Abstract
Experiments with the transmembrane (TM) domains of the glycoprotein (GP) Ib-IX complex have indicated that the associations between the TM domains of these subunits play an important role in the proper assembly of the complex. As a first step toward understanding these associations, we previously found that the Ibβ TM domain dimerized strongly in Escherichia coli cell membranes and led to Ibβ TM-CYTO (cytoplasmic domain) dimerization in the SDS-PAGE assay, while neither Ibα nor IX TM-CYTO was able to dimerize. In this study, we used the TOXCAT assay to probe the Ibβ TM domain dimerization interface by Ala- and Leu-scanning mutagenesis. Our results show that this interface is based on a leucine zipper-like heptad repeat pattern of amino acids. Mutating either one of polar residues Gln129 or His139 to Leu or Ala disrupted Ibβ TM dimerization dramatically, indicating that polar residues might form part of the leucine zipper-based dimerization interface. Furthermore, these specific mutational effects in the TOXCAT assay were confirmed in the thiol-disulfide exchange and SDS-PAGE assays. The computational modeling studies further revealed that the most likely leucine zipper interface involves hydrogen bonding of Gln129 and electrostatic interaction of the His139 side chain. Correlation of computer modeling results with experimental mutagenesis studies on the Ibβ TM domain may provide insights for understanding the role of the association of TM domains on the assembly of GP Ib-IX complex.
Keywords: GP Ib-IX complex, transmembrane domain, TOXCAT, dimerization, leucine zipper motif, polar residues
Introduction
Glycoprotein (GP) Ib-IX-V complex (CD42) is one of the major adhesion receptors expressed on the surface of circulating platelets, which mediates adhesion of platelets to the subendothelial matrix at sites of injury by binding von Willebrand factor (vWF).1 Once the vWF ligand binds to the extracellular domain of Ibα, the GP Ib-IX-V complex transmits a signal across the membrane that activates intracellular effectors and stimulates integrin αIIbβ3, leading to platelet activation and aggregation.2,3 Malfunction of this multisubunit receptor complex can lead to severe bleeding diathesis and contribute to many cardiovascular diseases.4 However, how this complex carries out its multifaceted functions is not clear, primarily due to poor understanding of its structure and organization. Elucidating the assembly principles of the GP Ib-IX-V complex will facilitate understanding of GP Ib-IX-V-mediated transmembrane (TM) processes. GP Ib-IX-V complex consists of four different polypeptides: Ibα, Ibβ, IX, and V with a 2:4:2:1 stoichiometry; these assemble into a functional receptor complex in the membrane.5,6 The importance of the GP Ib-IX-V complex is best exemplified by the most common form of the hereditary bleeding disorder Bernard-Soulier Syndrome (BSS), which results from the lack of the GP Ib-IX-V complex.4 Since abnormally low expression of the GP Ib-IX-V complex in human platelets can result from mutations in either Ibα, Ibβ, or IX, but not V, and since V is not required for efficient expression of the GP Ib-IX complex,7 research efforts have focused on the structure and function of the GP Ib-IX complex. Our previous studies show that the TM domain of Ibβ is critical for efficient expression of the GP Ib-IX complex in the plasma membrane, because replacing the TM with poly-Leu or poly-LeuAla diminished surface expression of the GP Ib-IX complex in the plasma membrane.8 Further studies showed that TM domain associations of Ibα, Ibβ, and IX domain favorably position the membrane-proximal Cys residues for disulfide bond formation, and this supports the αβ2 configuration.5,9 All these observations suggest that the TM domains of the GP Ib-IX complex are responsible for functionally important associations of the GP Ib-IX complex in membranes.
In general, a multitude of integral membrane proteins is simultaneously synthesized and integrated into various membranes followed by association into homo- or hetero-oligomeric complexes.10 Sequence-specific association between α-helical TM domains supports the proper assembly of many integral membrane proteins. Biochemical and functional analyses, molecular modeling, and structural studies have indicated that the association of TM helices is driven by a close packing of their characteristically shaped surfaces. These packing associations may result in pairs of α-helices with a right-handed twist as exemplified by glycophorin A.11 Other TM associations involve a leucine zipper type of side-chain packing, as is known to occur in certain membrane proteins such as phospholamban.12
Studies using TM peptides from the GP Ib-IX complex have demonstrated that only the Ibβ TM sequence, but not the Ibα and IX counterparts, can form homo-oligomers in the SDS-PAGE and TOXCAT assays.13 However, the specific residues and motifs that determine the association of the Ibβ TM domain have not been thoroughly examined. In this article, we used the TOXCAT assay combined with synthetic TM peptides to systematically analyze self-association of the Ibβ TM domain. Ala- and Leu-scanning mutagenesis show that Ibβ TM domain association is mediated by the polar residues within a leucine zipper motif. These findings may provide insights for understanding the role of TM–TM associations in the assembly of the GP Ib-IX complex.
Results
Optimizing the orientation of the Ibβ TM sequences within the TOXCAT system
The length of the inserted TM helix and the location of the junctions between the helix and MBP and ToxR are important in determining the degree of CAT expression levels in the TOXCAT assay.14 Therefore, we identified the optimal length for the Ibβ TM helix in this assay by incrementally deleting single residues from its C-terminal end (Fig. 1). Figure 2(A) shows that the CAT activity was very sensitive to the length of the Ibβ TM helix inserted in TOXCAT chimeric protein. Nonetheless, CAT activity of each Ibβ TM helix was significantly higher than that of the poorly dimerizing GpA mutant G83I. Control experiments showed that five out of eight Ibβ TM sequence fused to ToxR were expressed at comparable levels as demonstrated by Western blotting of whole-cell lysates [Fig. 2(A)]. Furthermore, MBP complementation assays confirmed that the topologies of these five ToxR-TM-MBP fusions were as expected [Fig. 2(B)].15 The spheroplast proteolysis assay showed the almost complete digestion of chimeric proteins in E. coli spheroplasts by proteinase K [Fig. 2(C)], indicating that the MBP domain of the chimera was located in the periplasm, consistent with its correct membrane insertion and topology. Finally, the chimera Ibβ-143L had reasonable CAT activity, a comparable expression level, and the correct topology, and was therefore chosen for subsequent experiments.
