Abstract
To measure molecular changes underlying pathogen adaptation, we generated a searchable dataset of more than 12,000 mass spectrometry events, corresponding to lipids and small molecules that constitute a lipidome for Mycobacterium tuberculosis. Iron is essential for M. tuberculosis survival, and the organism imports this metal using mycobactin and carboxymycobactin siderophores. Detection of an unexpected siderophore variant and deletions of genes for iron scavenging has led to a revised mycobactin biosynthesis model. An organism-wide search of the M. tuberculosis database for hypothetical compounds predicted by this model led to the discovery of two families of previously unknown lipids, designated monodeoxymycobactins and monodeoxycarboxymycobactins. These molecules suggest a revised biosynthetic model that alters the substrates and order of action of enzymes through the mycobactin biosynthetic pathway. We tested this model genetically by solving M. tuberculosis lipidomes after deletion of the iron-dependent regulator (ideR), mycobactin synthase B (mbtB), or mycobactin synthase G (mbtG). These studies show that deoxymycobactins are actively regulated during iron starvation, and also define essential roles of MbtG in converting deoxymycobactins to mycobactin and in promoting M. tuberculosis growth. Thus, lipidomics is an efficient discovery tool that informs genetic relationships, leading to a revised general model for the biosynthesis of these virulence-conferring siderophores.
Infection with Mycobacterium tuberculosis often involves decades of persistence in the host, during which time the organism sets up supply lines for importing host nutrients. Using two structurally related lipids, mycobactin and carboxymycobactin (also known as exochelin and exomycobactin), M. tuberculosis scavenges host iron for the reactions of DNA synthesis and respiration (1–3). Studies of human patients (4), genetic deletion of mycobactin synthases (5, 6), and the evolution of host siderocalin (7) indicate that the struggle between mycobacteria and hosts for capture of limited iron stores can determine the outcome of natural and experimental tuberculosis infection. Mycobactin and carboxymycobactin serve two functions. First, they are high-affinity, hexadentate iron ligands. Second, they neutralize the Fe3+ cation and tether it to a lipid tail, promoting iron transport across the hydrophobic membranes (8–10). Both lipids contain the same peptide. Mycobactin has a long fatty acid tail (C16–22), whereas carboxymycobactin has a shorter, dicarboxyl fatty acid tail (C6–C10), whose less hydrophobic character facilitates dispersion after secretion into the aqueous phagosome.
Mycobactin siderophores play an essential role in scavenging nonheme iron, as shown by growth attenuation after deletion of mycobactin synthases (5, 6). Whereas mycobactin biosynthesis is a known virulence-conferring pathway, the precise mechanisms by which mycobactins are produced and cycled through the cell wall are incompletely understood. Seminal work showed that mycobactin synthases, named MbtA-N, are encoded at two loci (6, 9, 10). IdeR controls siderophore biosynthesis by repressing transcription of mbtA-N in iron-replete conditions (9–11). The predicted functions of MbtA-N fit well into widely accepted theoretical models for the production of mycobactin, but the functions of most genes and intermediates in mycobactin biosynthesis have not been directly established (5, 6, 9, 10). Here we describe the development and application of mycobacterial lipidomics to identify deoxysiderophore intermediates that are inconsistent with the current model of mycobactin biosynthesis and lead to a new general mechanism of biosynthesis.
Solving the M. tuberculosis genome was a landmark study because it organized the ∼4,100 genes from one of the world's most deadly bacterial pathogens (12). Based on this gene catalog, transcriptomics and proteomics provide unbiased ways to measure mycobacterial responses, including enzymes that produce the lipid-laden cell wall (12–16). The unusually lipophilic cell wall regulates nutrient uptake, drug transport, and antigen shedding into the host. An unbiased, systematic profile of lipids in M. tuberculosis and its cell wall could have a broad impact on studies of nutrition, antigenicity, and drug uptake. However, existing methods for global M. tuberculosis lipid analysis were inadequate, for two reasons. Lipidomic databases contain masses of lipids in eukaryotes or model bacteria, rather than mycobactins (3), mycoketides (17), long-chain polyketides (18, 19), mycolic acids (20), and other lipids found only in mycobacteria. In addition, existing methods for extraction and separation of water-soluble cytosolic compounds cannot be used for the unusually hydrophobic neutral lipids of M. tuberculosis (13, 21).
