Abstract
Tuberculosis (TB) is a major public health problem. One-third of the world's population is estimated to be infected with Mycobacterium tuberculosis (MTB), the etiological agent causing TB, and active disease kills nearly 2 million individuals worldwide every year. Several lines of evidence indicate that interindividual variation in susceptibility to TB has a heritable component, yet we still know little about the underlying genetic architecture. To address this, we performed a genome-wide mapping study of loci that are associated with functional variation in immune response to MTB. Specifically, we characterized transcript and protein expression levels and mapped expression quantitative trait loci (eQTL) in primary dendritic cells (DCs) from 65 individuals, before and after infection with MTB. We found 198 response eQTL, namely loci that were associated with variation in gene expression levels in either untreated or MTB-infected DCs, but not both. These response eQTL are associated with natural regulatory variation that likely affects (directly or indirectly) host interaction with MTB. Indeed, when we integrated our data with results from a genome-wide association study (GWAS) for pulmonary TB, we found that the response eQTL were more likely to be genetically associated with the disease. We thus identified a number of candidate loci, including the MAPK phosphatase DUSP14 in particular, that are promising susceptibility genes to pulmonary TB.
Tuberculosis (TB) is among the oldest diseases recorded to affect humans. Skeletal remains show that prehistoric humans (7000 BC) had already suffered from TB (1) and tubercular decay has been found in the spines of mummies from 3000 to 2400 BC (2). Today, despite considerable efforts to fight the disease, TB remains a major public health problem, with 1.7 million deaths occurring annually worldwide and up to one-third of the global population estimated to be carrying latent Mycobacterium tuberculosis (MTB) infection (3). A striking feature of TB is that only 10% of individuals infected with MTB develop the disease (4, 5). Although a significant proportion of interindividual variation in susceptibility to TB can be attributed to environmental factors such as malnutrition or poor hygienic conditions, a substantial portion is thought to be due to host genetic factors (6–8). The strongest evidence for this probably comes from twin studies showing that the rate of TB in monozygotic twins is more than twice that observed among dizygotic twins (8). In addition, studies on Mendelian susceptibility to mycobacterial disease (MSMD) have identified multiple rare single-gene mutations linked with susceptibility to mycobacteria (7, 9, 10). Although studies on MSMD have played a key role in identifying important pathways involved in protective immunity against TB, such as the IL12/23- and IFN-γ pathways (7, 9–14), the mutations identified are too rare to have a significant impact on the overall variation in susceptibility to TB in the wider population.
The quest of the genetic determinants of susceptibility to TB at the population level has so far been primarily driven by case control studies of candidate genes. Studies based on such approaches identified over 20 genes associated with susceptibility to TB (reviewed in refs. 6, 7). However, limited samples sizes and difficulties in validating the majority of these findings across populations raised doubts about many of these associations (15). Recently, Thye et al. (16), reported the first genome-wide association study (GWAS) for host susceptibility to pulmonary TB (using 2,237 cases and 3,122 controls from West Africa). Despite the relatively large sample size, only a single locus in a gene desert region was associated with a significant genome-wide modest effect size (odds ratio = 1.2). Thus, to date, little is known about the underlying genetic determinants or mechanisms contributing to differences in susceptibility to TB at the population level.
In the context of an infectious disease such as TB, the most important molecular networks affecting disease susceptibility are probably those involved in mechanisms of immune defense. We thus reasoned that genetic variants that are associated with variation in immune response to MTB infection would be highly promising genetic candidates for susceptibility to TB. To identify such variants, we characterized genome-wide gene expression levels and mapped expression quantitative trait loci (eQTL) in untreated and MTB-infected monocyte-derived dendritic cells (DCs), from a panel of 65 healthy individuals of European descent. We chose to work with DCs because they play a central role in bridging innate and adaptive immunity. Upon MTB infection, DCs migrate from the lungs to the draining lymph nodes where they present antigens to naive T cells, orchestrating antimycobacterial immunity and ultimately determining disease outcome (17). We thus provide a comprehensive view of regulatory mechanisms underlying variation in immune response to MTB infection.
Results
Transcriptional Response of DCs to MTB Infection.
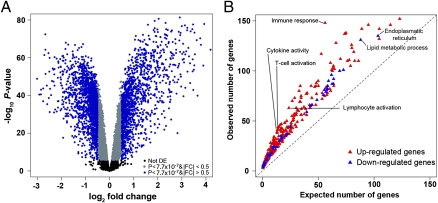
We infected DCs from 65 healthy individuals with a virulent strain of MTB. Following infection, we extracted RNA from the untreated and infected DCs at the same time, 18 h after the infection. We then characterized genome-wide gene expression profiles in all samples using the Illumina HT-12 expression arrays. After excluding data from poorly annotated array probes and from genes that were classified as not expressed, we normalized the expression data for the remaining 12,958 genes (details in Materials and Methods). We then analyzed the gene expression data using a linear model with a fixed effect for the treatment (infection with MTB; Materials and Methods). Using a moderated t test (18), we classified 2,948 and 4,055 genes as up- or down-regulated postinfection, respectively (P < 10−6, false discovery rate (FDR) < 10−4). The difference in expression level between the untreated and infected DCs was greater than 0.5-fold for 3,040 (43%) of these genes (Fig. 1A and Dataset S1).