Figure 1.

Ibβ TM domain sequences inserted into the TOXCAT assay. The eight Ibβ TM domain sequences tested in the TOXCAT assay are aligned and identified by their starting and ending residue numbers as in the mature protein. The amino acids that occupied a and d position in heptad leucine zipper motif were highlighted in the square brackets.
Figure 2.
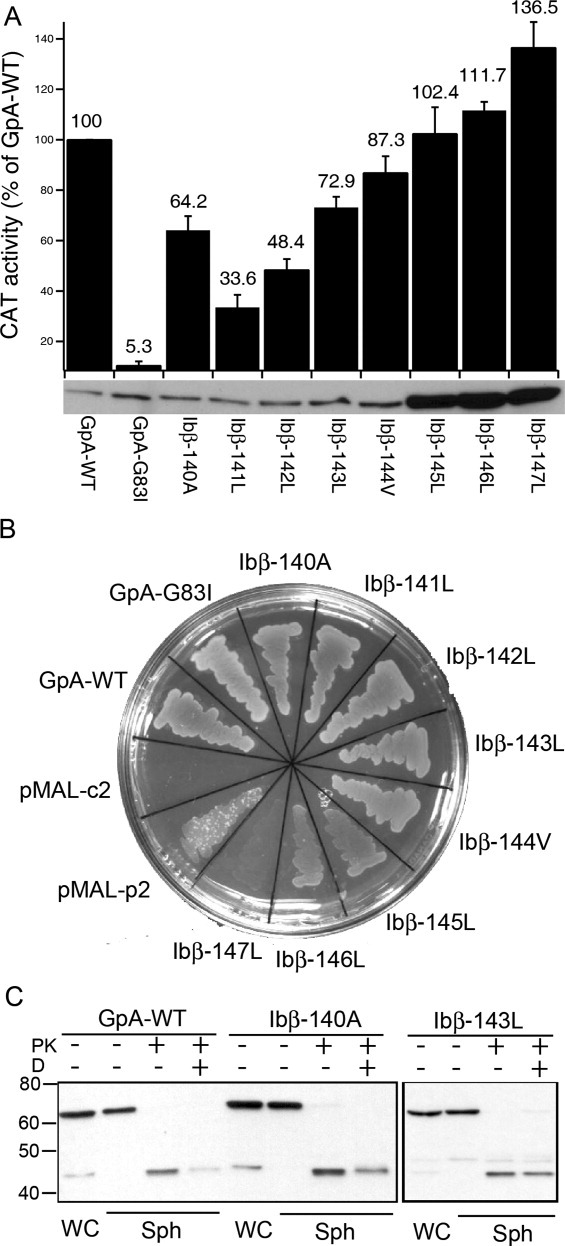
Ibβ TM domain dimerized in the E. coli cell membrane. (A) The enzymatic activity of CAT induced by self-association of the target TM domain and expressed as the percentage of that induced by the wild-type glycophorin A TM domain (GpA-WT). The GpA-WT and GpA-G83I constructs were used as positive and negative controls, respectively. The data are shown as mean ± SD (n = 3). The lower panel shows the expression levels of chimeric ToxR-TM-MBP proteins probed by Western blot. (B) MalE complementation assay of the Ibβ TM constructs to test the topology of ToxR′-TM-MBP chimeric proteins. On M9 minimum media, where maltose is the only carbon source, only cells with MBP expressed in the periplasm survive. MalE-deficient MM39 E. coli cells transformed with pMAL-p2 and pMAL-c2 vectors, which express MBP in the periplasm or in cytoplasm, were included as positive and negative controls, respectively. (C) Proteinase digestion of the spheroplasts of Ibβ-140A and Ibβ-143L constructs. For each construct, the spheroplast was prepared and subjected to proteinase K (PK) digestion in the absence and presence of detergent (D).
Identification of TM domain interface residues in the Ibβ TM helix
By inspecting on the Ibβ TM sequence and comparing it with previously reported association motifs,16,17 the Ibβ TM domain contains a leucine zipper motif and the polar residues Gln129 and His139; both these features are thought to mediate TM domain dimerization in the membrane. Ala-scanning mutagenesis can be used for the identification of interfacial residues because the mutation of most residues to Ala creates voids resulting in the incremental reduction of protein–protein associations, and thereby the critical residues can be identified.18 Thus, we performed site-directed mutagenesis, scanning the Ibβ TM helix with Ala at every position, unless the wild-type residue was already Ala. Complementation assays showed that the mutations did not affect the topology of the chimeric proteins in the bacterial membrane. Further, Western blot results indicated that most mutant constructs were expressed at similar levels except for mutants in the polar residues Gln129 and His139. The CAT activities were further corrected for construct expression as mutations can modulate the TOXCAT signal.19 Therefore, the effects of individual mutations on CAT levels provided insights into the physical basis for dimerization. Our TOXCAT results showed that mutation of each Leu residue to Ala decreased CAT activity to some extent [Fig. 3(A)], suggesting that the Ibβ TM dimerization motif may be a leucine zipper-based motif, which was previously thought to support TM dimerization in the bacterial membrane. Moreover, when either of the polar residues Gln129 or His139 was individually mutated to Ala, the signal dropped dramatically by over 50%; in contrast, mutation of Gly136 to Ala resulted in a significant increase in CAT activity. Interestingly, all these sensitive residues (such as Ala125, Gln129, Leu132, Gly136, His139, and Leu143) occupy a and d positions of (abcdefg)n leucine zipper motif,16,20 and this motif is also characteristic of leucine zipper interaction domains.