We have developed a lipidomics profiling platform for mycobacteria based on high performance liquid chromatography-mass spectrometry (HPLC-MS), which resolves diverse compounds to generate a catalog of more than 12,000 molecules organized by retention time and mass-to-charge ratio (m/z). After introducing nutritional stress or gene deletion, changes in ions of known mass are measured, and collisional MS solves the structure of biologically interesting molecules. By combining genetics and lipidomics, we have revealed deoxysiderophore intermediates of mycobactin/carboxymycobactin and identified the function of mbtG and other genes in the mycobactin biosynthetic pathway, leading to a unique stepwise pathway of mycobactin biosynthesis.
Results
HPLC-MS Analysis of M. tuberculosis Lipids Identifies 12,135 Events.
To generate MS datasets describing M. tuberculosis lipids, we developed three chromatographic methods for mycobacterial cell-intrinsic lipids and lipids released from the cell. Cell pellet extraction with chloroform:methanol followed by acetone precipitation yielded two fractions: acetone-insoluble lipids and acetone-soluble lipids, which entered into normal and reversed-phase HPLC, respectively. Lipids released from bacteria were collected by filtering supernatant from M. tuberculosis culture, then performing extraction with ethyl acetate. These were then entered into a third reversed-phase HPLC system. HPLC-MS methods were optimized based on the polarity of their lipidic constituents, taking into account ionization and detection requirements for the unusually hydrophobic neutral lipids in mycobacteria, ionization energy, countercurrent gas pressure, and trace cations added to solvents (22). We found that M. tuberculosis lipids are broadly assessed by detection of adducts in the positive mode (Fig. 1) (22), in contrast to anionic phospholipids, which dominate the lipid profiles of Gram-negative bacteria and eukaryotes.
Fig. 1.
Lipidomic analysis of M. tuberculosis by HPLC-MS. (A) Overview of the conversion of detected ions into molecular events. A total of 42,147 ions passed through five filters (gray box) to remove ions corresponding to inorganic compounds from non–M. tuberculosis sources. The 36,094 remaining ions were condensed to events by collapsing isotope windows and H+, Na+, or NH4+ adducts of one neutral M (black box). (B) Overview of the chromatography strategy. Each colored chromatogram includes all isotopes and adduct ions of an event. All events are listed in Dataset S1.
We applied a series of tests to convert 42,147 ions into 12,135 reproducible “events,” demonstrating chromatographic, mass spectral, and ionic characteristics of individual lipids present in M. tuberculosis (Fig. 1A). The conversion of ions to events involved excluding ions present in solvents or media, as well as ions with mass defects and isotope patterns indicating low carbon content. Isotopes of one neutral value M, as well as H+, Na+, or NH4+ adducts, were also grouped into a single event. The number of events served as an estimate of the number of lipids in the M. tuberculosis lipidome, but one M with varying charge states could generate multiple events, and some lipids might produce in-source fragments. Conversely, some lipids might have been nonionizing or present below detection thresholds.
This approach yielded one M. tuberculosis lipidome in three datasets, searchable by mass and retention time: released lipids (4,100 events), acetone-insoluble lipids (1,433 events), and acetone-soluble lipids (6,602 events) (Fig. 1B and Dataset S1). Separately plotting chromatograms of high-, intermediate-, and low-intensity events demonstrated a dynamic range of detection exceeding 4 log10 orders of magnitude (Fig. S1A). This wide dynamic range allowed nearly simultaneous detection of lipids at high intensities (mycolates, phthiocerol dimycocerosates, mycobactins) and low intensities (diacyl sulfoglycolipids) in a single injection (Fig. S1B). Other applications have sought to catalog known mycobacterial lipids at the outset of the experiment and globally track many named molecules (untargeted lipidomics) (22, 23). Our targeted lipidomics approach ranks unknown lipids according to a biological criterion (lipids consistent with mycobactin metabolites), and then solves their structures through collisional MS.