Fig. 1.
Functional characterization of immune responses to MTB infection. (A) Volcano plot showing differentially expressed genes after infection of DCs with MTB for 18 h. The negative log10 transformed P values test the null hypothesis of no difference in expression levels between untreated and infected DCs (y axis) and are plotted against the average log2 fold changes in expression (x axis). Data for genes that were not classified as differentially expressed are plotted in black. In gray and blue, we plotted data for genes that are differentially expressed after infection with MTB (P value <7.7 × 10−7; Bonferroni corrected P value <0.01) with an absolute log2 fold change (|FC|) less than or equal to 0.5 or greater than 0.5, respectively. (B) Gene ontology (GO) enrichment analysis for genes that were classified as up- (red) or down- (blue) regulated following infection of DCs with MTB. Only significant enrichments at an FDR <1% are plotted (complete results in Dataset S2).
Changes in the transcriptome following MTB infection are expected to reflect the transition of DCs from antigen-capturing cells to potent antigen-presenting cells and T-cell activators (19, 20). Consistent with this notion, we found that the maturation of DCs after MTB infection was accompanied by the strong up-regulation of genes involved in immune responses (FDR < 10−26), including cytokine signaling, T-cell activation, and antigen presentation (FDR < 0.006; Fig. 1B and Dataset S2). Conversely, down-regulated genes were significantly enriched for genes involved in metabolic pathways (FDR < 10−6) and in the reorganization of the cytoplasmic membrane (FDR < 10−6, Fig. 1B and Dataset S2). This observation probably reflects a reduced capacity of mature DCs to endocytose/phagocytose and the expected reduction in trafficking between the mycobacterial phagosome and the host cell recycling and biosynthetic pathways (20, 21).
Identification of Immune Response eQTL.
We next aimed to identify eQTL in both infected and noninfected DCs. To do so, we genotyped each individual at 970,287 single nucleotide polymorphisms (SNPs) using the Illumina Omni1-Quad BeadChip. Genotypes for 873,973 SNPs passed our quality checks (Materials and Methods). We used a linear regression model to test for an association between the expression levels of 11,996 autosomal genes and genotype at all SNPs with a minor allele frequency greater than 10% (SI Materials and Methods includes specific details on data processing and modeling). We mapped eQTL separately in infected and noninfected DCs. In agreement with previous studies (e.g., refs. 22, 23), we found that most SNPs strongly associated with gene expression levels lie near the corresponding gene (putatively acting in cis; Fig. S1). Thus, we focused our analysis on putative cis-regulatory variants defined as SNPs located in a 200-kb window centered on a gene's transcription starting site (TSS). At an FDR of 1% (at least one SNP with P < 1.4 × 10−5; more details in Materials and Methods) we found 720 and 756 genes with cis-eQTL in the infected and noninfected DCs, respectively (Fig. 2A and Dataset S3). Interestingly, genes whose expression levels were altered following MTB infection were 1.6 times more likely to be associated with cis-eQTL than the genome-wide average (14% compared with the 9% that are expected by chance alone; P = 1.6 × 10−16; Fig. 2B). Our results thus suggest that genes involved in immune response to MTB have increased levels of functional diversity in the population. Such functional variation is likely to underlie interindividual differences in susceptibility to infectious diseases in general and MTB infection in particular.
Fig. 2.
Deciphering the genetic basis of interindividual variation in immune response to MTB infection. (A) Plot contrasting the evidence for cis-eQTL in the untreated and infected DCs. For every gene we plotted the additive model P values (−log10 transformed) for the most strongly associated cis-SNP (defined as SNPs located in 200-kb window centered on the TSS of a proximal gene) with gene expression levels in the untreated (x axis) or infected (y axis) DCs, respectively. The red dashed lines specify the P values corresponding to an FDR of 1%. The blue dashed lines specify the second, more relaxed, cutoff (∼50% FDR) used to confidently classify response eQTL. Only genes with strong evidence of a cis-eQTL in at least one of the conditions (FDR of 1%) are plotted. (B) Proportion of cis-eQTL (y axis) observed among all tested genes and among genes that were classified as differentially expressed (DEG) following infection with MTB. (C) Example of a response eQTL found only in the untreated samples. (D) Example of a response eQTL found only in the infected samples.
