Abstract
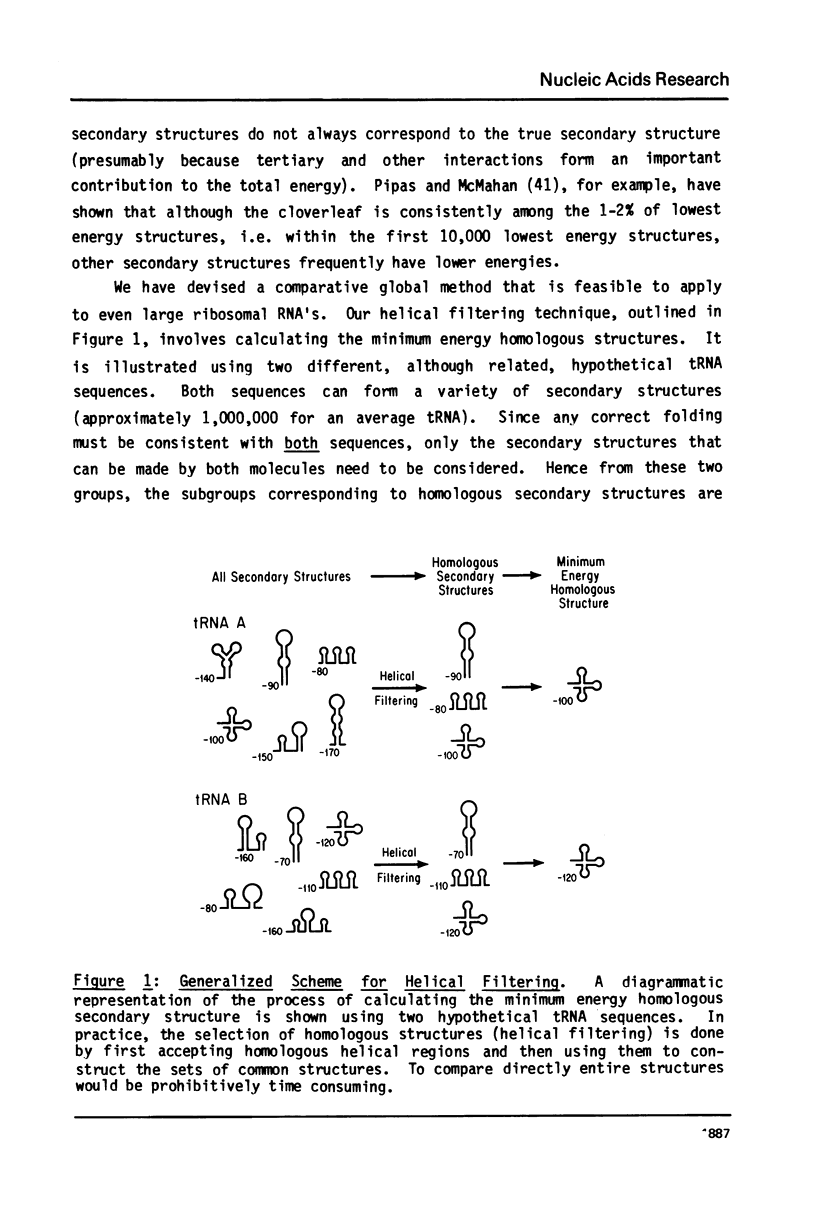
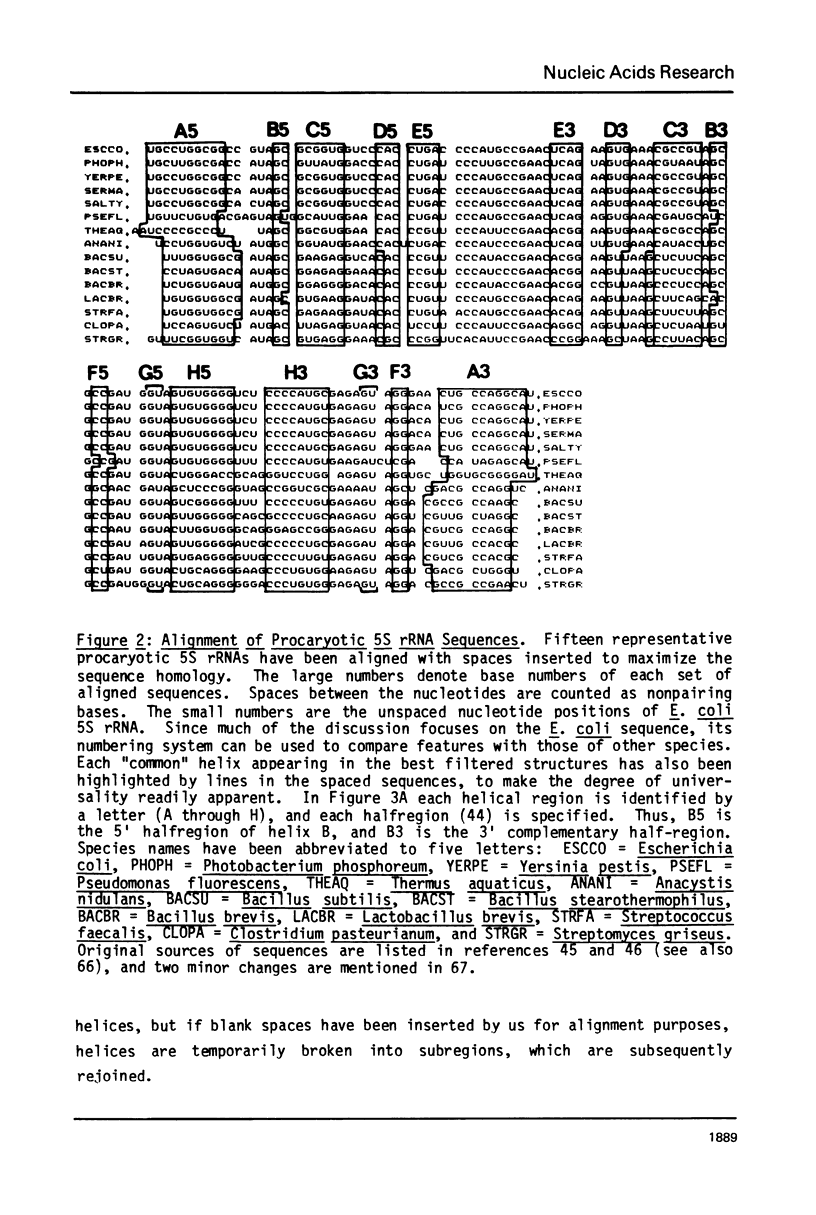
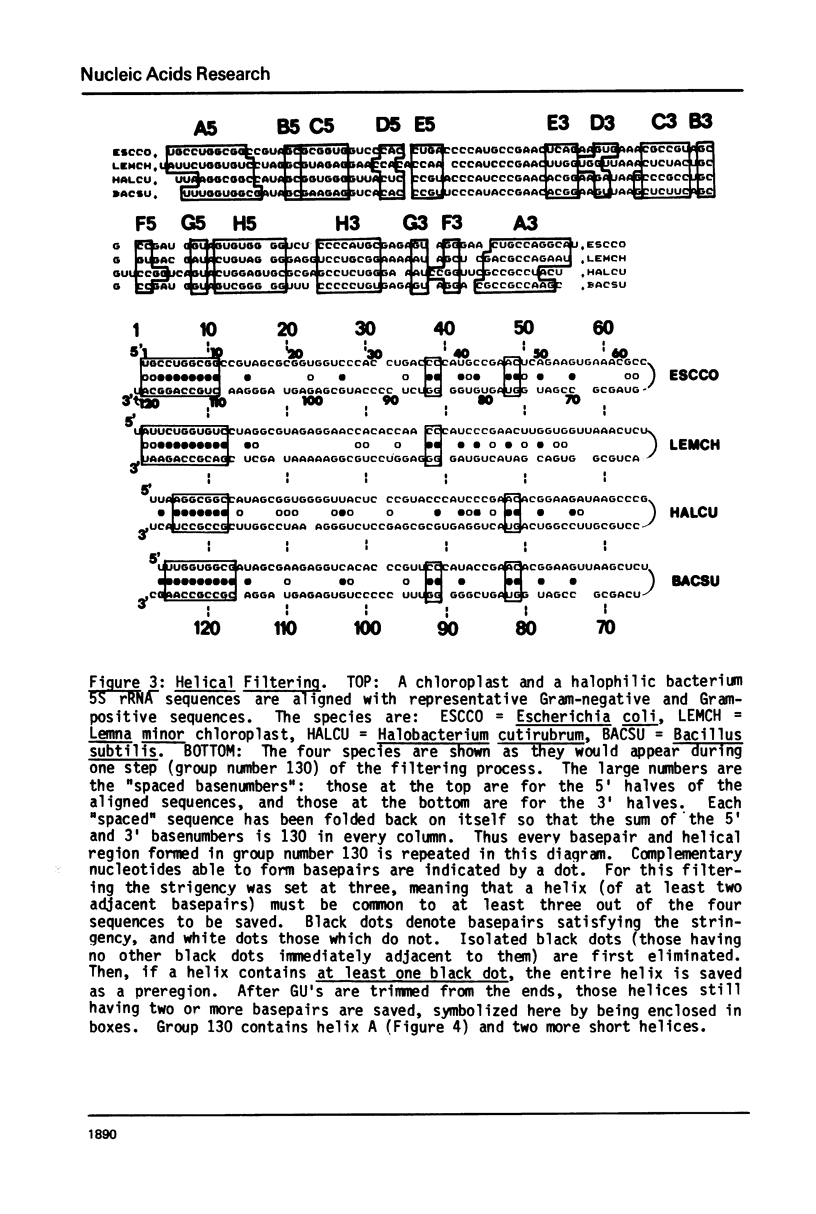
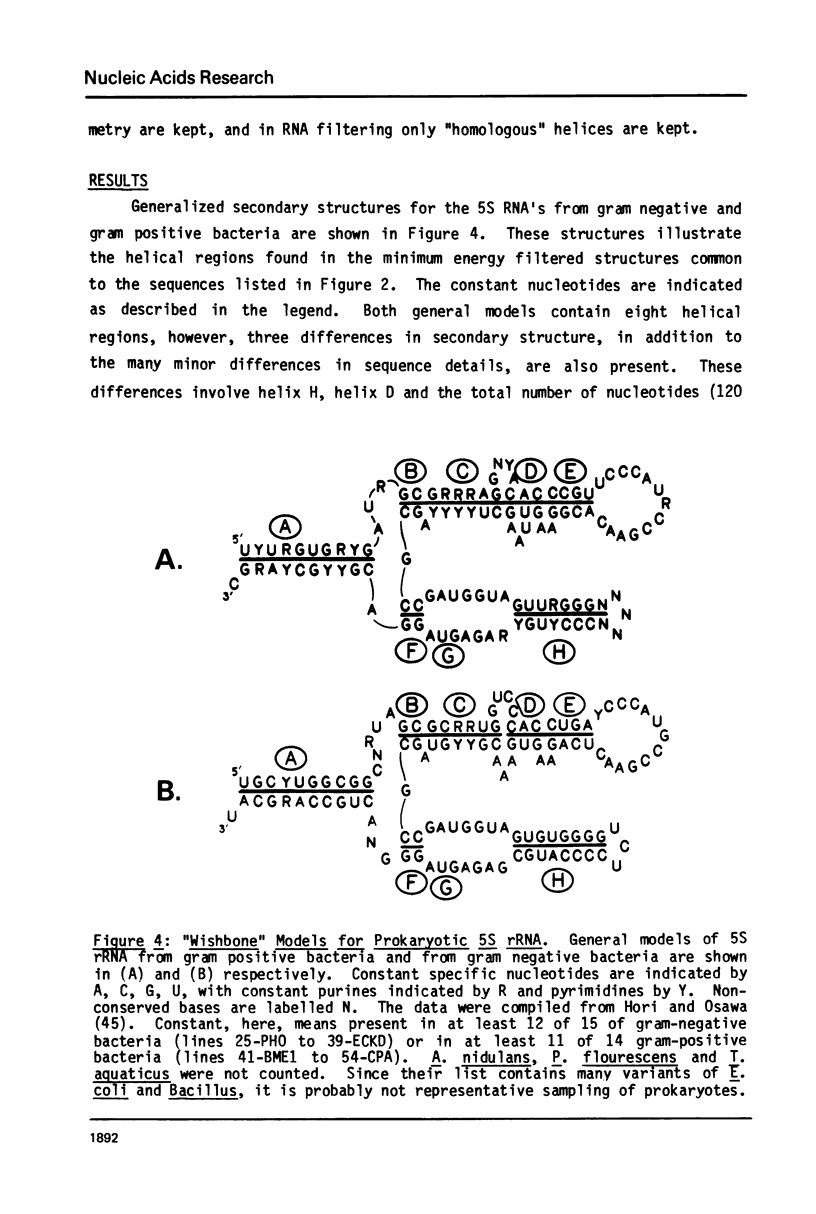
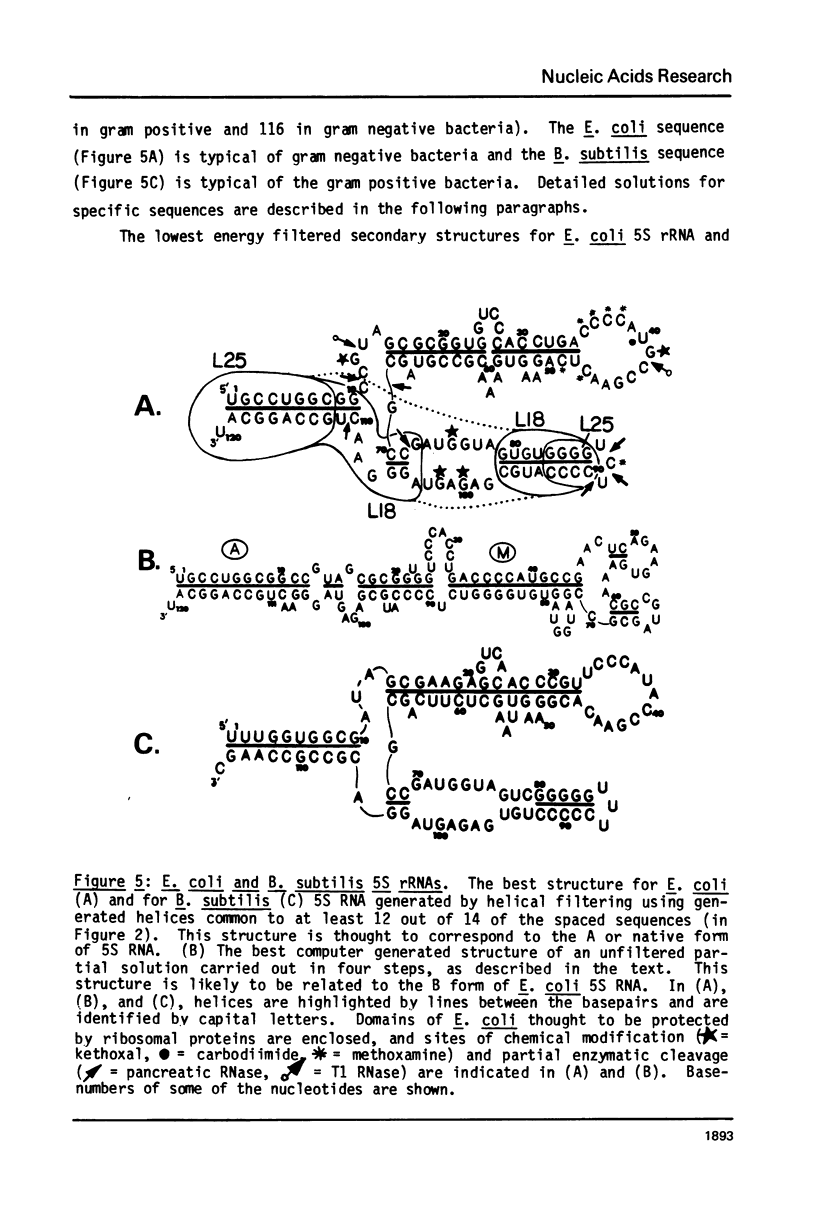
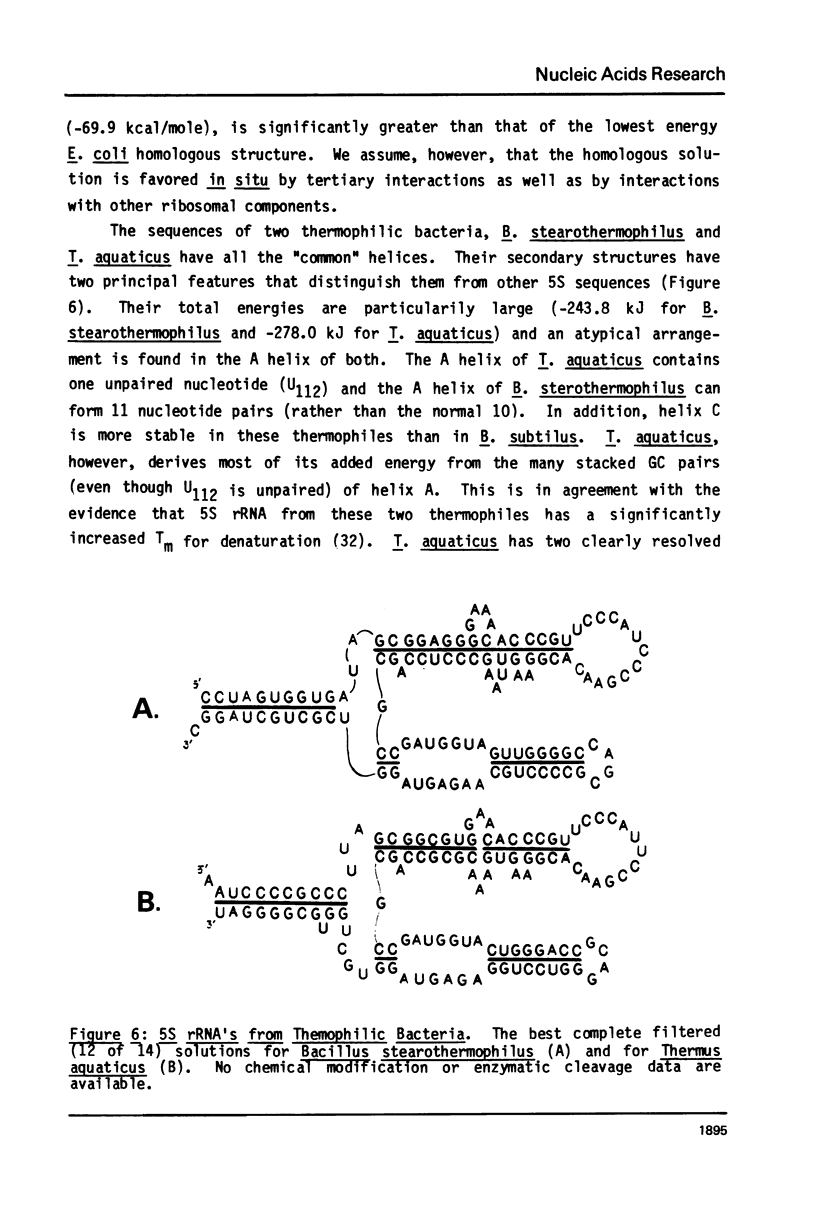
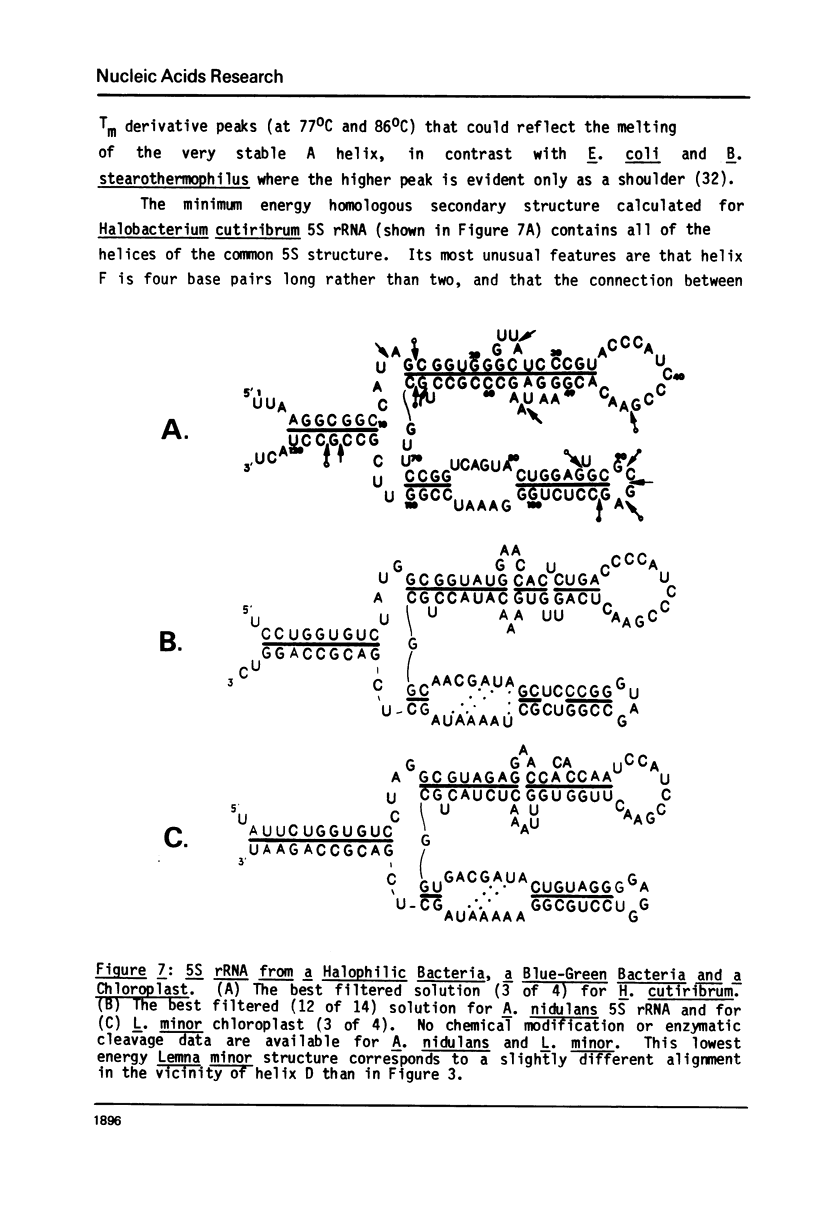
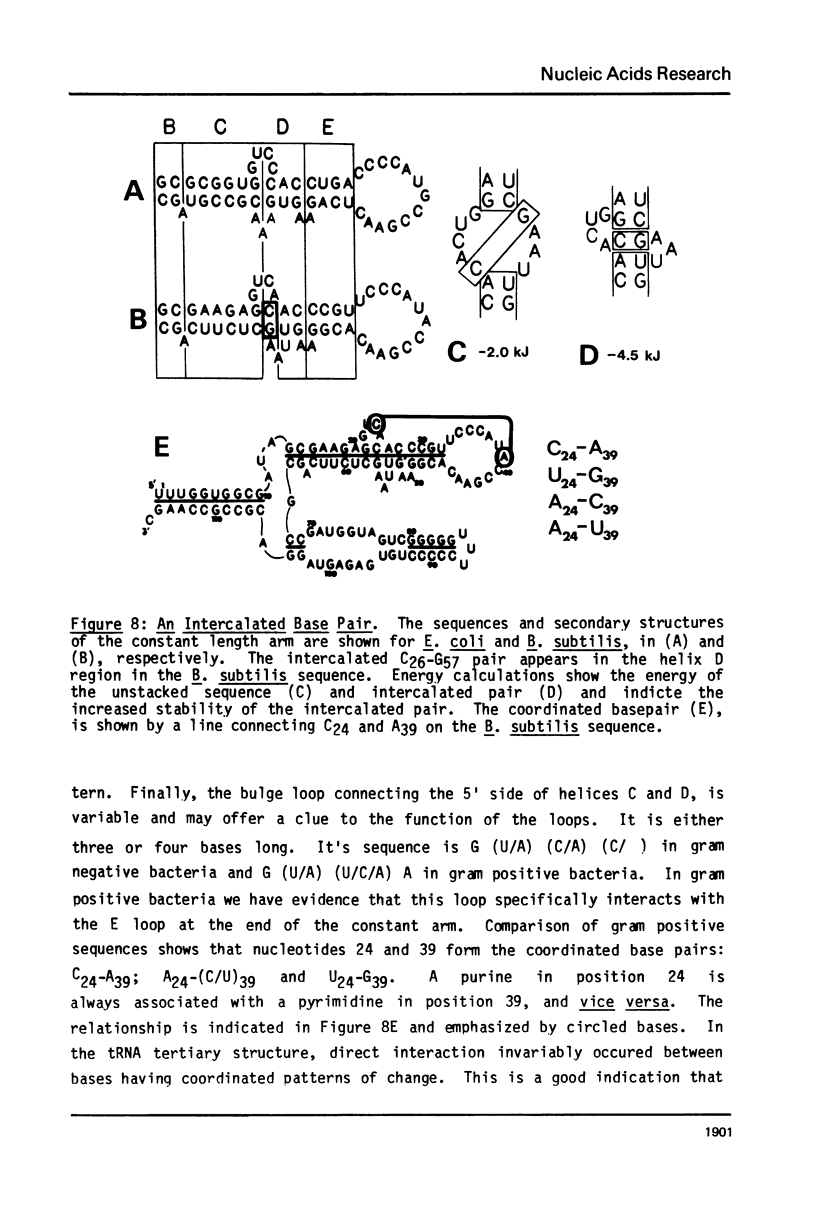
A general secondary structure is proposed for the 5S RNA of prokaryotic ribosomes, based on helical energy filtering calculations. We have considered all secondary structures that are common to 17 different prokaryotic 5S RNAs and for each 5S sequence calculated the (global) minimum energy secondary structure (300,000 common structures are possible for each sequence). The 17 different minimum energy secondary structures all correspond, with minor differences, to a single, secondary structure model. This is strong evidence that this general 5S folding pattern corresponds to the secondary structure of the functional 5S rRNA. The general 5S secondary structure is forked and in analogy with the cloverleaf of tRNA is named the "wishbone" model. It constant 8 double helical regions; one in the stem, four in the first, or constant arm, and three in the second arm. Four of these double helical regions are present in a model earlier proposed (1) and four additional regions not proposed by them are presented here. In the minimum energy general structure, the four helices in the constant arm are exactly 15 nucleotide pairs long. These helices are stacked in the sequences from gram-positive bacteria and probably stacked in gram-negative sequences as well. In sequences from gram-positive bacteria the length of the constant arm is maintained at 15 stacked pairs by an unusual minimum energy interaction involving a C26-G57 base pair intercalated between two adjacent helical regions.