M. tuberculosis Regulates Dideoxymycobactin During Iron Starvation.
General mechanisms of mycobactin biosynthesis have been proposed (Fig. 2, Upper) (6, 9, 10, 24). However, the discovery of antigens presented by CD1a identified dideoxymycobactin (DDM), an apparent mycobactin variant lacking hydroxyl groups on two lysines (Fig. 3A) (24). Existing models of mycobactin biosynthesis postulate that lysine hydroxylation occurs early in biosynthesis, so that a deoxy variant like DDM is not produced (Fig. 2, Upper). Therefore, we considered that DDM might be an unnatural product formed ex vivo. However, T cells recognized myeloid cells infected with M. tuberculosis (24), specifically recognizing purified DDM instead of purified mycobactin and indicating that DDM is produced during infection (Fig. 3A).
Fig. 2.
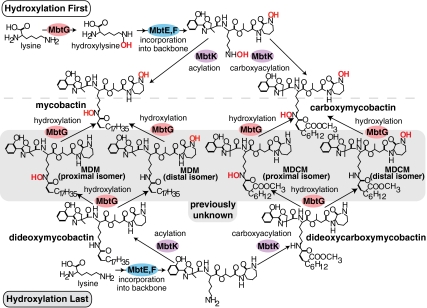
A new mycobactin biosynthesis model. The conventional biosynthesis model, designated hydroxylation first (Upper), predicts lysine hydroxylation before assembly of the peptide. Our proposed hydroxylation last model (Lower) predicts that peptide synthesis and acylation precede hydroxylation by MbtG. Enzymes for hydroxylation, lysine incorporation, and acylation are shown in red, blue, and purple, respectively.
Fig. 3.
Iron starvation regulates DDM. (A) CD1a-restricted T-cell secretion of IL-2 in response to dendritic cells treated with purified, M. tuberculosis-derived DDM (Left; m/z 840.5) or mycobactin (Right; m/z 925.5) shows that mycobactin does not trigger T-cell activation and provides a bioassay of DDM production. (B) Acetone-soluble lipids from WT M. tuberculosis or ΔmbtB show natural DDM-838 at 16.2 min or the synthetic DDM-784 internal standard at 8.3 min. (C) Acetone soluble lipids from WT M. tuberculosis or ΔideR grown with and without added iron were analyzed as in B.
To investigate the genetic regulation of DDM during the M. tuberculosis iron- scavenging response, we detected DDM within acetone-soluble lipid extracts from WT M. tuberculosis and mutants under high-iron and low-iron growth conditions. Loading and detection of natural DDM (m/z 838.5) was confirmed using an internal synthetic DDM standard of distinct mass (m/z 784.5) and T-cell bioassays for DDM (Fig. 3 B and C). Cells transfected with the α and β chains from the CD8-2 DDM-specific T-cell line stimulated IL-2 release from monocyte-derived dendritic cells when treated with DDM (24). This served as a bioassay for DDM production and verification of MS results. Acetone-soluble lipids from a mycobactin synthase B mutant (ΔmbtB) (6) yielded trace or absent activation in T-cell bioassays and DDM-838 signals (Fig. 3B). IdeR, an iron-binding transcription factor, represses mycobactin synthase transcription (11). Adding iron to growth medium strongly inhibited DDM production, an effect absent in bacteria lacking IdeR (ΔideR) (11) (Fig. 3C). This indicates that DDM production requires MbtB and is inhibited by iron in an IdeR-dependent manner, demonstrating that M. tuberculosis regulates DDM during iron starvation.