As expected, there is a large overlap of eQTL identified independently in the noninfected and infected DCs (Pearson correlation of eQTL association P values in the two classes of DCs is 0.68; P < 10−15). In the context of susceptibility to TB, however, the most interesting eQTL are arguably those with a different effect on gene expression levels before and after infection with MTB, which we term “response eQTL.” Response eQTL likely interact with MTB (directly or indirectly) and thus may account for interindividual variation in immune response to MTB infection. We classified response eQTL by using highly conservative criteria to minimize the probability of false positives (e.g., when true eQTL in both untreated and infected DCs are only classified as such in one class because of incomplete power). Specifically, we defined response eQTL when we found strong evidence for a cis-eQTL for a gene in either untreated or infected DCs at an FDR of 1% (P < 1.4 × 10−5) and no statistical evidence supporting a cis-eQTL for the same gene (in the entire tested 200-kb region) in the other condition at a very relaxed FDR threshold of 50% (more details in Materials and Methods). Using this approach, we identified 198 genes with strong evidence of being associated with at least one response eQTL (102 and 96 eQTL in untreated and infected DCs, respectively; Fig. 2 A, C, and D and Dataset S3).
Mapping Cytokine and Chemokine Secretion Levels.
In addition to characterizing transcriptional profiles following MTB infection, we measured the levels of 19 cytokines in the supernatants of untreated and infected DCs (Fig. 3 and Fig. S2; Materials and Methods for details). With the exception of IL-17, the protein measurements of all cytokines/chemokines showed a significant correlation with estimates of the corresponding transcript expression levels (P < 0.01; median Spearman correlations = 0.6; Fig. 3A). As expected, the expression levels of all tested proteins were significantly elevated after MTB infection (Wilcoxon signed-rank test, FDR < 0.01, Fig. S2), and we observed strong induction of all of the cytokines presently known to play a critical role in protective immunity against TB (24–26) (Fig. 3B), including TNF-α (460-fold induction), IFN-γ (24-fold induction), and IL-12 (8-fold induction).
Fig. 3.
Association between cis-eQTL and protein secretion levels. (A) Boxplot of Spearman correlations between mRNA and protein expression levels. (B) Examples of data from TNF-α, IFN-γ, and IL-12 (from Left to Right). Protein level measurements from untreated (green) and infected (red) DCs from each of the 65 individuals are plotted. Results for the remaining proteins can be found in Fig. S2. P values were obtained using a nonparametric Wilcoxon test, which takes into account the paired nature of our data. (C) Manhattan plot showing the negative log10 transformed P values (y axis) for the association between all SNPs classified as cis-eQTL and the secretion levels of IL-1Ra measured in the supernatant of infected DCs. (D) Correlation between genotypes at rs11960575 and the relative secretion levels of IL-1Ra. In addition to the association between rs11960575 and IL-1Ra secretion levels, we also found a significant association (Bonferroni P < 1.7 × 10−6) between rs854100 and the secretion levels of IL-15, although the secretion levels of IL-15 after infection were very low (Figs. S2 and S3).
Using the protein secretion data we also looked for protein QTL (pQTL), namely loci that are associated with cytokines/chemokines’ secretion levels measured after infection of the DCs. We did not find any significant association, either genome-wide or when we restricted the analysis to SNPs within 200 kb of the TSSs of the genes encoding the measured proteins. However, when we only considered the SNPs that were previously identified as cis-eQTL (in the analysis of transcript levels above) we found a clear enrichment in the association of genotypes with protein levels (Fig. S3). This observation is tentative and requires further validation and replication, but it suggests that the secretion of a subset of these cytokines is regulated in trans by proteins whose expression levels are, in turn, regulated in cis by the eQTL we found. In particular, in two cases (of the 19 tested), the association was significant even after the conservative Bonferroni correction for multiple testing. Specifically, rs854100 and rs11960575 are associated with the secretion levels of IL15 and IL-1Ra, respectively (Bonferroni P < 1.7 × 10−6; Fig. 3 C and D and Fig. S3). The genetic variation at rs11960575 may be of particular interest: This locus was classified as a cis-eQTL for the gene fibrillin 2 (FBN2; Pnoninfected DCs = 8.3 × 10−9; PMTB-infected DCs = 2 × 10−3), and a trans-pQTL for IL-1Ra (Bonferroni P < 1.7 × 10−6; Fig. 3 C and D). This observation may reflect a regulatory interaction between FBN2 and IL-1Ra. Consistent with this inference, the members of the fibrillin gene family are known to be involved in the storage and activation of TGF-β (27, 28), which in turn has been shown to regulate the production of IL-1Ra (29, 30). Because genetic variation in IL1Ra has previously been associated with susceptibility to TB (31, 32), rs11960575 (or another SNP in linkage disequilibrium with it) represents a strong candidate locus for impacting susceptibility to TB.
