeast mitochondria. Biochemistry. 1979;18:4600–4605. doi: 10.1021/bi00588a021. [DOI] [PubMed] [Google Scholar]
- [61].Akashi K, Takenaka M, Yamaoka S, Suyama Y, Fukuzawa H, Ohyama K. Coexistence of nuclear DNA-encoded tRNAVal(AAC) and mitochondrial DNA-encoded tRNAVal(UAC) in mitochondria of a liverwort Marchantia polymorpha. Nucleic acids research. 1998;26:2168–2172. doi: 10.1093/nar/26.9.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Rubio MA, Rinehart JJ, Krett B, Duvezin-Caubet S, Reichert AS, Soll D, Alfonzo JD. Mammalian mitochondria have the innate ability to import tRNAs by a mechanism distinct from protein import. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9186–9191. doi: 10.1073/pnas.0804283105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kren BT, Wong PY, Sarver A, Zhang X, Zeng Y, Steer CJ. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA biology. 2009;6:65–72. doi: 10.4161/rna.6.1.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Entelis NS, Kolesnikova OA, Dogan S, Martin RP, Tarassov IA. 5 S rRNA and tRNA import into human mitochondria. Comparison of in vitro requirements. The Journal of biological chemistry. 2001;276:45642–45653. doi: 10.1074/jbc.M103906200. [DOI] [PubMed] [Google Scholar]
- [65].Chang DD, Clayton DA. Mouse RNAase MRP RNA is encoded by a nuclear gene and contains a decamer sequence complementary to a conserved region of mitochondrial RNA substrate. Cell. 1989;56:131–139. doi: 10.1016/0092-8674(89)90991-4. [DOI] [PubMed] [Google Scholar]
- [66].Salinas T, Duchene AM, Marechal-Drouard L. Recent advances in tRNA mitochondrial import. Trends in biochemical sciences. 2008;33:320–329. doi: 10.1016/j.tibs.2008.04.010. [DOI] [PubMed] [Google Scholar]
- [67].Tarassov I, Entelis N, Martin RP. Mitochondrial import of a cytoplasmic lysine-tRNA in yeast is mediated by cooperation of cytoplasmic and mitochondrial lysyl-tRNA synthetases. The EMBO journal. 1995;14:3461–3471. doi: 10.1002/j.1460-2075.1995.tb07352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kolesnikova OA, Entelis NS, Mireau H, Fox TD, Martin RP, Tarassov IA. Suppression of mutations in mitochondrial DNA by tRNAs imported from the cytoplasm. Science (New York, N.Y. 2000;289:1931–1933. doi: 10.1126/science.289.5486.1931. [DOI] [PubMed] [Google Scholar]
- [69].Entelis NS, Krasheninnikov IA, Martin RP, Tarassov IA. Mitochondrial import of a yeast cytoplasmic tRNA (Lys): possible roles of aminoacylation and modified nucleosides in subcellular partitioning. FEBS letters. 1996;384:38–42. doi: 10.1016/0014-5793(96)00259-1. [DOI] [PubMed] [Google Scholar]
- [70].Schmitz UK, Lonsdale DM. A yeast mitochondrial presequence functions as a signal for targeting to plant mitochondria in vivo. The Plant cell. 1989;1:783–791. doi: 10.1105/tpc.1.8.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Brandina I, Graham J, Lemaitre-Guillier C, Entelis N, Krasheninnikov I, Sweetlove L, Tarassov I, Martin RP. Enolase takes part in a macromolecular complex associated to mitochondria in yeast. Biochimica et biophysica acta. 2006;1757:1217–1228. doi: 10.1016/j.bbabio.2006.07.001. [DOI] [PubMed] [Google Scholar]
- [72].Alfonzo JD, Soll D. Mitochondrial tRNA import--the challenge to understand has just begun. Biological chemistry. 2009;390:717–722. doi: 10.1515/BC.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Schneider A, Marechal-Drouard L. Mitochondrial tRNA import: are there distinct mechanisms? Trends in cell biology. 2000;10:509–513. doi: 10.1016/s0962-8924(00)01854-7. [DOI] [PubMed] [Google Scholar]
- [74].Salinas T, Duchene AM, Delage L, Nilsson S, Glaser E, Zaepfel M, Marechal-Drouard L. The voltage-dependent anion channel, a major component of the tRNA import machinery in plant mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18362–18367. doi: 10.1073/pnas.0606449103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Pusnik M, Charriere F, Maser P, Waller RF, Dagley MJ, Lithgow T, Schneider A. The single mitochondrial porin of Trypanosoma brucei is the main metabolite transporter in the outer mitochondrial membrane. Molecular biology and evolution. 2009;26:671–680. doi: 10.1093/molbev/msn288. [DOI] [PubMed] [Google Scholar]
- [76].Bouzaidi-Tiali N, Aeby E, Charriere F, Pusnik M, Schneider A. Elongation factor 1a mediates the specificity of mitochondrial tRNA import in T. brucei. The EMBO journal. 2007;26:4302–4312. doi: 10.1038/sj.emboj.7601857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Reimer G, Raska I, Scheer U, Tan EM. Immunolocalization of 7-2-ribonucleoprotein in the granular component of the nucleolus. Experimental cell research. 1988;176:117–128. doi: 10.1016/0014-4827(88)90126-7. [DOI] [PubMed] [Google Scholar]