Full text
PDF











Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Appel B., Erdmann V. A., Stulz J., Ackerman T. Determination of base pairing in Escherichia coli and Bacillus stearothermophilus 5S RNAs by infrared spectroscopy. Nucleic Acids Res. 1979 Oct 25;7(4):1043–1057. doi: 10.1093/nar/7.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert M., Bellemare G., Monier R. Selective reaction of glyoxal with guanine residues in native and denatured Escherichia coli 5S RNA. Biochimie. 1973;55(2):135–142. doi: 10.1016/s0300-9084(73)80385-2. [DOI] [PubMed] [Google Scholar]
- Aubert M., Scott J. F., Reynier M., Monier R. Rearrangement of the conformation of Escherichia coli 5S RNA. Proc Natl Acad Sci U S A. 1968 Sep;61(1):292–299. doi: 10.1073/pnas.61.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellemare G., Jordan B. R., Monier R. Demonstration of a highly exposed region in Escherichia coli 5 s RNA by partial hydrolysis with ribonuclease IV and sheep kidney nuclease. J Mol Biol. 1972 Nov 14;71(2):307–315. doi: 10.1016/0022-2836(72)90353-1. [DOI] [PubMed] [Google Scholar]
- Bellemare G., Jordan B. R., Rocca-Serra J., Monier R. Accessibility of Escherichia coli 5S RNA base residues to chemical reagents. Influence of chemical alterations on the affinity of 5S RNA for the 50S subunit structure. Biochimie. 1972;54(11):1453–1466. doi: 10.1016/s0300-9084(72)80087-7. [DOI] [PubMed] [Google Scholar]
- Bellemare G., Vigne R., Jordan B. R. Interaction between Escherichia coli ribosomal proteins and 5S RNA molecules: recognition of prokaryotic 5S RNAs and rejection of eukaryotic 5S RNAs. Biochimie. 1973;55(1):29–35. doi: 10.1016/s0300-9084(73)80233-0. [DOI] [PubMed] [Google Scholar]
- Boyle J., Robillard G. T., Kim S. H. Sequential folding of transfer RNA. A nuclear magnetic resonance study of successively longer tRNA fragments with a common 5' end. J Mol Biol. 1980 Jun 5;139(4):601–625. doi: 10.1016/0022-2836(80)90051-0. [DOI] [PubMed] [Google Scholar]
- Cantor C. R. The extent of base pairing in 5S ribosomal RNA. Proc Natl Acad Sci U S A. 1968 Feb;59(2):478–483. doi: 10.1073/pnas.59.2.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connors P. G., Beeman W. W. Size and shape of 5 S ribosomal RNA. J Mol Biol. 1972 Oct 28;71(1):31–37. doi: 10.1016/0022-2836(72)90398-1. [DOI] [PubMed] [Google Scholar]
- Delihas N., Dunn J. J., Erdmann V. A. The reaction of 5S RNA in 70S ribosomes with kethoxal. FEBS Lett. 1975 Oct 15;58(1):76–80. doi: 10.1016/0014-5793(75)80229-8. [DOI] [PubMed] [Google Scholar]
- Delisi C., Crothers D. M. Prediction of RNA secondary structure. Proc Natl Acad Sci U S A. 1971 Nov;68(11):2682–2685. doi: 10.1073/pnas.68.11.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis H., Wegnez M. Evolution of the 5 S RNA genes in vertebrates. J Mol Evol. 1978 Oct 27;12(1):11–15. doi: 10.1007/BF01732542. [DOI] [PubMed] [Google Scholar]
- Dohme F., Nierhaus K. H. Role of 5S RNA in assembly and function of the 50S subunit from Escherichia coli. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2221–2225. doi: 10.1073/pnas.73.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douthwaite S., Garrett R. A., Wagner R., Feunteun J. A ribonuclease-resistant region of 5S RNA and its relation to the RNA binding sites of proteins L18 and L25. Nucleic Acids Res. 1979 Jun 11;6(7):2453–2470. doi: 10.1093/nar/6.7.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann V. A., Fahnestock S., Higo K., Nomura M. Role of 5S RNA in the functions of 50S ribosomal subunits. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2932–2936. doi: 10.1073/pnas.68.12.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann V. A., Sprinzl M., Pongs O. The involvement of 5S RNA in the binding of tRNA to ribosomes. Biochem Biophys Res Commun. 1973 Oct 1;54(3):942–948. doi: 10.1016/0006-291x(73)90785-7. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. The architecture of 5S rRNA and its relation to function. J Mol Evol. 1975 Oct 3;6(1):61–76. doi: 10.1007/BF01732674. [DOI] [PubMed] [Google Scholar]
- Gray P. N., Bellemare G., Monier R. Degradation of a specific 5 S RNA - 23 S RNA - protein complex by pancreatic ribonuclease. FEBS Lett. 1972 Aug 1;24(2):156–160. doi: 10.1016/0014-5793(72)80756-7. [DOI] [PubMed] [Google Scholar]
- HOLLEY R. W., APGAR J., EVERETT G. A., MADISON J. T., MARQUISEE M., MERRILL S. H., PENSWICK J. R., ZAMIR A. STRUCTURE OF A RIBONUCLEIC ACID. Science. 1965 Mar 19;147(3664):1462–1465. doi: 10.1126/science.147.3664.1462. [DOI] [PubMed] [Google Scholar]