Immune Response eQTL Are Enriched for Susceptibility Genes for TB.
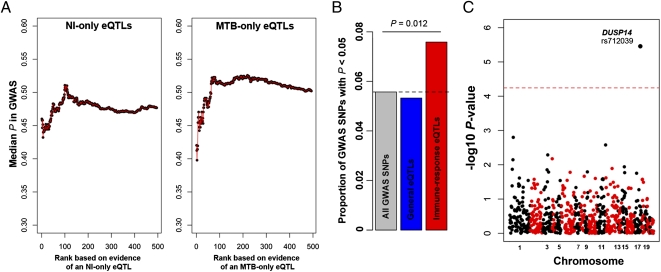
The immune response eQTL as a class are strong candidates for affecting susceptibility to TB. To test this possibility, we integrated our data with results from the only GWAS for host susceptibility to pulmonary TB published to date, which included 2,237 cases and 3,122 controls from Ghana and Gambia (16). In the original GWAS analysis, only a single locus, in a gene desert region on chromosome 18q11, reached genome-wide significance (16). When we focused on the response eQTL identified in this study, however, we found that these loci were more likely to be associated with TB than expected by chance (Fig. 4). Specifically, we observed more than 1.4-fold enrichment of SNPs with a nominal GWAS P value <0.05 among response eQTL, compared with genome-wide expectations (Fig. 4B, χ2 test, P = 0.012). Importantly, we did not observe a similar enrichment when we considered eQTL that are common to untreated and infected DCs (Fig. 4B; this observation serves as a control for a possible bias in the power to detect an association in the GWAS when only eQTL are considered).
Fig. 4.
MTB-response eQTL are strong candidates to impact susceptibility to pulmonary TB. (A) The median GWAS P value for an expanding window of genes is plotted. We used the GWAS P values obtained when combining the Ghana and Gambia cohorts (16). Genes are ordered by the strength of evidence supporting an association with an eQTL only in the untreated (Left) or the infected (Right) DCs, respectively. To avoid positional biases, we restricted our analyses to the set of cis-SNPs that was tested in our study (i.e., SNPs located in 200-kb window centered on the TSS of proximal genes). (B) Histogram of the proportion of GWAS SNPs with nominal P values <0.05 among all GWAS SNPs (gray), among SNPs that were classified as eQTL in both untreated and infected DCs (blue), and among response eQTL (red). (C) Manhattan plot showing the negative log10 transformed P values (y axis) for the association between the response eQTL identified in this study and susceptibility to pulmonary TB. The dashed line corresponds to the genome-wide significance cutoff after a conservative Bonferroni correction.
We note that our observations are robust to the particular method used to identify response eQTL. Indeed, an alternative approach used to identify cis-interactions (i.e., response-eQTL) is to consider variation in the magnitude of the shifts in gene expression after a treatment, in our case MTB infection, as the quantitative trait to be mapped (33, 34). Supporting our previous findings, MTB-response eQTL identified using this approach (Dataset S3) were also significantly enriched for GWAS P values <0.05 (1.8-fold enrichment, χ2 test, P = 0.01; Fig. S4).
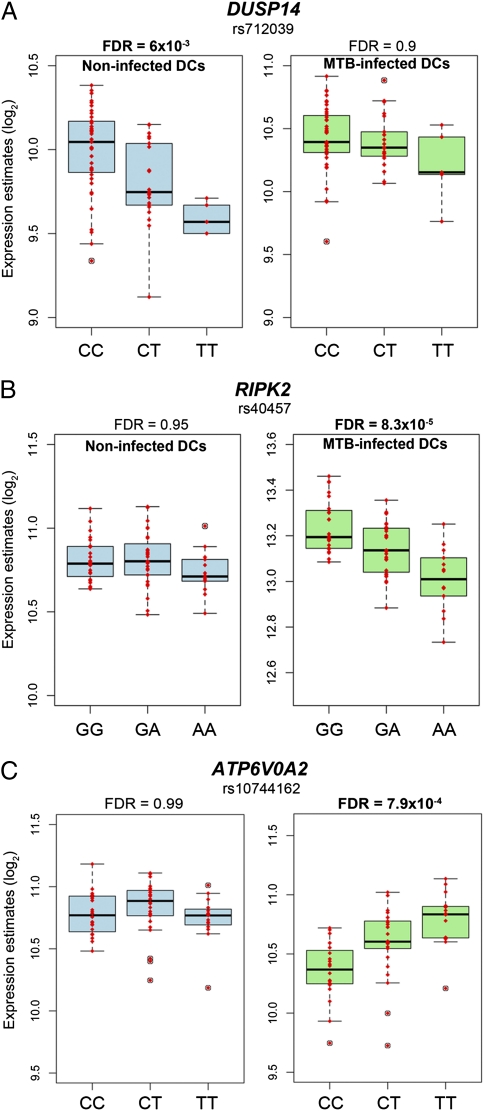
Taken together, our results indicate that response eQTL are likely enriched for susceptibility alleles for TB. In particular, our data strongly support the notion that dual-specificity phosphatase 14 (DUSP14) is a new susceptibility gene for TB. Indeed, we found strong evidence that rs712039 is an immune response eQTL associated with variation in the expression levels of DUSP14 exclusively in noninfected DCs (Pnoninfected DCs = 9.6 × 10−6 (FDR = 6 × 10−3); PMTB-infected DCs = 0.09 (FDR = 0.9); Fig. 5A). In addition, in our focused GWAS analysis, which was restricted to the set of SNPs we classified as immune response eQTL (as the set of most likely candidates), the strength of the association between rs712039 and pulmonary TB is significant even after a conservative Bonferroni correction (Fig. 4D). This SNP was not discussed in the original GWAS paper, probably because it was not significant at a genome-wide threshold, yet it shows one of the strongest genetic associations with pulmonary TB, in both the Ghana cohort (P = 9.8 × 10−4) and the Gambia replication cohort (P = 2.3 × 10−3, combined P = 3.3 × 10−6; Table S1).
