Abstract
Somatic and oocyte 5S rRNAs from the liver and unfertilized eggs of the loach (Misgurnus fossilis have been sequenced and found to differ in six nucleotides. All the substitutions are confined to the 5'-half of the molecules; 4 of them are pyrimidine-pyrimidine substitutions, and 2 are purine-pyrimidine ones. Considerable differences, both in the position and the character of substitutions, have been established when these 5S rRNAs were compared with somatic and oocyte 5S rRNAs from Xenopus borealis and Xenopus laevis. Among the known primary structures, somatic 5S rRNA of M. fossilis is most similar to trout 5S rRNA.
Full text
PDF




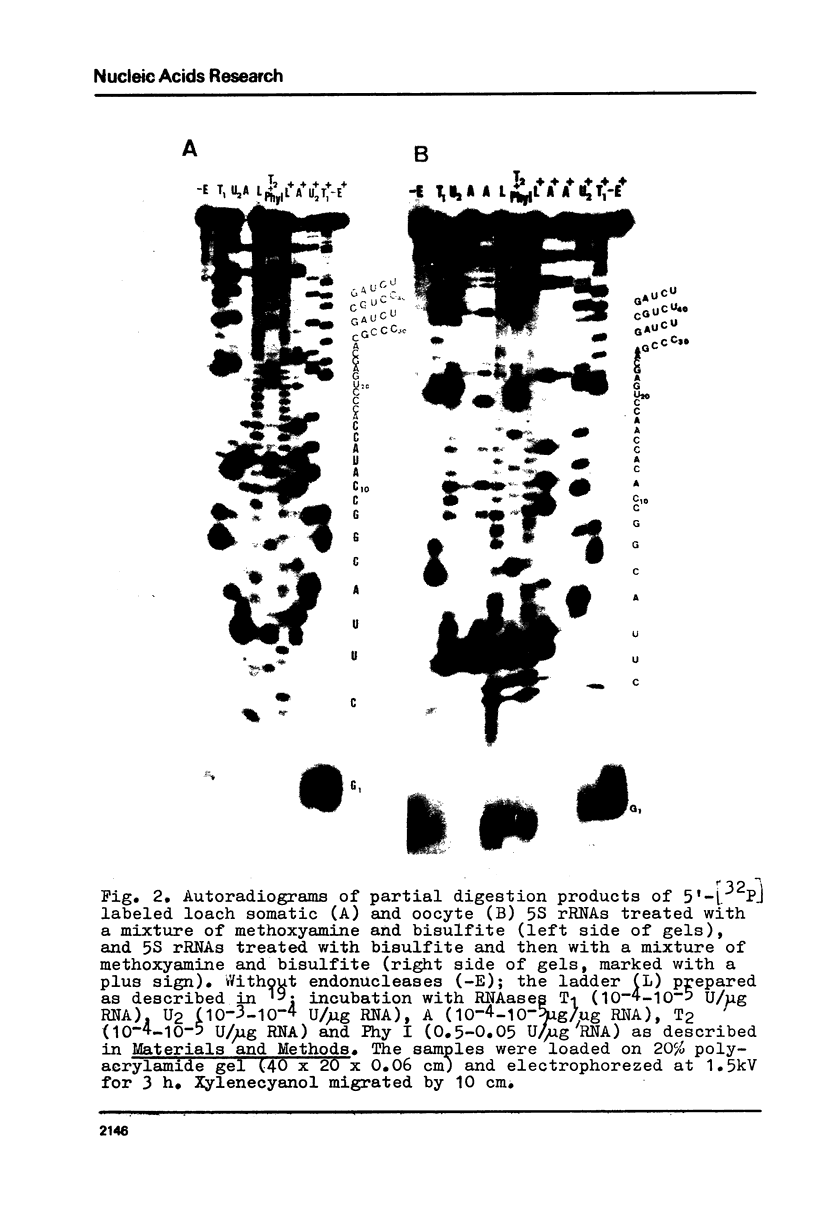

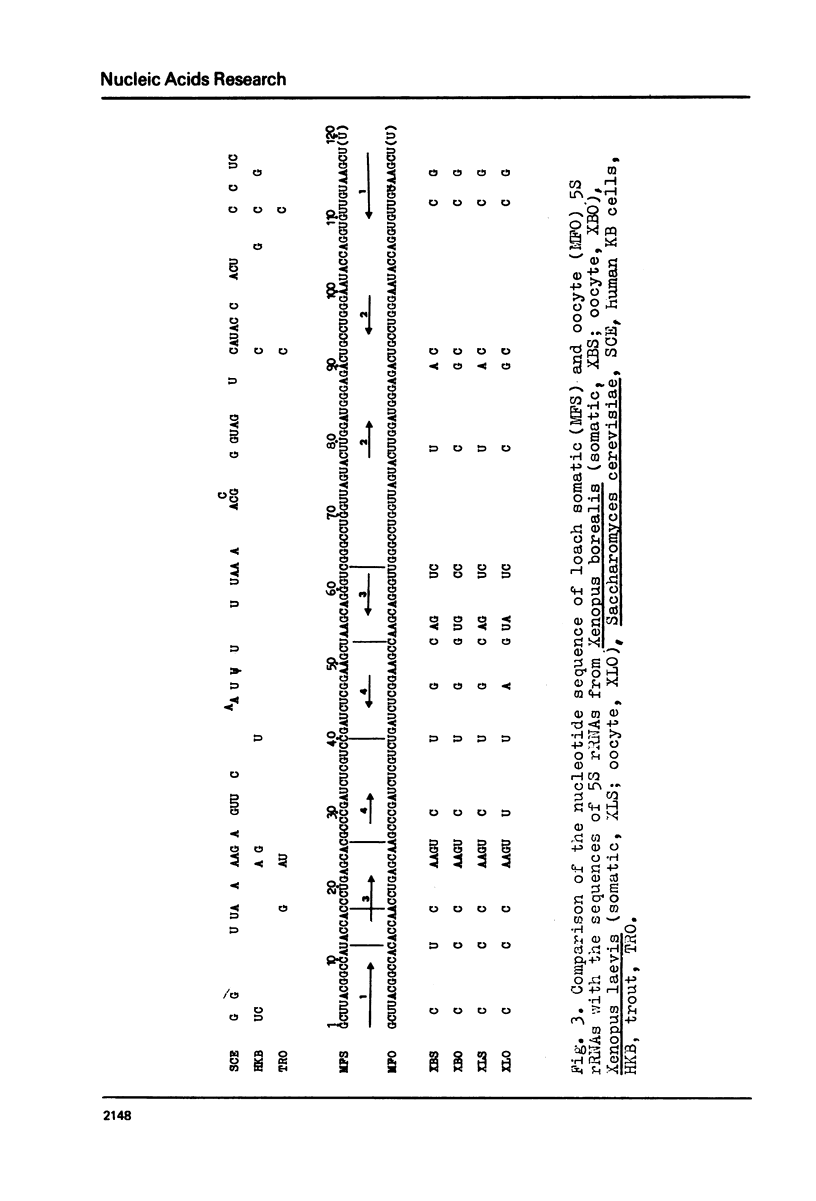



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aloni Y., Hatlen L. E., Attardi G. Studies of fractionated HeLa cell metaphase chromosomes. II. chromosomal distribution of sites for transfer RNA and 5 s RNA. J Mol Biol. 1971 Mar 28;56(3):555–563. doi: 10.1016/0022-2836(71)90401-3. [DOI] [PubMed] [Google Scholar]
- Brown D. D., Carrol D., Brown R. D. The isolation and characterization of a second oocyte 5s DNA from Xenopus laevis. Cell. 1977 Dec;12(4):1045–1056. doi: 10.1016/0092-8674(77)90168-4. [DOI] [PubMed] [Google Scholar]
- Brown D. D., Sugimoto K. 5 S DNAs of Xenopus laevis and Xenopus mulleri: evolution of a gene family. J Mol Biol. 1973 Aug 15;78(3):397–415. doi: 10.1016/0022-2836(73)90464-6. [DOI] [PubMed] [Google Scholar]
- Brown D. D., Wensink P. C., Jordan E. Purification and some characteristics of 5S DNA from Xenopus laevis. Proc Natl Acad Sci U S A. 1971 Dec;68(12):3175–3179. doi: 10.1073/pnas.68.12.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis H., Wegnez M. Biochemical research on oogenesis. Oocytes and liver cells of the teleost fish Tinca tinca contain different kinds of 5S RNA. Dev Biol. 1977 Sep;59(2):228–236. doi: 10.1016/0012-1606(77)90256-1. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H. Phy M: an RNase activity specific for U and A residues useful in RNA sequence analysis. Nucleic Acids Res. 1980 Jul 25;8(14):3133–3142. doi: 10.1093/nar/8.14.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England T. E., Uhlenbeck O. C. Enzymatic oligoribonucleotide synthesis with T4 RNA ligase. Biochemistry. 1978 May 30;17(11):2069–2076. doi: 10.1021/bi00604a008. [DOI] [PubMed] [Google Scholar]
- Fedoroff N. V. On spacers. Cell. 1979 Apr;16(4):697–710. doi: 10.1016/0092-8674(79)90086-2. [DOI] [PubMed] [Google Scholar]
- Ford P. J., Southern E. M. Different sequences for 5S RNA in kidney cells and ovaries of Xenopus laevis. Nat New Biol. 1973 Jan 3;241(105):7–12. doi: 10.1038/newbio241007a0. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Herr W., Noller H. F. A fragment of 23S RNA containing a nucleotide sequence complementary to a region of 5S RNA. FEBS Lett. 1975 May 1;53(2):248–252. doi: 10.1016/0014-5793(75)80030-5. [DOI] [PubMed] [Google Scholar]
- Hori H. Molecular evolution of 5S RNA. Mol Gen Genet. 1976 May 7;145(2):119–123. doi: 10.1007/BF00269583. [DOI] [PubMed] [Google Scholar]
- Hori H., Osawa S. Evolutionary change in 5S RNA secondary structure and a phylogenic tree of 54 5S RNA species. Proc Natl Acad Sci U S A. 1979 Jan;76(1):381–385. doi: 10.1073/pnas.76.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacq C., Miller J. R., Brownlee G. G. A pseudogene structure in 5S DNA of Xenopus laevis. Cell. 1977 Sep;12(1):109–120. doi: 10.1016/0092-8674(77)90189-1. [DOI] [PubMed] [Google Scholar]
- Kimura M., Ohta T. Eukaryotes-prokaryotes divergence estimated by 5S ribosomal RNA sequences. Nat New Biol. 1973 Jun 13;243(128):199–200. doi: 10.1038/newbio243199a0. [DOI] [PubMed] [Google Scholar]