A Revised Model for Mycobactin Biosynthesis.
This evidence for DDM as a natural target of the human T-cell response (24) and an element of the M. tuberculosis iron-starvation response forced consideration of an alternative model for mycobactin biosynthesis. The simplest model for mycobactin biosynthesis, designated “hydroxylation first,” predicts that mycobactin synthase G (MbtG) converts free lysine to hydroxylysine, followed by mycobactin synthases C, D, E, and F forming the peptide before acylation by MbtK (Fig. 2, Upper). In this model, DDM is never formed. In contrast, our proposed “hydroxylation last” model accounts for DDM production, with MbtG hydroxylating DDM rather than free lysine (Fig. 2, Lower). Although conservative, this model necessarily revises the structures of intermediates and the order of activity of most enzymes in the pathway. Our hydroxylation last model predicts that monodeoxymycobactins, monodeoxycarboxymycobactins, and dideoxycarboxymycobactins should exist; however, such lipids have not been detected during decades of M. tuberculosis investigation (Fig. 2, gray).
Mining the M. tuberculosis Lipidome for Hypothetical Molecules.
To search for hypothetical deoxysiderophores, we calculated their ion masses by assuming that they differed from known mycobactins and carboxymycobactins only in hydroxylation state (Fig. 3A and Dataset S2). With this mass list, we searched the lipidomic datasets (Dataset S1) for events matching calculated masses within 10 ppm of mass accuracy and similar elution to siderophore standards (Fig. 4A). This search returned 38 events matching calculated masses of known siderophores, including ferri-mycobactin (m/z 923.4724), ferri-carboxymycobactin (m/z 799.2752), and apo-DDM (m/z 838.5708) (Fig. 4D).
Fig. 4.
Lipidomic discovery of monodeoxymycobactins and monodeoxycarboxy mycobactins. (A) Overview of the approach to detecting previously undetected deoxysiderophores. (B and C) Comparison of lipidomic events (Dataset S1) (B) and calculated masses of known and hypothetical siderophores (Dataset S2) (C). (D) Events matched calculated masses (≤10 ppm error) of known lipids ferri-mycobactin, apo-DDM, ferri-carboxymycobactin, and apo-carboxymycobactin. (E) Events matched calculated masses of 21 hypothetical deoxysiderophores, including series members (smaller boxes) with variations in acyl chain length/unsaturation, oxazoline ring amino acid, and carboxy methylation.
Ions initially matching masses of known compounds were subjected to five tests to confirm their chemical identity (Fig. 4A). First, because 56Fe is 15 times more naturally abundant than 54Fe, ferri-siderophores should show a characteristic ion doublet in which [M-2H+56Fe]+ is 15-fold more intense than [M-2H+54Fe]+ (m/z 905.4815 in Fig. 5A). Second, siderophores should be part of lipid families with predictable variations in alkyl carbons (C6–22) and unsaturation (0–1) (Figs. 4E and 5C and Dataset S2). Finally, we demonstrated near coelution with standards (Fig. 5B) and determined molecular structures by collisional MS (Fig. 5D and Fig. S2).
Fig. 5.
Hydroxylation on one of two lysines makes deoxysiderophores isomeric. (A) Ferri-monodeoxymycobactin-907 has a characteristic 56Fe ion (m/z 907.4780) and a lower-intensity 54Fe ion (905.4815; red). (B) Monodeoxymycobactin-907 nearly coelutes with synthetic DDM-838 standard (synthetic DDM), natural DDM-838, and natural mycobactin-937. (C) Chromatograms of monodeoxymycobactins, appearing as doublets, show later elution when containing increasing carbon numbers. (D) Collisional MS of two monodeoxymycobactin-907 isomers. The early eluting isomer fragmented to m/z 711.3540, isolating the hydroxyl group to the proximal lysine. The later eluting isomer fragmented to m/z 696.3665, isolating the hydroxyl group to the distal lysine.