Fig. 5.
Genes with response eQTL likely to impact susceptibility to pulmonary TB. (A) Response eQTL identified for DUSP14. (B) Response eQTL identified for RIPK2. (C) Response eQTL identified for ATP6V0A2.
Discussion
We studied variation in the regulatory response to MTB infection of DCs. We chose to focus on DCs because they have been shown to play an essential, nonredundant role in protective immunity to TB (35). DCs exhibit the unique ability to migrate to secondary lymphoid organs where they present mycobacteria-derived antigens to naive T cells (17). In the absence of DCs, the response induced by CD4+ T cells is impaired and bacterial load uncontrolled (36–38). Accordingly, individuals with a deficiency of DCs, monocytes, and B and natural killer (NK) cells (DCML deficiency) show increased susceptibility to poorly virulent strains of Mycobaterium spp (39). Similarly, mutations in the IFN regulatory factor 8, which have recently been shown to be associated with a selective loss of CD11c+ CD1c+ myeloid DCs, result in a remarkable increased susceptibility to disseminated bacillus Calmette–Guérin-osis, arguing for a specific role for DCs in resistance to mycobacteria (40).
It is well established that macrophages (Mφ) also play a central role in TB pathogenesis (25–26). They are one of the primary cell targets of MTB, which have developed different strategies to survive and to multiply inside the Mφ phagosome such as prevention of phagosome acidification (41) and inhibition of phagolysosomal fusion (42). In addition, macrophages play an important role in determining the outcome of infection by orchestrating the formation of granulomas and by killing the bacillus upon IFN-γ activation. Thus, it would be of great interest to also study the transcriptional response and map response eQTL to MTB infection of Mφ. Extending this approach to additional relevant cell types, such as Mφ, should greatly increase our understanding of the genetic architecture of TB response, with the ultimate aim of deciphering the genetic factors controlling human susceptibility to this ancient disease.
Our results indicate that variation in the regulatory immune response to MTB infection of DCs is often associated with genetic factors. Indeed, at least 14% of differentially expressed genes following infection with MTB are associated with an eQTL (Fig. 2B). This observation strongly supports the hypothesis that genetic susceptibility to TB is likely to be accounted for by the combination of many genetic variants with small effects. Under this model, classical GWAS approaches alone may be insufficient for fully characterizing the multitude of genetic associations with TB susceptibility (a challenge that is shared with a large number of other complex phenotypes, including many other infectious diseases). Our findings indicate that in vitro experimental studies of the eQTL response to infection represent a powerful approach that can help overcome this challenge, as well as point to the likely regulatory mechanisms that affect disease susceptibility.
Our data revealed DUSP14 as a susceptibility gene to pulmonary TB. The DUSP14 gene is a member of a large family of phosphatases that specifically dephosporylate threonine and tyrosine residues on mitogen-activated protein kinases (MAPKs), rendering them inactive (43). These phosphatases therefore ultimately control the levels of proinflammatory cytokines released after infection (43). Accordingly, and further supporting a potential role of DUSP14 in susceptibility to TB, we found that the response eQTL we identified for DUSP14, rs712039, is also associated with the secretion levels of TNF-α (P = 0.02) and IFN-γ (P = 0.03). These proteins are probably the two most important cytokines known to be involved in immune protection against TB. These genes participate in the formation and maintenance of the granulomes (25) and activate the bactericidal activity of phagocytes (26).