- [78].Gold HA, Topper JN, Clayton DA, Craft J. The RNA processing enzyme RNase MRP is identical to the Th RNP and related to RNase P. Science (New York, N.Y. 1989;245:1377–1380. doi: 10.1126/science.2476849. [DOI] [PubMed] [Google Scholar]
- [79].Schmitt ME, Clayton DA. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Molecular and cellular biology. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Chu S, Archer RH, Zengel JM, Lindahl L. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Lygerou Z, Allmang C, Tollervey D, Seraphin B. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science (New York, N.Y. 1996;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- [82].Cai T, Aulds J, Gill T, Cerio M, Schmitt ME. The Saccharomyces cerevisiae RNase mitochondrial RNA processing is critical for cell cycle progression at the end of mitosis. Genetics. 2002;161:1029–1042. doi: 10.1093/genetics/161.3.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Gill T, Cai T, Aulds J, Wierzbicki S, Schmitt ME. RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: novel method of mRNA degradation. Molecular and cellular biology. 2004;24:945–953. doi: 10.1128/MCB.24.3.945-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Lu Q, Wierzbicki S, Krasilnikov AS, Schmitt ME. Comparison of mitochondrial and nucleolar RNase MRP reveals identical RNA components with distinct enzymatic activities and protein components. RNA (New York, N.Y. 2010;16:529–537. doi: 10.1261/rna.1893710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- [86].Hollingsworth MJ, Martin NC. RNase P activity in the mitochondria of Saccharomyces cerevisiae depends on both mitochondrion and nucleus-encoded components. Molecular and cellular biology. 1986;6:1058–1064. doi: 10.1128/mcb.6.4.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Smirnov AV, Entelis NS, Krasheninnikov IA, Martin R, Tarassov IA. Specific features of 5S rRNA structure - its interactions with macromolecules and possible functions. Biochemistry (Mosc) 2008;73:1418–1437. doi: 10.1134/s000629790813004x. [DOI] [PubMed] [Google Scholar]
- [88].Kouvela EC, Gerbanas GV, Xaplanteri MA, Petropoulos AD, Dinos GP, Kalpaxis DL. Changes in the conformation of 5S rRNA cause alterations in principal functions of the ribosomal nanomachine. Nucleic acids research. 2007;35:5108–5119. doi: 10.1093/nar/gkm546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Lang BF, Goff LJ, Gray MW. A 5 S rRNA gene is present in the mitochondrial genome of the protist Reclinomonas americana but is absent from red algal mitochondrial DNA. Journal of molecular biology. 1996;261:407–413. doi: 10.1006/jmbi.1996.0486. [DOI] [PubMed] [Google Scholar]
- [90].Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- [91].Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature reviews. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- [92].Meyers BC, Souret FF, Lu C, Green PJ. Sweating the small stuff: microRNA discovery in plants. Current opinion in biotechnology. 2006;17:139–146. doi: 10.1016/j.copbio.2006.01.008. [DOI] [PubMed] [Google Scholar]
- [93].Behm-Ansmant I, Rehwinkel J, Izaurralde E. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harbor symposia on quantitative biology. 2006;71:523–530. doi: 10.1101/sqb.2006.71.013. [DOI] [PubMed] [Google Scholar]
- [94].Reeves MB, Davies AA, McSharry BP, Wilkinson GW, Sinclair JH. Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science (New York, N.Y. 2007;316:1345–1348. doi: 10.1126/science.1142984. [DOI] [PubMed] [Google Scholar]
- [95].Bandiera S, Ruberg S, Girard M, Cagnard N, Hanein S, Chretien D, Munnich A, Lyonnet S, Henrion-Caude A. Nuclear outsourcing of RNA interference components to human mitochondria. PloS one. 2011;6:e20746. doi: 10.1371/journal.pone.0020746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, Filipovska A, Mattick JS. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- [98].Tarassov I, Entelis N, Martin RP. An intact protein translocating machinery is required for mitochondrial import of a yeast cytoplasmic tRNA. Journal of molecular biology. 1995;245:315–323. doi: 10.1006/jmbi.1994.0026. [DOI] [PubMed] [Google Scholar]