- Hatlen L. E., Amaldi F., Attardi G. Oligonucleotide pattern after pancreatic ribonuclease digestion and the 3' and 5' termini of 5S ribonucleic acid from HeLa cells. Biochemistry. 1969 Dec;8(12):4989–5005. doi: 10.1021/bi00840a048. [DOI] [PubMed] [Google Scholar]
- Hori H. Evolution of 5sRNA. J Mol Evol. 1975 Dec 31;7(1):75–86. doi: 10.1007/BF01732181. [DOI] [PubMed] [Google Scholar]
- Hori H. Molecular evolution of 5S RNA. Mol Gen Genet. 1976 May 7;145(2):119–123. doi: 10.1007/BF00269583. [DOI] [PubMed] [Google Scholar]
- Hori H., Osawa S. Evolutionary change in 5S RNA secondary structure and a phylogenic tree of 54 5S RNA species. Proc Natl Acad Sci U S A. 1979 Jan;76(1):381–385. doi: 10.1073/pnas.76.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne J. R., Erdmann V. A. Isolation and characterization of 5S RNA-protein complexes from Bacillus stearothermophilus and Escherichia coli ribosomes. Mol Gen Genet. 1972;119(4):337–344. doi: 10.1007/BF00272091. [DOI] [PubMed] [Google Scholar]
- Jordan B. R. Computer generation of pairing schemes for RNA molecules. J Theor Biol. 1972 Feb;34(2):363–378. doi: 10.1016/0022-5193(72)90168-3. [DOI] [PubMed] [Google Scholar]
- Jordan B. R. Studies on 5 s RNA conformation by partial ribonuclease hydrolysis. J Mol Biol. 1971 Feb 14;55(3):423–439. doi: 10.1016/0022-2836(71)90327-5. [DOI] [PubMed] [Google Scholar]
- Kao T. H., Crothers D. M. A proton-coupled conformational switch of Escherichia coli 5S ribosomal RNA. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3360–3364. doi: 10.1073/pnas.77.6.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns D. R., Wong Y. P. Investigation of the secondary structure of Escherichia coli 5 S RNA by high-resolution nuclear magnetic resonance. J Mol Biol. 1974 Aug 25;87(4):755–774. doi: 10.1016/0022-2836(74)90083-7. [DOI] [PubMed] [Google Scholar]
- Larrinúa I., Delihas N. Modification of the guanine in the invariant sequence 5' CCG44AAC3' of the Escherichia coli 5 S RNA in solution by kethoxal. FEBS Lett. 1979 Dec 1;108(1):181–184. doi: 10.1016/0014-5793(79)81205-3. [DOI] [PubMed] [Google Scholar]
- Lecanidou R., Richards E. G. The thermodynamics and kinetics of conformational changes in 5-S RNA from Escherichia coli. Eur J Biochem. 1975 Sep 1;57(1):127–133. doi: 10.1111/j.1432-1033.1975.tb02283.x. [DOI] [PubMed] [Google Scholar]
- Lee J. C., Ingram V. M. Reaction of 5 s RNA with a radioactive carbodiimide. J Mol Biol. 1969 May 14;41(3):431–441. doi: 10.1016/0022-2836(69)90286-1. [DOI] [PubMed] [Google Scholar]
- Lewis J. B., Doty P. Derivation of the secondary structure of 5S RNA from its binding of complementary oligonucleotides. Nature. 1970 Feb 7;225(5232):510–512. doi: 10.1038/225510a0. [DOI] [PubMed] [Google Scholar]
- Luoma G. A., Marshall A. G. Lasar Raman evidence for a new cloverleaf secondary structure for eucaryotic 5 S RNA. J Mol Biol. 1978 Oct 15;125(1):95–105. doi: 10.1016/0022-2836(78)90256-5. [DOI] [PubMed] [Google Scholar]
- Luoma G. A., Marshall A. G. Laser Raman evidence for new cloverleaf secondary structures for eukaryotic 5.8S RNA and prokaryotic 5S RNA. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4901–4905. doi: 10.1073/pnas.75.10.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison J. T. Primary structure of RNA. Annu Rev Biochem. 1968;37:131–148. doi: 10.1146/annurev.bi.37.070168.001023. [DOI] [PubMed] [Google Scholar]
- Nazar R. N., Sprott G. D., Matheson A. T., Van N. T. An enhanced thermostability in thermophilic 5-S ribonucleic acids under physiological salt conditions. Biochim Biophys Acta. 1978 Nov 21;521(1):288–294. doi: 10.1016/0005-2787(78)90271-x. [DOI] [PubMed] [Google Scholar]
- Nazar R. N., Willick G. E., Matheson A. T. The 5 S RNA.protein complex from an extreme halophile, Halobacterium cutirubrum. Studies on the RNA-protein interaction. J Biol Chem. 1979 Mar 10;254(5):1506–1512. [PubMed] [Google Scholar]
- Nichols J. L., Welder L. S1 nuclease as a probe of yeast ribosomal 5 S RNA conformation. Biochim Biophys Acta. 1979 Feb 27;561(2):445–451. doi: 10.1016/0005-2787(79)90152-7. [DOI] [PubMed] [Google Scholar]
- Nishikawa K., Takemura S. Structure and function of 5S ribosomal ribonucleic acid from Torulopsis utilis. II. Partial digestion with ribonucleases and derivation of the complete sequence. J Biochem. 1974 Nov;76(5):935–947. [PubMed] [Google Scholar]