Other novel strong candidates to affect susceptibility to pulmonary TB include ATP6V0A2 and RIPK2. Both genes are associated with response eQTL in MTB-infected DCs (Fig. 5 B and C), and the same genetic variants are nominally associated with pulmonary TB in the GWAS data (Table S1). RIPK2 encodes an adapter protein of the NOD2-dependent signaling pathway, which is a key pathway involved in the regulation of host responses to MTB infection (44–46). ATP6V0A2 encodes a subunit of the vesicular proton-ATPase (v-ATPase) (47). Interestingly, one major virulence feature of the tubercle bacillus is its ability to parasitize macrophages through the exclusion of v-ATPase from phagosomes, preventing them from maturing (41, 48). Interindividual variation in the levels of ATP6V0A2 is therefore likely to impact the ability of the bacillus to grow and survive inside phagocytic cells, ultimately impacting susceptibility to TB.
It is important to note that our approach does not allow us to distinguish between response eQTL that are specific to MTB infection and those shared with other infectious diseases. Because we show that the response eQTL we identified are enriched for genetic variants associated with susceptibility to TB, it is reasonable that at least a subset of these eQTL have specific roles in the immune response to MTB infection. We expect that our findings will stimulate future work aimed at characterizing response eQTL associated with other infectious diseases, and that such work will allow us to further classify specific MTB-response eQTL on the one hand and genetic variation that is associated with more general variation in the response to infectious agents on the other hand.
Materials and Methods
Details of the experimental and statistical procedures can be found in SI Materials and Methods. Blood samples from 68 healthy donors were obtained from Research Blood Components. All samples were collected with informed consent under the company's independent ethics committee approval. All individuals recruited in this study were healthy Caucasian males between the ages of 21 and 55 y old. Genotyping of all individuals was performed using the Illumina's Omni1-Quad BeadChip array. After a series of quality checks (SI Materials and Methods), genotyping data from 65 samples were kept for downstream analyses. Blood mononuclear cells from each donor were isolated by Ficoll-Paque centrifugation and blood monocytes were purified from peripheral blood mononuclear cells (PBMCs) by positive selection with magnetic CD14 MicroBeads (Miltenyi Biotec). Monocytes were then derived into DCs as previously described (20) and subsequently infected with MTB for 18 h at a multiplicity of infection of 1-to-1. Genome-wide gene expression profiles of untreated and infected DCs were obtained by hybridizing the RNA to the Illumina HumanHT-12 v4 Expression BeadChips arrays. Cytokine and chemokine levels were measured using the Bio-Plex Pro Human Cytokine 27-plex, according to the manufacturer's recommendations. To identify genes whose expression levels were altered following MTB infection, we used the linear modeling-based approach implemented in the Bioconductor limma package (18). We used GeneTrail (49) to test for enrichment of functional annotations among differentially expressed genes after MTB infection. Association between SNP genotypes and either transcript or protein expression levels were examined by using a linear regression model where each phenotype was regressed against genotype. In all cases, we assumed that SNPs affecting the phenotype did so in an additive manner. We mapped infected and noninfected DCs separately. All regressions were performed using a Python script, whereas downstream analyses were carried out using the R statistical framework.
Supplementary Material
Acknowledgments
We thank M. Stephens for helpful discussions; Z. Gauhar, J. Tung, D. Cusanovich, L. Quintana-Murci, B. Jabri, J. Maranville, and F. Luca for comments on the manuscript; G. Stewart for a gift of the MTB strain used in this study; and T. Thye for sharing the GWAS data with us. This study was funded by National Institutes of Health Grant AI087658 (to Y.G. and L.T.). L.B.B. was supported by an Human Frontier Science Program postdoctoral fellowship. A.A.P. was supported by an American Heart Association fellowship.
Footnotes
The authors declare no conflict of interest.
Data deposition: The gene expression and genotype data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database (accession nos. GSE34151 and GSE34588, respectively).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115761109/-/DCSupplemental.