Figure 3.
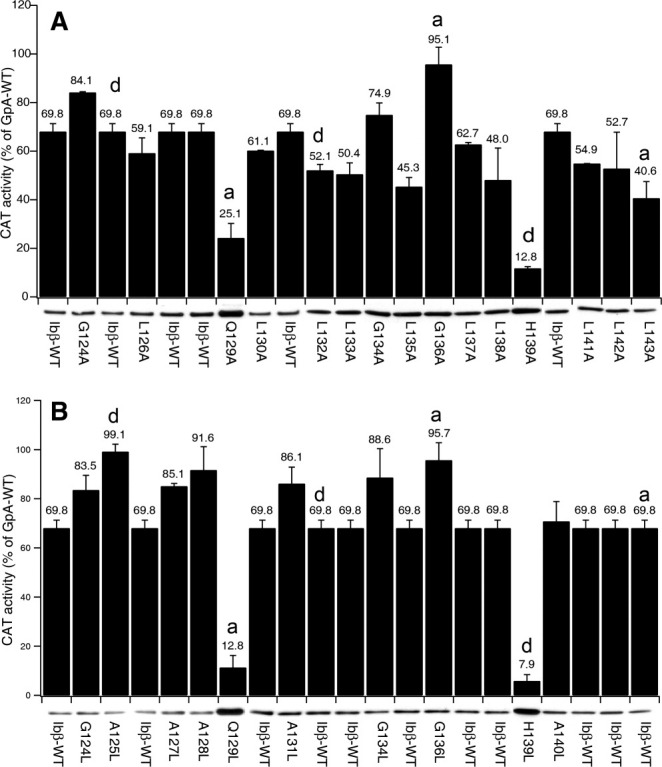
Effect of scanning mutagenesis of the Ibβ TM domain on CAT activity in the TOXCAT assay. The effect of single-site mutations at consecutive positions in the Ibβ TM domain was quantitated by CAT assays, corrected for construct expression, and compared with the signal from the GpA-WT. Black bars represent the CAT activity quantified from cleared lysates, and error bars represent the standard error for three measurements of each lysate. Bands below the bars are details from Western blots of the ToxR fusions; the intensities of these bands were quantified by photon counting of chemiluminescence. Each residue was mutated to either Ala (A) or Leu (B).
To confirm that the dimerization interface of Ibβ TM helices is based on the leucine zipper motif; we mutated each residue to Leu and tested the CAT activity in the TOXCAT assay [Fig. 3(B)]. Our results showed that mutating any residue except for the two polars residue to Leu increased the CAT activity, further suggesting that the dimerization interface of the Ibβ TM helices may be mediated by a leucine zipper-based dimerization motif. It is noteworthy that mutation of either His139 or Gln129 to Leu abolished dimerization despite enhanced formation of leucine zipper motif. This indicates that in addition to the leucine zipper motif, side-chain packing of the polar residues might play a critical role in forming leucine zipper-based dimerization of Ibβ TM domain in membrane.
Characterization of mutant Ibβ peptides in the thiol-disulfide exchange and SDS-PAGE assays
To replicate mutational effects on the dimerization of the Ibβ TM domain in the E. coli cell membrane, we monitored dimmer formation of Ibβ TM peptides directly in detergent solution using thiol-disulfide exchange and SDS-PAGE assays. SDS-PAGE provides a simple, direct, and quick method to detect mutational effects on the TM–TM association.10,17,21 As shown in Figure 4(A), the wild-type Ibβ TM peptide (termed “Ibβ TMpep”) and three Ibβ mutant TM peptides, mutation of Ala128 to Leu (IbβA128L TMpep), mutation of Gly136 to Leu (IbβG136L TMpep), and mutation of His139 to Leu (IbβH139L TMpep), were expressed in E. coli and purified by HPLC. Mutation of Gln129 to Leu (IbβQ129L TMpep) was not included in the SDS-PAGE and thiol-disulfide exchange assays due to the low expression yield of IbβQ129L TMpep in our E. coli expression system. In SDS-PAGE, Ibβ TMpep migrated as two bands corresponding to monomer and dimer species, which was consistent with our previous studies [Fig. 4(B)].13 The disruptive mutation IbβH139L TMpep, which has a disruptive mutation, migrated only as a monomer, whereas mutants with an enhanced leucine zipper motif, IbβG136L TMpep and IbβA128L TMpep, migrated as dimers and tetramers.
Figure 4.