Discovery of Monodeoxymycobactins.
In addition to the 38 known siderophores, 21 lipidomic events matched masses of hypothetical deoxysiderophores. For example, events corresponding to a hypothetical monodeoxymycobactin with a C20:1 fatty acyl unit were detected with Fe3+ (m/z 907.4820) and without Fe3+ (m/z 854.5663). Only the former exhibited the iron doublet ion (Figs. 4E and 5A). Other events matched calculated masses with shorter or saturated fatty acids, including ferri-monodeoxymycobactins with C18:1 and C20:1 fatty acids and apo-monodeoxymycobactins with C16:1, C18:1, and C20:1 fatty acids (Fig. 4E). Monodeoxymycobactin nearly coeluted with synthetic DDM standard, natural DDM, and mycobactin (Fig. 5B). Monodeoxymycobactins with increasing carbon numbers eluted in reverse order to their carbon number, as expected for a lipid series in reversed-phase HPLC (Fig. 5C). Finally, collisional MS detected the expected fragments of monodeoxymycobactins for every member of the monodeoxymycobactin family (Fig. 5D, Fig. S2, and Fig. S6).
Proximal and Distal Isomers of Monodeoxymycobactins.
Whereas mycobactin and DDM eluted in one peak, monodeoxymycobactins unexpectedly eluted in two peaks. Because ions were detected in narrow mass windows, the two molecules likely represent isomers (Fig. 5 B and C). DDM is likely converted to mycobactin through a two-step process involving similar reactions on lysines located proximal or distal to the acyl chain (Fig. 2, Lower). If the hydroxylase prefers one lysine, then only one form of monodeoxymycobactin is produced. The unexpected doublets seen only in monodeoxy forms could be explained if hydroxylation occurred at either lysine with similar efficiency, resulting in distinct proximal and distal isomers. Monodeoxymycobactin-907 collided to give fragments of m/z 711.3540 or m/z 696.3665, indicating the early-eluting and late-eluting isomers carried the hydroxyl group on the proximal and distal lysines, respectively (Fig. 5D). Thus, monodeoxymycobactins are produced as pairs of isomers, providing insight into their two-step conversion from DDM to mycobactin.
Discovery of Monodeoxycarboxymycobactins.
After identifying monodeoxymycobactins, we used a similar approach to discover monodeoxycarboxymycobactins. Using screening based on mass (Fig. 4, Right) and the five confirmatory tests (Fig. 4A), we identified apo-monodeoxycarboxymycobactins in the released lipid dataset (Fig. 4E and Dataset S2). Thus, lipidomic profiling rapidly identified two previously unknown classes of deoxysiderophores as 42 distinct molecules, demonstrating the surprising efficiency of the platform for molecular discovery (Fig. S3). Although we detected every theoretical monodeoxysiderophore predicted by the hydroxylation last model, we detected no dideoxycarboxymycobactins (Fig. 2, Lower). Monodeoxycarboxymycobactins are detected at 20-fold lower levels than monodeoxymycobactins, which is just above the limit of detection (Fig. 5); thus, if dideoxycarboxymycobactins exist, they are likely below the detection threshold. Only the hydroxylation last model predicts the existence of DDM, monodeoxymycobactin, and monodeoxycarboxymycobactin; thus, the lipidomic data strongly support the more complex hydroxylation last biosynthetic pathway.
MbtG Is Necessary for Growth and Mycobactin Production.
Previous homology analyses (9) and experimental data (10, 25) have shown that recombinant MbtG hydroxylates free lysine and acylated lysine peptides. We deleted mbtG through homologous recombination (ΔmbtG) to determine whether this gene is essential to mycobactin production and growth of M. tuberculosis. In low-iron media, ΔmbtG failed to grow unless treated with exogenous mycobactin (Fig. 6B and Fig. S4C). Formal testing of growth revealed that mycobactin, but not iron, fully complemented ΔmbtG growth to WT rates (Fig. 6B and Fig. S4B).