- Kimura M., Ota T. On the stochastic model for estimation of mutational distance between homologous proteins. J Mol Evol. 1972 Dec 29;2(1):87–90. doi: 10.1007/BF01653945. [DOI] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Mashkova T. D., Mazo A. M., Scheinker V. S., Beresten S. F., Bogdanova S. L., Avdonina T. A., Kisselev L. L. A rapid method for mapping exposed cytosines in polyribonucleotides. Application to tRNATrp (yeast, beef liver). Mol Biol Rep. 1980 Jul 31;6(2):83–87. doi: 10.1007/BF00778434. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazo A. M., Mashkova T. D., Avdonina T. A., Ambartsumyan N. S., Kisselev L. L. An improved rapid enzymatic method of RNA sequencing using chemical modification. Nucleic Acids Res. 1979 Dec 20;7(8):2469–2482. doi: 10.1093/nar/7.8.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue M. L., Brown D. D., Birnstiel M. L. Location of the genes for 5S ribosomal RNA in Xenopus laevis. Chromosoma. 1973;42(2):191–203. doi: 10.1007/BF00320940. [DOI] [PubMed] [Google Scholar]
- Richardson C. C. Phosphorylation of nucleic acid by an enzyme from T4 bacteriophage-infected Escherichia coli. Proc Natl Acad Sci U S A. 1965 Jul;54(1):158–165. doi: 10.1073/pnas.54.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoncsits A., Brownlee G. G., Brown R. S., Rubin J. R., Guilley H. New rapid gel sequencing method for RNA. Nature. 1977 Oct 27;269(5631):833–836. doi: 10.1038/269833a0. [DOI] [PubMed] [Google Scholar]
- Singhal R. P. Chemical probe of structure and function of transfer ribonucleic acids. Biochemistry. 1974 Jul 2;13(14):2924–2932. doi: 10.1021/bi00711a023. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Uhlenbeck O. C., Levine M. D. Estimation of secondary structure in ribonucleic acids. Nature. 1971 Apr 9;230(5293):362–367. doi: 10.1038/230362a0. [DOI] [PubMed] [Google Scholar]
- Wegnez M., Denis H., Mazabraud A., Clérot J. C. Biochemical research on oogenesis. RNA accumulation during oogenesis of the dogfish Scyliorhinus caniculus. Dev Biol. 1978 Jan;62(1):99–111. doi: 10.1016/0012-1606(78)90095-7. [DOI] [PubMed] [Google Scholar]
- Wegnez M., Monier R., Denis H. Sequence heterogeneity of 5 S RNA in Xenopus laevis. FEBS Lett. 1972 Sep 1;25(1):13–20. doi: 10.1016/0014-5793(72)80443-5. [DOI] [PubMed] [Google Scholar]