- Nishikawa K., Takemura S. Structure and function of 5S ribosomal ribonucleic acid from Torulopsis utilis. IV. Detection of exposed guanine residues by chemical modification with kethoxal. J Biochem. 1978 Aug;84(2):259–266. doi: 10.1093/oxfordjournals.jbchem.a132126. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Garrett R. A. Structure of 5 S ribosomal RNA from Escherichia coli: identification of kethoxal-reactive sites in the A and B conformations. J Mol Biol. 1979 Aug 25;132(4):621–636. doi: 10.1016/0022-2836(79)90378-4. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Garrett R. A. Structure of 5 S ribosomal RNA from Escherichia coli: identification of kethoxal-reactive sites in the A and B conformations. J Mol Biol. 1979 Aug 25;132(4):621–636. doi: 10.1016/0022-2836(79)90378-4. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Herr W. Letters to the editor: Accessibility of 5 S RNA in 50 S ribosomal subunits. J Mol Biol. 1974 Nov 25;90(1):181–184. doi: 10.1016/0022-2836(74)90266-6. [DOI] [PubMed] [Google Scholar]
- Osterberg R., Sjöberg B., Garrett R. A. Molecular model for 5-S RNA. A small-angle x-ray scattering study of native, denatured and aggregated 5-S RNA from Escherichia coli ribosomes. Eur J Biochem. 1976 Sep 15;68(2):481–487. doi: 10.1111/j.1432-1033.1976.tb10835.x. [DOI] [PubMed] [Google Scholar]
- Pipas J. M., McMahon J. E. Method for predicting RNA secondary structure. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2017–2021. doi: 10.1073/pnas.72.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards E. G., Lecanidou R., Geroch M. E. The kinetics of renaturation of 5-S RNA from Escherichia coli in the presence of Mg 2+ ions. Eur J Biochem. 1973 Apr;34(2):262–267. doi: 10.1111/j.1432-1033.1973.tb02755.x. [DOI] [PubMed] [Google Scholar]
- Richter D., Erdmann V. A., Sprinzl M. Specific recognition of GTpsiC loop (loop IV) of tRNA by 50S ribosomal subunits from E. coli. Nat New Biol. 1973 Dec 5;246(153):132–135. doi: 10.1038/newbio246132a0. [DOI] [PubMed] [Google Scholar]
- Ross A., Brimacombe R. Experimental determination of interacting sequences in ribosomal RNA. Nature. 1979 Sep 27;281(5729):271–276. doi: 10.1038/281271a0. [DOI] [PubMed] [Google Scholar]
- Schwarz U., Menzel H. M., Gassen H. G. Codon-dependent rearrangement of the three-dimensional structure of phenylalanine tRNA, exposing the T-psi-C-G sequence for binding to the 50S ribosomal subunit. Biochemistry. 1976 Jun 1;15(11):2484–2490. doi: 10.1021/bi00656a035. [DOI] [PubMed] [Google Scholar]
- Scott J. F., Monier R., Aubert M., Reynier M. Some optical properties of 5S-RNA from E. coli. Biochem Biophys Res Commun. 1968 Dec 9;33(5):794–800. doi: 10.1016/0006-291x(68)90230-1. [DOI] [PubMed] [Google Scholar]
- Simoncsits A. 3' Terminal labelling of RNA of RNA with beta-32P-pyrophosphate group and its application to the sequence analysis of 5S RNA from Streptomyces griseus. Nucleic Acids Res. 1980 Sep 25;8(18):4111–4124. doi: 10.1093/nar/8.18.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Wagner T., Lorenz S., Erdmann V. A. Regions of tRNA important for binding to the ribosomal A and P sites. Biochemistry. 1976 Jul 13;15(14):3031–3039. doi: 10.1021/bi00659a015. [DOI] [PubMed] [Google Scholar]
- Studnicka G. M., Rahn G. M., Cummings I. W., Salser W. A. Computer method for predicting the secondary structure of single-stranded RNA. Nucleic Acids Res. 1978 Sep;5(9):3365–3387. doi: 10.1093/nar/5.9.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigne R., Jordan B. R., Monier R. A common conformational feature in several prokaryotic and eukaryotic 5 S RNAs: a highly exposed, single-stranded loop around position 40. J Mol Biol. 1973 May 15;76(2):303–311. doi: 10.1016/0022-2836(73)90393-8. [DOI] [PubMed] [Google Scholar]
- Vigne R., Jordan B. R. Partial enzyme digestion studies on Escherichia coli, Pseudomonas, Chlorella, Drosophila, HeLa and yeast 5S RNAs support a general class of 5S RNA models. J Mol Evol. 1977 Sep 20;10(1):77–86. doi: 10.1007/BF01796136. [DOI] [PubMed] [Google Scholar]
- Wrede P., Erdmann V. A. Activities of B. stearothermophilus 50 S ribosomes reconstituted with prokaryotic and eukaryotic 5 S RNA. FEBS Lett. 1973 Jul 15;33(3):315–319. doi: 10.1016/0014-5793(73)80219-4. [DOI] [PubMed] [Google Scholar]
- Wrede P., Erdmann V. A. Escherichia coli 5S RNA binding proteins L18 and L25 interact with 5.8S RNA but not with 5S RNA from yeast ribosomes. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2706–2709. doi: 10.1073/pnas.74.7.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrede P., Pongs O., Erdmann V. A. Binding oligonucleotides to Escherichia coli and Bacillus stearothermophilus 5 S RNA. J Mol Biol. 1978 Mar 25;120(1):83–96. doi: 10.1016/0022-2836(78)90296-6. [DOI] [PubMed] [Google Scholar]