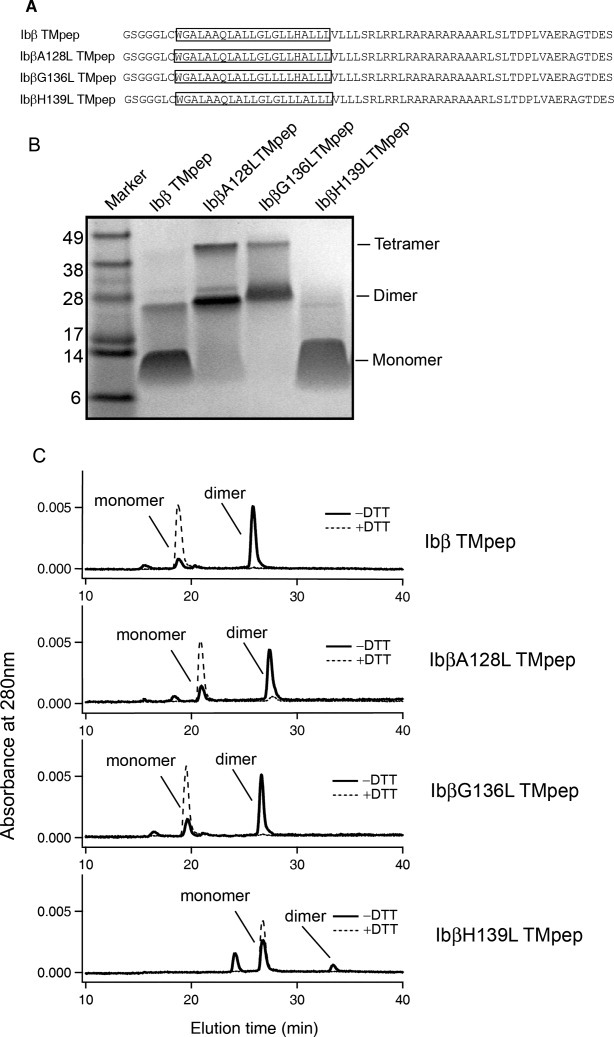
Studies of the oligomerization of wild-type and mutant Ibβ TMpep peptides by SDS-PAGE and thiol-disulfide exchange assays. (A) Sequences of Ibβ TMpep and mutant peptides. The TM domain in each construct is boxed. A GGG sequence was added between the thrombin cleavage site and the Ibβ sequence to facilitate thrombin digestion. (B) SDS-PAGE of Ibβ TMpep peptide and IbβA128L TMpep, IbβG136L TMpep, and IbβH1139L TMpep mutants. (C) The thiol-disulfide exchange reaction of Ibβ TMpep peptide and the IbβA128L TMpep, IbβG136L TMpep, and IbβH1139L TMpep mutants.
The thiol-disulfide exchange assay has been used to measure protein associations, protein folding, and protein stability.22 It can also be used to monitor the association of TM helices in detergents and lipid bilayers.23 Ibβ TMpep contains a membrane-proximal Cys residue and the formation of a disulfide bond depends on the TM–TM association.5,13 In dodecylphosphocholine (DPC) micelles, >80% of the Ibβ TMpep formed disulfide-linked dimers [Fig. 4(C)], demonstrating that the Ibβ TM domain strongly self-associates to help formation of the disulfide-linked dimer in the DPC micelle environment. As we expected, the disulfide-linked dimers were the dominant products for both IbβA128L TMpep and IbβG136L TMpep; by contrast, a significantly lower proportion of the IbβH139L TMpep mutant formed dimers. These data show that the H139L mutation substantially decreased Ibβ TM domain dimerization and that A128L and G136L mutations substantially increased Ibβ TM domain dimerization.
Computer modeling of the Ibβ TM homodimer
Molecular dynamics (MD) simulations provide a computational tool for probing the structure and dynamics of membrane proteins in detergent micelles and lipid bilayers.24,25 Knowing that the Ibβ TM domain dimerizes allows us to take advantage of MD simulation strategies that analyze interhelical interactions to predict the most probable structure of Ibβ TM helix–helix dimers. Residues 147–178 of the Ibβ TM domain were assigned an α-helical structure by using PyMol,26 and hydrogen atoms were also added to the structure. The structural models generated were further optimized by nanosecond-scale MD simulations, and an explicit membrane–water environment was considered. The pre-equilibrated dipalmitoylphosphatidylcholine (DPPC) bilayer (http://moose.bio.ucalgary.ca) with extended water layers was used in all dynamic simulations.
Rigid-body protein–protein dockings were performed using the structures obtained from the MD simulations.25 Next, the docking poses were clustered to different groups and filtered. Finally, the structure from the largest group that had the best Memtop Score27 was used to identify the possible contact pattern of the Ibβ TM helix. Examination of representative structures of the Ibβ TM dimer revealed that the helices were packed in a left-handed fashion [Fig. 5(A)] with Ala125, Gln129, Leu132, Gly136, His139, Leu143, and Leu146 residues forming the helix–helix interface [Fig. 5(B)], as confirmed by mutagenesis data. A series of Leu residues along the interface were found to pack in a leucine zipper-like motif, suggesting that these structures are largely stabilized by hydrophobic interactions. As shown in Figure 5(C), the highly polar amide group on the side chain of Gln129 could form a hydrogen bond to the carbonyl oxygen of the Gln129 on a different helix, and the imidazole side chain of His139 forms a weak electrostatic interaction across the dimer interface to the other His139 residue. The correlation between computer modeling and experimental data is readily apparent, indicating that side-chain packing of polar residues plays a critical role in the leucine zipper-based dimerization of the Ibβ TM domain.
Figure 5.