References
- 1.Hershkovitz I, et al. Detection and molecular characterization of 9,000-year-old Mycobacterium tuberculosis from a Neolithic settlement in the Eastern Mediterranean. PLoS ONE. 2008;3:e3426. doi: 10.1371/journal.pone.0003426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zink AR, et al. Characterization of Mycobacterium tuberculosis complex DNAs from Egyptian mummies by spoligotyping. J Clin Microbiol. 2003;41:359–367. doi: 10.1128/JCM.41.1.359-367.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization . WHO Report 2009: Global Tuberculosis Control: Epidemiology, Strategy, and Financing. Geneva: WHO; 2009. [Google Scholar]
- 4.North RJ, Jung YJ. Immunity to tuberculosis. Annu Rev Immunol. 2004;22:599–623. doi: 10.1146/annurev.immunol.22.012703.104635. [DOI] [PubMed] [Google Scholar]
- 5.Young DB, Gideon HP, Wilkinson RJ. Eliminating latent tuberculosis. Trends Microbiol. 2009;17:183–188. doi: 10.1016/j.tim.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Berrington WR, Hawn TR. Mycobacterium tuberculosis, macrophages, and the innate immune response: Does common variation matter? Immunol Rev. 2007;219:167–186. doi: 10.1111/j.1600-065X.2007.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casanova JL, Abel L. Genetic dissection of immunity to mycobacteria: The human model. Annu Rev Immunol. 2002;20:581–620. doi: 10.1146/annurev.immunol.20.081501.125851. [DOI] [PubMed] [Google Scholar]
- 8.Comstock GW. Tuberculosis in twins: A re-analysis of the Prophit survey. Am Rev Respir Dis. 1978;117:621–624. doi: 10.1164/arrd.1978.117.4.621. [DOI] [PubMed] [Google Scholar]
- 9.Casanova JL, Abel L. Primary immunodeficiencies: A field in its infancy. Science. 2007;317:617–619. doi: 10.1126/science.1142963. [DOI] [PubMed] [Google Scholar]
- 10.Dorman SE, et al. Clinical features of dominant and recessive interferon gamma receptor 1 deficiencies. Lancet. 2004;364:2113–2121. doi: 10.1016/S0140-6736(04)17552-1. [DOI] [PubMed] [Google Scholar]
- 11.Altare F, et al. Impairment of mycobacterial immunity in human interleukin-12 receptor deficiency. Science. 1998;280:1432–1435. doi: 10.1126/science.280.5368.1432. [DOI] [PubMed] [Google Scholar]
- 12.Altare F, et al. Inherited interleukin 12 deficiency in a child with bacille Calmette-Guérin and Salmonella enteritidis disseminated infection. J Clin Invest. 1998;102:2035–2040. doi: 10.1172/JCI4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jouanguy E, et al. Interferon-gamma-receptor deficiency in an infant with fatal bacille Calmette-Guérin infection. N Engl J Med. 1996;335:1956–1961. doi: 10.1056/NEJM199612263352604. [DOI] [PubMed] [Google Scholar]
- 14.Newport MJ, et al. A mutation in the interferon-gamma-receptor gene and susceptibility to mycobacterial infection. N Engl J Med. 1996;335:1941–1949. doi: 10.1056/NEJM199612263352602. [DOI] [PubMed] [Google Scholar]
- 15.de Bakker PI, Telenti A. Infectious diseases not immune to genome-wide association. Nat Genet. 2010;42:731–732. doi: 10.1038/ng0910-731. [DOI] [PubMed] [Google Scholar]
- 16.Thye T, et al. African TB Genetics Consortium. Wellcome Trust Case Control Consortium Genome-wide association analyses identifies a susceptibility locus for tuberculosis on chromosome 18q11.2. Nat Genet. 2010;42:739–741. doi: 10.1038/ng.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper AM. Cell-mediated immune responses in tuberculosis. Annu Rev Immunol. 2009;27:393–422. doi: 10.1146/annurev.immunol.021908.132703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 19.Huang Q, et al. The plasticity of dendritic cell responses to pathogens and their components. Science. 2001;294:870–875. doi: 10.1126/science.294.5543.870. [DOI] [PubMed] [Google Scholar]
- 20.Tailleux L, et al. Constrained intracellular survival of Mycobacterium tuberculosis in human dendritic cells. J Immunol. 2003;170:1939–1948. doi: 10.4049/jimmunol.170.4.1939. [DOI] [PubMed] [Google Scholar]
- 21.Banchereau J, et al. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- 22.Pickrell JK, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stranger BE, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berry MP, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flynn JL, Chan J. Immunology of tuberculosis. Annu Rev Immunol. 2001;19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- 26.Korbel DS, Schneider BE, Schaible UE. Innate immunity in tuberculosis: Myths and truth. Microbes Infect. 2008;10:995–1004. doi: 10.1016/j.micinf.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 27.Brinckmann J, et al. Enhanced fibrillin-2 expression is a general feature of wound healing and sclerosis: Potential alteration of cell attachment and storage of TGF-beta. Lab Invest. 2010;90:739–752. doi: 10.1038/labinvest.2010.49. [DOI] [PubMed] [Google Scholar]