Fig. 6.
MbtG acts downstream of DDM but upstream of monodeoxymycobactin and mycobactin. (A) Schematic representation of the strategy used to generate ΔmbtG with the counterselectable plasmid pJM1::ΔmbtG. (B) WT or ΔmbtG M. tuberculosis was subcultured in low-iron medium, then inoculated into low-iron, high-iron (50 μM ferric chloride), or 7H9 media containing ferri-mycobactin J (2 μg/mL). Growth was measured by absorbance at 600 nm. (C) Acetone-soluble lipids from plates show mycobactin-923 (m/z 923.4701; Top), monodeoxymycobactin-907 (m/z 907.4752; Middle), and DDM-838 (m/z 838.5688; Bottom).
Furthermore, lipidomic profiling of ΔmbtG could test specific predictions of the hydroxylation first and hydroxylation last models. Monodeoxymycobactin and mycobactin should be absent from ΔmbtG. Whereas the hydroxylation first model predicts the complete loss of DDM, the hydroxylation last model predicts preservation or even an increase in DDM if MbtG is the key enzyme mediating DDM turnover to mycobactin. Lipidomic analysis showed that ΔmbtG failed to produce mycobactin and monodeoxymycobactin in low-iron media, indicating MbtG is upstream of mycobactin and essential for its biosynthesis (Fig. 6C and Fig. S5). Importantly, DDM accumulated in ΔmbtG (Fig. 6C), placing MbtG downstream of DDM. This finding suggests that all other mycobactin synthases required to make DDM are functional in ΔmbtG. Therefore, the loss of mycobactin in ΔmbtG was likely due to the downstream position of MbtG in the biosynthetic pathway, not to an unexpected interruption of mbt genes near the mbtG locus, global alterations in metabolism, or unforeseen feedback loops. This result demonstrates a specific and essential function of MbtG as the hydroxylase that converts DDM to mycobactin and the necessity of fully hydroxylated mycobactin for M. tuberculosis growth. Overall, these genetic and lipidomic results strongly support the hydroxylation last model as the sole or predominant pathway for mycobactin biosynthesis (Fig. 2, Lower).
Discussion
Existing models of mycobactin biosynthesis are based on predicted but largely untested functions of 14 mycobactin synthase genes. Although the hydroxylation first model is not formally ruled out, no data distinguishing between the two models support its existence. In addition, all data strongly suggest that the hydroxylation last model is the quantitatively dominant mechanism of mycobactin production during M. tuberculosis iron starvation. These data include direct evidence for iron, MbtB and IdeR governing DDM synthesis; production of three classes of deoxysiderophores predicted only by this model (DDM, monodeoxymycobactins, and monodeoxycarboxymycobactins); and absence of monodeoxymycobactin and mycobactin with accumulation of DDM in ΔmbtG.
Our proposed model has general implications for the study of M. tuberculosis lipids with systems biology methods, as well as practical information for targeting mycobactins during human infection. Furthermore, our experiments illustrate that targeted lipidomics provides a reliable means of lipid discovery, as illustrated by the rapid detection of 42 compounds in two classes. The value of this targeted approach is that it bypasses the need for compound databases that identify molecules based on mass.
Metabolomics is the youngest systems biology discipline, and its relationships to more established organism-wide profiling methods and to conventional single enzyme-substrate methods are only now emerging. Although enzymology usually emphasizes characterizing the reactions for which an enzyme is sufficient, gene deletion followed by organism-wide lipidomics broadly reveals the functions for which an enzyme is necessary. A combination of these methods has proven useful. Lipidomic profiling of ΔmbtG establishes MbtG as the essential enzyme in conversion of DDM to mycobactin, and places this reaction as the final step in the pathway.