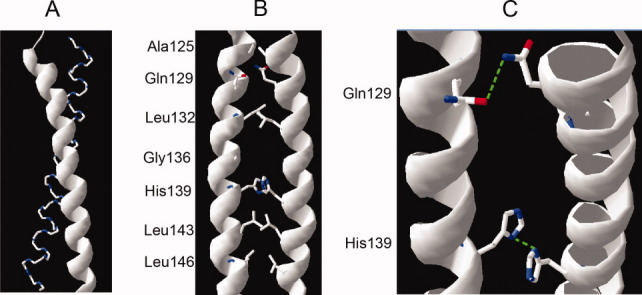
Average molecular structures for the group with the largest population illustrating the dimer interface of Ibβ TM domain. (A) The helices in the Ibβ TM dimer form left-handed coiled coils. (B) The Leu side chains of one helix pack alongside the side chains of the opposing helix. (C) The side-chain NH2 group of Gln129 is hydrogen bonded across the dimer interface to the side-chain C=O group of Gln129, and the imidazole side chain of His139 forms a weak electrostatic interaction across the dimer interface with its counterpart in the other TM helix. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Discussion
The TM domain of Ibβ has been suspected of playing an important role in the assembly of the GP Ib-IX complex in the plasma membrane. Specific residues in the TM domain, namely, Gln129 and His139, have been shown to be crucial in the functional assembly of the GP Ib-IX complex, possibly by stabilizing TM–TM associations.9,13 Our previous studies showed that only Ibβ, and not Ibα or IX, was able to self-associate in the SDS detergent micelle and E. coli cell membrane.13 However, the specific residues or motifs that determine the association of the Ibβ TM domain have not been thoroughly examined. In this article, we have systematically analyzed self-association of the Ibβ TM domain using different assays and characterized the specific dimerization motifs that are responsible for the dimerization of the Ibβ TM helix. First, we explored the sequence dependence of Ibβ TM domain dimerization by Ala- and Leu-scanning mutagenesis using the TOXCAT assay. Differences in the TOXCAT signals can arise from differences in the self-association of the membrane-inserted fusion proteins but may also reflect variations in the total amounts of fusion protein expressed.19 In our experimental data, the CAT activities were corrected for construct expression and only reflect the self-association of the membrane-inserted fusion proteins. We found that mutating most of the residues to Ala decreased the CAT activity, whereas mutating each nonpolar residue to Leu increased the CAT activity, which suggested that the Ibβ TM domain is dimerized via a leucine zipper-like heptad repeat pattern of amino acids. The leucine zipper motif is a common TM helix sequence motif that governs associations between two TM proteins. Such leucine zipper motifs have been shown to form the interfaces of many TM proteins such as phospholamban28 and the M2 proton channel.29 Compared to the GxxxG motif, the leucine zipper motif has one important feature: the replaceability of any Leu residues in the sequence motif. Because the leucine zipper motif often spans many turns in the TM helix and covers 6–10 leucine and similar residues, replacing any one of them with Ala is not expected to totally abolish the packing of all the Leu residues. Thus, the mild, but, reproducible mutational effects reported in our manuscript are entirely consistent with the leucine zipper motif. The residues involved form a repeating heptad (abcdefg) pattern, wherein residues at the a and d positions constitute the core of the interfaces.30,31 Although CAT activity was decreased or increased to a variable extent in Ala- and Leu-scanning mutagenesis methods, the extent of the variability correlated well with the distance from the potential dimerization interface. Consistent with this, our results showed that the sensitive Ibβ TM residues (Ala125, Gln129, Leu132, Gly136, and His139) also occupied a and d positions of an (abcdefg)n leucine zipper motif. Second, mutational effects (seen with the mutant peptides IbβA128L TMpep, IbβG136L TMpep, and IbβH139L TMpep) on the dimerization of the Ibβ TM domain in the E. coli cell membrane were replicated by SDS-PAGE and thiol-disulfide exchange assays. Consistent with the TOXCAT results, SDS-PAGE results showed that mutants with an enhanced leucine zipper motif (IbβA128L TMpep and IbβG136L TMpep) migrated as high oligomer dimer, even tetramer. The possible explanation is that these mutations disrupt the specific dimerization association and form nonspecific higher order oligomers.
The hydrophobic TM domains of membrane proteins contain fewer polar residues compared with soluble proteins. However, despite the predominance of hydrophobic residues within the lipid bilayer, analyses of TM domains reveal that about 25% of all TM domains from single-spanning membrane proteins contain a strongly polar residue.32–34 Our results are consistent with previous studies that polar residues could provide stability and specificity to TM–TM helix association; for example, mutations at polar residues Gln129 and His139 dramatically decrease CAT activities in the TOXCAT assay. The ability of a polar residue to promote helix association appears to be related to the side chain's potential to be both a good hydrogen bond (H-bond) donor and acceptor. It is known that the strength of an H-bond depends on both the orientation and angle of the polar groups. If the bond angle falls between 120 and180 with an average distance of 2 Å, the C=O· · ·H–N intermolecular interaction will result in a strong H-bond.32 Once the parameters of an H-bond move beyond these ranges, it loses its H-bonding character and becomes a weaker electrostatic interaction. Our computer modeling results suggested that interhelical hydrogen bonds of Gln129 could drive TM helix association. As shown in Figure 5(C), the distance (1.81 Å) and angle (170.88°) of the C=O· · ·H–N interaction of Gln129 in the Ibβ TM helix agrees with the geometrical H-bond criteria. Our computing data combined with experimental data demonstrated that the interhelix hydrogen bonding may be an important role of the polar residues in Ibβ TM domain dimerization. Essentially, the imidazole side chain of His has two nitrogen atoms with different properties: one is bound to a hydrogen atom and donates its lone pair of electrons to the aromatic ring and as such is slightly acidic. The long pair of the other nitrogen atom is not donated to the ring and is basic. Therefore, because of its unique side chain, His can act as both an acid and a base, both donating and accepting protons. In the GP Ib-IX complex TM domain, the computer modeling results revealed that one of these two nitrogen atoms from the TM helix forms a weak electrostatic interaction across the dimer interface with its counterpart in the other TM helix. The other nitrogen atom is oriented away from the interface, in a position where it can form electrostatic or hydrogen-bonding interactions with other proteins (such as Ibα and IX) to stabilize heteromeric TM–TM associations in the GP Ib-IX complex. Furthermore, the lack of reactivity of Cys122 in our computer-generated model of the homodimer will promote disulfide bonding between Ibα and Ibβ heterodimer for the formation of the GP Ib-IX complex. In light of the above considerations, we speculate that TM portion of the Ibβ dimer may serve as a scaffold for TM domain heterotetramerization of the GP Ib-IX complex. Future experiments designed to correlate the full-length GP Ib-IX complex with the TM tetramer structure will further enhance our understanding of the assembly of the GP Ib-IX complex and transmembrane signaling.