- [99].Kolesnikova OA, Entelis NS, Jacquin-Becker C, Goltzene F, Chrzanowska-Lightowlers ZM, Lightowlers RN, Martin RP, Tarassov I. Nuclear DNA-encoded tRNAs targeted into mitochondria can rescue a mitochondrial DNA mutation associated with the MERRF syndrome in cultured human cells. Human molecular genetics. 2004;13:2519–2534. doi: 10.1093/hmg/ddh267. [DOI] [PubMed] [Google Scholar]
- [100].Rinehart J, Krett B, Rubio MA, Alfonzo JD, Soll D. Saccharomyces cerevisiae imports the cytosolic pathway for Gln-tRNA synthesis into the mitochondrion. Genes & development. 2005;19:583–592. doi: 10.1101/gad.1269305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Smirnov A, Comte C, Mager-Heckel AM, Addis V, Krasheninnikov IA, Martin RP, Entelis N, Tarassov I. Mitochondrial enzyme rhodanese is essential for 5 S ribosomal RNA import into human mitochondria. The Journal of biological chemistry. 2010;285:30792–30803. doi: 10.1074/jbc.M110.151183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Smirnov A, Tarassov I, Mager-Heckel AM, Letzelter M, Martin RP, Krasheninnikov IA, Entelis N. Two distinct structural elements of 5S rRNA are needed for its import into human mitochondria. RNA (New York, N.Y. 2008;14:749–759. doi: 10.1261/rna.952208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Portnoy V, Palnizky G, Yehudai-Resheff S, Glaser F, Schuster G. Analysis of the human polynucleotide phosphorylase (PNPase) reveals differences in RNA binding and response to phosphate compared to its bacterial and chloroplast counterparts. RNA (New York, N.Y. 2008;14:297–309. doi: 10.1261/rna.698108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- [105].Tomita K, Ueda T, Watanabe K. RNA editing in the acceptor stem of squid mitochondrial tRNA(Tyr) Nucleic acids research. 1996;24:4987–4991. doi: 10.1093/nar/24.24.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Yokobori S, Paabo S. Polyadenylation creates the discriminator nucleotide of chicken mitochondrial tRNA(Tyr) Journal of molecular biology. 1997;265:95–99. doi: 10.1006/jmbi.1996.0728. [DOI] [PubMed] [Google Scholar]
- [107].Slomovic S, Laufer D, Geiger D, Schuster G. Polyadenylation and degradation of human mitochondrial RNA: the prokaryotic past leaves its mark. Molecular and cellular biology. 2005;25:6427–6435. doi: 10.1128/MCB.25.15.6427-6435.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Temperley RJ, Seneca SH, Tonska K, Bartnik E, Bindoff LA, Lightowlers RN, Chrzanowska-Lightowlers ZM. Investigation of a pathogenic mtDNA microdeletion reveals a translation-dependent deadenylation decay pathway in human mitochondria. Human molecular genetics. 2003;12:2341–2348. doi: 10.1093/hmg/ddg238. [DOI] [PubMed] [Google Scholar]
- [109].Nagaike T, Suzuki T, Ueda T. Polyadenylation in mammalian mitochondria: insights from recent studies. Biochimica et biophysica acta. 2008;1779:266–269. doi: 10.1016/j.bbagrm.2008.02.001. [DOI] [PubMed] [Google Scholar]
- [110].Slomovic S, Schuster G. Stable PNPase RNAi silencing: its effect on the processing and adenylation of human mitochondrial RNA. RNA (New York, N.Y. 2008;14:310–323. doi: 10.1261/rna.697308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Tomecki R, Dmochowska A, Gewartowski K, Dziembowski A, Stepien PP. Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic acids research. 2004;32:6001–6014. doi: 10.1093/nar/gkh923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Wallace DC. Mitochondrial DNA sequence variation in human evolution and disease. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:8739–8746. doi: 10.1073/pnas.91.19.8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Wallace DC. Mitochondrial diseases in man and mouse. Science (New York, N.Y. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- [114].Fan W, Waymire KG, Narula N, Li P, Rocher C, Coskun PE, Vannan MA, Narula J, Macgregor GR, Wallace DC. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science (New York, N.Y. 2008;319:958–962. doi: 10.1126/science.1147786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Paradies G, Petrosillo G, Paradies V, Ruggiero FM. Mitochondrial dysfunction in brain aging: role of oxidative stress and cardiolipin. Neurochemistry international. 2010;58:447–457. doi: 10.1016/j.neuint.2010.12.016. [DOI] [PubMed] [Google Scholar]
- [116].Paradies G, Ruggiero FM, Petrosillo G, Quagliariello E. Age-dependent impairment of mitochondrial function in rat heart tissue. Effect of pharmacological agents. Annals of the New York Academy of Sciences. 1996;786:252–263. doi: 10.1111/j.1749-6632.1996.tb39068.x. [DOI] [PubMed] [Google Scholar]