A key goal of lipidomics is to provide insight into the general structure of a biochemical pathway in ways that cannot be known from studies of individual genes, transcripts, or proteins. It is unlikely the hydroxylation last model could have been derived from studies of individual mycobactin synthases studied in isolation. The pathway revisions proposed here alter the order of action of most mycobactin synthases, specifically MbtE, MbtF, MbtG, MbtK, MbtL, MbtM, and MbtN (Fig. 2). Although we have not detected all intermediates in the revised pathway, the discovery of DDM, monodeoxymycobactin and monodeoxycarboxymycobactin implies that many intermediates are produced in the deoxy form, and that each enzyme's substrate specificities must be reconsidered. Furthermore, this finding suggests that natural substrates for many mycobactin synthases should be reconsidered, including MbtE, MbtF, MbtG, and MbtK. The importance of mycobactins in M. tuberculosis virulence led Quadri (26) to propose pharmacologic interruption of mycobactin biosynthesis as a tuberculosis treatment. Moreover, carboxymycobactins are present in the tissues of patients with tuberculosis, and thus are candidate biomarkers of M. tuberculosis infection (27, 28). Understanding the precise chemical structures of mycobactin intermediates supports both goals.
Do deoxysiderophores also provide information about the basic cellular mechanisms of M. tuberculosis iron scavenging? Our analysis of deoxysiderophores (Figs. 3A and 4) suggests they have lower iron affinity than mycobactin and carboxymycobactin, which could allow MbtG hydroxylation to regulate siderophore iron binding at the final step of production. Future studies are needed to examine the possibility that apo-deoxysiderophores can be selectively positioned and activated within the mycobacterium, facilitating one-way iron transport toward M. tuberculosis, the world's most successful bacterial pathogen.
Methods
Detailed information is provided in SI Methods. In brief, 100-mL lipidomic cultures were grown to OD600 = 0.4. Supernatant was collected by centrifugation, filtered through a 0.22-μm filter, and treated with 300 μL of 6 M hydrochloric acid. After the addition of 1.4 volumes of ethyl acetate and mixing for >1 h, the ethyl acetate was removed and dried to yield released lipids. To extract cell lipids, bacteria scraped from plates were treated with 60 mL of 2:1 chloroform:methanol, then 1:1, then 1:2 for 1 h each. Extractable lipids were separated by centrifugation, collected, and dried. Acetone-insoluble cell lipids were made by collecting precipitates from 30 mg of total lipids per 0.21 mL of 4 °C acetone on ice for 1 h, followed by washing with 4 °C acetone. Acetone precipitates were washed three times with 4 °C acetone, and supernatants and washes were collected and dried to yield acetone-soluble cell lipids. MS analysis was performed with an Agilent 6520 Accurate Mass QtoF mass spectrometer.
Supplementary Material
Acknowledgments
This work was supported by National Institute of Allergy and Infectious Diseases Grants U19 AI076217, R01 AI071155, and AR048632 (to D.B.M.), Grant R01 AI071881 (to E.J.R.), the Intramural Research Program (C.E.B.), the Mark and Lisa Schwartz Foundation (to D.B.M. and E.J.R.), and the National Science Foundation (to C.A.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/10.1073/pnas.1109958109/-/DCSupplemental.