Materials and Method
Vector and strains
The expression vectors pccKAN, pccGpA-wt, pccGpA-G83I, and Escherichia coli strain MM39 were kindly provided by Dr. Donald M. Engelman. DNA coding for Ibβ TM was generated by PCR and ligated in-frame to the NheI and BamHI sites of pccKAN using XbaI and BamHI sites in the PCR products. Subsequent Leu and Ala scanning and other point mutants were generated using a QuikChange mutagenesis kit (Stratagene, La Jolla, CA). The sequences of the wild-type and mutant Ibβ TM regions were confirmed by DNA sequencing. The resulting plasmids were transformed into E. coli MM39 cells for further analysis.
Expression of the chimeric protein in MM39
A freshly streaked single colony was inoculated into 5 mL of LB broth containing 50 μg/mL ampicillin and grown to A420 of 1 at 37°C with vigorous shaking. 6.0 A420 units of cells were harvested by centrifugation and washed with 0.5 mL of sonication buffer (25 mM Tris-HCl, 2 mM EDTA, pH 8.0). Cells were then resuspended in 2 mL of sonication buffer and lysed by probe sonication. Thirteen microliters of bacterial lysates was mixed with 5 μL of 4× LDS buffer and 2 μL 10× reducing agent, then vortexed, boiled, and loaded onto a 4–12% NuPAGE Bis-Tris precast gel in MOPS buffer system (Invitrogen, Carlsbad, CA). The separate proteins were transferred to nitrocellulose paper and immunoblotted with an anti-maltose-binding protein (MBP) monoclonal antibody (Sigma, St. Louis, MO). The remaining lysate was clarified by centrifugation at 13,000g, and the supernatant was stored on ice until the spectrophotometric assay was performed.
Spectrophotometric CAT assay
Twelve micoliters of lysate was mixed with 300 μL of reaction buffer (0.1 mM acetyl-CoA, 0.4 mg/mL 5,5′-dithiobis-(2-nitrobenzoic acid), 0.1M Tris-HCl, pH 7.8), and the absorbance at 412 nm was acquired at 3-s intervals for a minimum of 4 min to establish a background rate of acetyl-CoA hydrolysis. Twelve micoliters of 2.5 mM chloramphenicol was then added with mixing, and the absorbance was followed for 1 min. Each lysate was assayed in triplicate, and the slopes of the recorded data were converted to enzyme activity units (ɛ = 13,600 at 412 nm). Linear least-squares fit to the data with high correlation coefficients and low variance indicated that the data being fit corresponded to regions for initial rates. All kinetics traces were acquired on a UV-1700 spectrophotometer (SHIMADZU). Results were reported as a percentage of GpA wild-type activity and normalized to protein expression levels on the immunoblot.
MalE complementation test
The glucose in M9 minimal medium plates was replaced by 0.4% maltose as the only carbon source. Transformed MM39 cells were streaked onto these plates containing ampicillin and incubated for 2 days at 37°C.
Protease digestion of spheroplasts
Preparation of spheroplasts was carried out following the protocol of Mendrola et al.35 Proteinase K (Sigma) was then added to a final concentration of 50 μg/mL, with or without 1% Nonidet P-40 (Sigma).
Production Ibβ TM peptides
The His-GST-TM fusion protein was overexpressed in the inclusion body of E. coli and purified according to published protocols.5 Briefly, the fusion protein was first separated from soluble cytosolic proteins and membrane fractions, solubilized in 50 mM Tris·HCl, pH 8.0, 100 mM NaCl, 0.2% N-lauroyl sarcosine, and 5 mM β-mercaptoethanol, and purified by Ni-affinity chromatography. The purified protein was then cleaved by thrombin (10–20 U/mg of protein) overnight at room temperature in 50 mM Tris·HCl, pH 8.0, buffer containing 0.1% N-lauroyl sarcosine and 200 mM imidazole. The TM peptides were separated from the His-GST fragment by preparative reverse-phase HPLC. The purity and identity of Ibβ TMpep, Ibβ A128L TMpep, Ibβ G136L TMpep, and Ibβ H139L TMpep peptides were confirmed by SDS-PAGE, analytical HPLC, and MALDI-TOF mass spectrometry.
SDS-PAGE
Ethanol was removed from a stock solution of protein, and the protein was dissolved in 20 μL of LDS sample buffer. After mixing for half an hour, the protein sample was heated to 100°C for 10 min and loaded onto a 4–12% NuPAGE Bis-Tris precast gel in MES running buffer (Invitrogen, Carlsbad, CA). The sample was electrophoresed at 4°C for 4 h at a constant current of 20 mA before being stained by Coomassie Blue.
In vitro thiol-disulfide exchange assay
Purified TM peptides were mixed with DPC (Avanti Polar Lipids, Alabaster, AL) in ethanol at a peptide/detergent molar ratio of 1:200. The solution was dried by Argon stream to a thin film and placed under vacuum overnight. The dried protein/detergent mixture was hydrated in 40 μL reaction buffer (100 mM Tris·HCl, pH 8.0, 100 mM KCl, 2 mM EDTA, 1 mM oxidized glutathione, and 1 mM reduced glutathione) purged with helium gas, topped with argon gas, sealed, and incubated at room temperature for 6 h to allow the thiol-disulfide exchange reaction to reach equilibrium. In the control reaction, 2.5 mM freshly prepared dithiothreitol was included. The final peptide concentration in the reaction was 25 μM. To quench the reaction, 6.7 μL 0.5M HCl, followed by 33.3 μL organic solvent containing 0.1% TFA, was added to the glass vial. The reaction products were analyzed by reverse-phase HPLC in a C4 or phenyl analytical column (Grace Vydac) using a linear gradient of buffer A (0.1% TFA) and buffer B (6:3:1 v/v 2-propanol/acetonitrile/water).