- 28.Ramirez F, Rifkin DB. Extracellular microfibrils: Contextual platforms for TGFbeta and BMP signaling. Curr Opin Cell Biol. 2009;21:616–622. doi: 10.1016/j.ceb.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner M, et al. Induction of the interleukin 1 receptor antagonist protein by transforming growth factor-beta. Eur J Immunol. 1991;21:1635–1639. doi: 10.1002/eji.1830210708. [DOI] [PubMed] [Google Scholar]
- 30.Muzio M, Sironi M, Polentarutti N, Mantovani A, Colotta F. Induction by transforming growth factor-beta 1 of the interleukin-1 receptor antagonist and of its intracellular form in human polymorphonuclear cells. Eur J Immunol. 1994;24:3194–3198. doi: 10.1002/eji.1830241242. [DOI] [PubMed] [Google Scholar]
- 31.Bellamy R, et al. Assessment of the interleukin 1 gene cluster and other candidate gene polymorphisms in host susceptibility to tuberculosis. Tuber Lung Dis. 1998;79:83–89. doi: 10.1054/tuld.1998.0009. [DOI] [PubMed] [Google Scholar]
- 32.Wilkinson RJ, et al. Influence of polymorphism in the genes for the interleukin (IL)-1 receptor antagonist and IL-1beta on tuberculosis. J Exp Med. 1999;189:1863–1874. doi: 10.1084/jem.189.12.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maranville JC, et al. Interactions between glucocorticoid treatment and cis-regulatory polymorphisms contribute to cellular response phenotypes. PLoS Genet. 2011;7:e1002162. doi: 10.1371/journal.pgen.1002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smirnov DA, Morley M, Shin E, Spielman RS, Cheung VG. Genetic analysis of radiation-induced changes in human gene expression. Nature. 2009;459:587–591. doi: 10.1038/nature07940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schreiber HA, Sandor M. The role of dendritic cells in mycobacterium-induced granulomas. Immunol Lett. 2010;130:26–31. doi: 10.1016/j.imlet.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiao X, et al. Dendritic cells are host cells for mycobacteria in vivo that trigger innate and acquired immunity. J Immunol. 2002;168:1294–1301. doi: 10.4049/jimmunol.168.3.1294. [DOI] [PubMed] [Google Scholar]
- 37.Tian T, Woodworth J, Sköld M, Behar SM. In vivo depletion of CD11c+ cells delays the CD4+ T cell response to Mycobacterium tuberculosis and exacerbates the outcome of infection. J Immunol. 2005;175:3268–3272. doi: 10.4049/jimmunol.175.5.3268. [DOI] [PubMed] [Google Scholar]
- 38.Wolf AJ, et al. Mycobacterium tuberculosis infects dendritic cells with high frequency and impairs their function in vivo. J Immunol. 2007;179:2509–2519. doi: 10.4049/jimmunol.179.4.2509. [DOI] [PubMed] [Google Scholar]
- 39.Bigley V, et al. The human syndrome of dendritic cell, monocyte, B and NK lymphoid deficiency. J Exp Med. 2011;208:227–234. doi: 10.1084/jem.20101459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hambleton S, et al. IRF8 mutations and human dendritic-cell immunodeficiency. N Engl J Med. 2011;365:127–138. doi: 10.1056/NEJMoa1100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sturgill-Koszycki S, et al. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science. 1994;263:678–681. doi: 10.1126/science.8303277. [DOI] [PubMed] [Google Scholar]
- 42.Armstrong JA, Hart PD. Phagosome-lysosome interactions in cultured macrophages infected with virulent tubercle bacilli. Reversal of the usual nonfusion pattern and observations on bacterial survival. J Exp Med. 1975;142:1–16. doi: 10.1084/jem.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salojin K, Oravecz T. Regulation of innate immunity by MAPK dual-specificity phosphatases: Knockout models reveal new tricks of old genes. J Leukoc Biol. 2007;81:860–869. doi: 10.1189/jlb.1006639. [DOI] [PubMed] [Google Scholar]
- 44.Brooks MN, et al. NOD2 controls the nature of the inflammatory response and subsequent fate of Mycobacterium tuberculosis and M. bovis BCG in human macrophages. Cell Microbiol. 2011;13:402–418. doi: 10.1111/j.1462-5822.2010.01544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Divangahi M, et al. NOD2-deficient mice have impaired resistance to Mycobacterium tuberculosis infection through defective innate and adaptive immunity. J Immunol. 2008;181:7157–7165. doi: 10.4049/jimmunol.181.10.7157. [DOI] [PubMed] [Google Scholar]
- 46.Pandey AK, et al. NOD2, RIP2 and IRF5 play a critical role in the type I interferon response to Mycobacterium tuberculosis. PLoS Pathog. 2009;5:e1000500. doi: 10.1371/journal.ppat.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith AN, et al. Revised nomenclature for mammalian vacuolar-type H+ -ATPase subunit genes. Mol Cell. 2003;12:801–803. doi: 10.1016/s1097-2765(03)00397-6. [DOI] [PubMed] [Google Scholar]
- 48.Xu S, et al. Intracellular trafficking in Mycobacterium tuberculosis and Mycobacterium avium-infected macrophages. J Immunol. 1994;153:2568–2578. [PubMed] [Google Scholar]
- 49.Backes C, et al. GeneTrail—advanced gene set enrichment analysis. Nucleic Acids Res. 2007;35:W186–192. doi: 10.1093/nar/gkm323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.