Computer modeling of Ibβ TM homodimer
Residues 147–178 of Ibβ TM were assigned a α-helical structure by using PyMol, and hydrogens were also added to the structure. The pre-equilibrated DPPC bilayer with extended water layers was used in all dynamic simulations. MD simulations were performed using the GROMACS package,36 using NPT and periodic boundary conditions. A modification of GROMOS87 force field37 was applied for protein, and the lipid parameters adopted were the same as the previous MD studies of lipid bilayers.38–40 A twin cutoff of 9 Å was used for the short-range interaction and a cutoff of 12 Å was used for the Lennard–Jones interaction. Particle mesh Ewald algorithm41 was used for the calculation of electrostatic contributions to energies and forces. Prediction of homodimer structures was done by means of the rigid-body docking program ZDOCK3.0.42 We collected 160 structural conformations from the MDS. A rotational sampling interval of 60 was used and the top 4000 solutions, ranked by ZDOCK score, were survived for each run. The filtering of the models was performed by FiPD according to MemTop definition,26 with the thresholds of 0.4 rad for the tilt angle and 6.0 Å for the z-offset, respectively. Automatic clusters were then performed, divided the remaining structures to different groups. The group with the largest population was taken into further analysis.
Acknowledgments
The authors thank Dr. Renhao Li for sharing the reagents.
Glossary
Abbreviations
- BSS
Bernard-Soulier Syndrome
- CYTO
cytoplasmic domain
- GP
glycoprotein
- TM
transmembrane
References
- 1.Andrews RK, Gardiner EE, Shen Y, Whisstock JC, Berndt MC. Glycoprotein Ib-IX-V. Intl J Biochem Cell Biol. 2003;35:1170–1174. doi: 10.1016/s1357-2725(02)00280-7. [DOI] [PubMed] [Google Scholar]
- 2.Du X. Signaling and regulation of the platelet glycoprotein Ib-IX-V complex. Curr Opin Hematol. 2007;14:262–269. doi: 10.1097/MOH.0b013e3280dce51a. [DOI] [PubMed] [Google Scholar]
- 3.Berndt MC, Shen Y, Dopheide SM, Gardiner EE, Andrews RK. The vascular biology of the glycoprotein Ib-IX-V complex. Thromb Haemost. 2001;86:178–188. [PubMed] [Google Scholar]
- 4.Lopez JA, Andrews RK, Afshar-Kharghan V, Berndt MC. Bernard-Soulier syndrome. Blood. 1998;91:4397–4418. [PubMed] [Google Scholar]
- 5.Luo SZ, Mo X, Afshar-Kharghan V, Srinivasan S, Lopez JA, Li R. Glycoprotein Ibalpha forms disulfide bonds with 2 glycoprotein Ibbeta subunits in the resting platelet. Blood. 2007;109:603–609. doi: 10.1182/blood-2006-05-024091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solum NO, Clemetson KJ. The discovery and characterization of platelet GPIb. J Thromb Haemost. 2005;3:1125–1132. doi: 10.1111/j.1538-7836.2005.01072.x. [DOI] [PubMed] [Google Scholar]
- 7.Lopez JA, Leung B, Reynolds CC, Li CQ, Fox JE. Efficient plasma membrane expression of a functional platelet glycoprotein Ib-IX complex requires the presence of its three subunits. J Biol Chem. 1992;267:12851–12859. [PubMed] [Google Scholar]
- 8.Mo X, Lu N, Padilla A, Lopez JA, Li R. The transmembrane domain of glycoprotein Ibbeta is critical to efficient expression of glycoprotein Ib-IX complex in the plasma membrane. J Biol Chem. 2006;281:23050–23059. doi: 10.1074/jbc.M600924200. [DOI] [PubMed] [Google Scholar]
- 9.Luo SZ, Mo X, Lopez JA, Li R. Role of the transmembrane domain of glycoprotein IX in assembly of the glycoprotein Ib-IX complex. J Thromb Haemost. 2007;5:2494–2502. doi: 10.1111/j.1538-7836.2007.02785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemmon MA, Flanagan JM, Treutlein HR, Zhang J, Engelman DM. Sequence specificity in the dimerization of transmembrane alpha-helices. Biochemistry. 1992;31:12719–12725. doi: 10.1021/bi00166a002. [DOI] [PubMed] [Google Scholar]
- 11.MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 12.Oxenoid K, Chou JJ. The structure of phospholamban pentamer reveals a channel-like architecture in membranes. Proc Natl Acad Sci USA. 2005;102:10870–10875. doi: 10.1073/pnas.0504920102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo SZ, Li R. Specific heteromeric association of four transmembrane peptides derived from platelet glycoprotein Ib-IX complex. J Mol Biol. 2008;382:448–457. doi: 10.1016/j.jmb.2008.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li R, Gorelik R, Nanda V, Law PB, Lear JD, DeGrado WF, Bennett JS. Dimerization of the transmembrane domain of Integrin alphaIIb subunit in cell membranes. J Biol Chem. 2004;279:26666–26673. doi: 10.1074/jbc.M314168200. [DOI] [PubMed] [Google Scholar]
- 15.Russ WP, Engelman DM. TOXCAT: a measure of transmembrane helix association in a biological membrane. Proc Natl Acad Sci USA. 1999;96:863–868. doi: 10.1073/pnas.96.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurezka R, Laage R, Brosig B, Langosch D. A heptad motif of leucine residues found in membrane proteins can drive self-assembly of artificial transmembrane segments. J Biol Chem. 1999;274:9265–9270. doi: 10.1074/jbc.274.14.9265. [DOI] [PubMed] [Google Scholar]
- 17.Lemmon MA, Flanagan JM, Hunt JF, Adair BD, Bormann BJ, Dempsey CE, Engelman DM. Glycophorin A dimerization is driven by specific interactions between transmembrane alpha-helices. J Biol Chem. 1992;267:7683–7689. [PubMed] [Google Scholar]
- 18.Plotkowski ML, Kim S, Phillips ML, Partridge AW, Deber CM, Bowie JU. Transmembrane domain of myelin protein zero can form dimers: possible implications for myelin construction. Biochemistry. 2007;46:12164–12173. doi: 10.1021/bi701066h. [DOI] [PubMed] [Google Scholar]
- 19.Duong MT, Jaszewski TM, Fleming KG, MacKenzie KR. Changes in apparent free energy of helix-helix dimerization in a biological membrane due to point mutations. J Mol Biol. 2007;371:422–434. doi: 10.1016/j.jmb.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noordeen NA, Carafoli F, Hohenester E, Horton MA, Leitinger B. A transmembrane leucine zipper is required for activation of the dimeric receptor tyrosine kinase DDR1. J Biol Chem. 2006;281:22744–22751. doi: 10.1074/jbc.M603233200. [DOI] [PubMed] [Google Scholar]
- 21.Sulistijo ES, Jaszewski TM, MacKenzie KR. Sequence-specific dimerization of the transmembrane domain of the “BH3-only” protein BNIP3 in membranes and detergent. J Biol Chem. 2003;278:51950–51956. doi: 10.1074/jbc.M308429200. [DOI] [PubMed] [Google Scholar]
- 22.Creighton TE. Disulfide bonds as probes of protein folding pathways. Methods Enzymol. 1986;131:83–106. doi: 10.1016/0076-6879(86)31036-x. [DOI] [PubMed] [Google Scholar]
- 23.Cristian L, Lear JD, DeGrado WF. Use of thiol-disulfide equilibria to measure the energetics of assembly of transmembrane helices in phospholipid bilayers. Proc Natl Acad Sci USA. 2003;100:14772–14777. doi: 10.1073/pnas.2536751100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Psachoulia E, Marshall DP, Sansom MS. Molecular dynamics simulations of the dimerization of transmembrane alpha-helices. Acc Chem Res. 2010;43:388–396. doi: 10.1021/ar900211k. [DOI] [PubMed] [Google Scholar]
- 25.Dell'Orco D, De Benedetti PG, Fanelli F. In silico screening of mutational effects on transmembrane helix dimerization: insights from rigid-body docking and molecular dynamics simulations. J Phys Chem. 2007;111:9114–9124. doi: 10.1021/jp071383r. [DOI] [PubMed] [Google Scholar]
- 26.DeLano WL. The PyMOL molecular graphics system. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
- 27.Casciari D, Seeber M, Fanelli F. Quaternary structure predictions of transmembrane proteins starting from the monomer: a docking-based approach. BMC Bioinform. 2006;7:340. doi: 10.1186/1471-2105-7-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simmerman HK, Kobayashi YM, Autry JM, Jones LR. A leucine zipper stabilizes the pentameric membrane domain of phospholamban and forms a coiled-coil pore structure. J Biol Chem. 1996;271:5941–5946. doi: 10.1074/jbc.271.10.5941. [DOI] [PubMed] [Google Scholar]
- 29.Pinto LH, Dieckmann GR, Gandhi CS, Papworth CG, Braman J, Shaughnessy MA, Lear JD, Lamb RA, DeGrado WF. A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc Natl Acad Sci USA. 1997;94:11301–11306. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruan W, Becker V, Klingmuller U, Langosch D. The interface between self-assembling erythropoietin receptor transmembrane segments corresponds to a membrane-spanning leucine zipper. J Biol Chem. 2004;279:3273–3279. doi: 10.1074/jbc.M309311200. [DOI] [PubMed] [Google Scholar]
- 31.Ruan W, Lindner E, Langosch D. The interface of a membrane-spanning leucine zipper mapped by asparagine-scanning mutagenesis. Protein Sci. 2004;13:555–559. doi: 10.1110/ps.03357404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dawson JP, Melnyk RA, Deber CM, Engelman DM. Sequence context strongly modulates association of polar residues in transmembrane helices. J Mol Biol. 2003;331:255–262. doi: 10.1016/s0022-2836(03)00714-9. [DOI] [PubMed] [Google Scholar]
- 33.Eilers M, Shekar SC, Shieh T, Smith SO, Fleming PJ. Internal packing of helical membrane proteins. Proc Natl Acad Sci USA. 2000;97:5796–5801. doi: 10.1073/pnas.97.11.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Senes A, Gerstein M, Engelman DM. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J Mol Biol. 2000;296:921–936. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- 35.Mendrola JM, Berger MB, King MC, Lemmon MA. The single transmembrane domains of ErbB receptors self-associate in cell membranes. J Biol Chem. 2002;277:4704–4712. doi: 10.1074/jbc.M108681200. [DOI] [PubMed] [Google Scholar]
- 36.Berendsen HJ, Vanderspoel D, Vandrunen R. Gromacs—a message-passing parallel molecular-dynamics implementation. Comput Phys Commun. 1995;91:43–56. [Google Scholar]
- 37.Vanbuuren AR, Marrink SJ, Berendsen HJC. A molecular-dynamics study of the decane water interface. J Phys Chem. 1993;97:9206–9212. [Google Scholar]
- 38.Berger O, Edholm O, Jahnig F. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys J. 1997;72:2002–2013. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tieleman DP, Berendsen HJ, Sansom MS. An alamethicin channel in a lipid bilayer: molecular dynamics simulations. Biophys J. 1999;76:1757–1769. doi: 10.1016/s0006-3495(99)77337-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tieleman DP, Sansom MS, Berendsen HJ. Alamethicin helices in a bilayer and in solution: molecular dynamics simulations. Biophys J. 1999;76:40–49. doi: 10.1016/S0006-3495(99)77176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darden T, York D, Pedersen L. Particle mesh Ewald—an n.log(n) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 42.Chen R, Weng ZP. A novel shape complementarity scoring function for protein-protein docking. Proteins. 2003;51:397–408. doi: 10.1002/prot.10334. [DOI] [PubMed] [Google Scholar]