References
- 1.Schaible UE, Kaufmann SHE. Iron and microbial infection. Nat Rev Microbiol. 2004;2:946–953. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- 2.Ratledge C. Iron, mycobacteria and tuberculosis. Tuberculosis (Edinb) 2004;84:110–130. doi: 10.1016/j.tube.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 3.Francis J, MacTurk HM, Madinaveitia J, Snow GA. Mycobactin, a growth factor for Mycobacterium johnei, I: Isolation from Mycobacterium phlei. Biochem J. 1953;55:596–607. doi: 10.1042/bj0550596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray MJ, Murray AB, Murray MB, Murray CJ. The adverse effect of iron repletion on the course of certain infections. BMJ. 1978;2:1113–1115. doi: 10.1136/bmj.2.6145.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegrist MS, et al. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc Natl Acad Sci USA. 2009;106:18792–18797. doi: 10.1073/pnas.0900589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Voss JJ, et al. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc Natl Acad Sci USA. 2000;97:1252–1257. doi: 10.1073/pnas.97.3.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holmes MA, Paulsene W, Jide X, Ratledge C, Strong RK. Siderocalin (Lcn 2) also binds carboxymycobactins, potentially defending against mycobacterial infections through iron sequestration. Structure. 2005;13:29–41. doi: 10.1016/j.str.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 8.Gobin J, et al. Iron acquisition by Mycobacterium tuberculosis: Isolation and characterization of a family of iron-binding exochelins. Proc Natl Acad Sci USA. 1995;92:5189–5193. doi: 10.1073/pnas.92.11.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quadri LEN, Sello J, Keating TA, Weinreb PH, Walsh CT. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem Biol. 1998;5:631–645. doi: 10.1016/s1074-5521(98)90291-5. [DOI] [PubMed] [Google Scholar]
- 10.Krithika R, et al. A genetic locus required for iron acquisition in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103:2069–2074. doi: 10.1073/pnas.0507924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez GM, Voskuil MI, Gold B, Schoolnik GK, Smith I. ideR, An essential gene in Mycobacterium tuberculosis: Role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect Immun. 2002;70:3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole ST, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 13.Shi L, et al. Carbon flux rerouting during Mycobacterium tuberculosis growth arrest. Mol Microbiol. 2010;78:1199–1215. doi: 10.1111/j.1365-2958.2010.07399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu S, et al. Comprehensive proteomic profiling of the membrane constituents of a Mycobacterium tuberculosis strain. Mol Cell Proteomics. 2003;2:1284–1296. doi: 10.1074/mcp.M300060-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Park H-D, et al. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol. 2003;48:833–843. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schnappinger D, et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: Insights into the phagosomal environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsunaga I, et al. Mycobacterium tuberculosis pks12 produces a novel polyketide presented by CD1c to T cells. J Exp Med. 2004;200:1559–1569. doi: 10.1084/jem.20041429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cox JS, Chen B, McNeil M, Jacobs WR., Jr Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 19.Reed MB, et al. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature. 2004;431:84–87. doi: 10.1038/nature02837. [DOI] [PubMed] [Google Scholar]
- 20.Barry CE, 3rd, et al. Mycolic acids: Structure, biosynthesis and physiological functions. Prog Lipid Res. 1998;37:143–179. doi: 10.1016/s0163-7827(98)00008-3. [DOI] [PubMed] [Google Scholar]
- 21.de Carvalho LPS, et al. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem Biol. 2010;17:1122–1131. doi: 10.1016/j.chembiol.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 22.Layre E, et al. A comparative lipidomics platform for Mycobacterium tuberculosis provides chemotaxonomic analysis for biomarker discovery. Chem Biol. 2011;18:1537–1549. doi: 10.1016/j.chembiol.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sartain MJ, Dick DL, Rithner CD, Crick DC, Belisle JT. Lipidomic analyses of Mycobacterium tuberculosis based on accurate mass measurements and the novel Mtb LipidDB. J Lipid Res. 2011;52:861–872. doi: 10.1194/jlr.M010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moody DB, et al. T cell activation by lipopeptide antigens. Science. 2004;303:527–531. doi: 10.1126/science.1089353. [DOI] [PubMed] [Google Scholar]
- 25.Chavadi SS, et al. Mutational and phylogenetic analyses of the mycobacterial mbt gene cluster. J Bacteriol. 2011;193:5905–5913. doi: 10.1128/JB.05811-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quadri LEN. Strategic paradigm shifts in the antimicrobial drug discovery process of the 21st century. Infect Disord Drug Targets. 2007;7:230–237. doi: 10.2174/187152607782110040. [DOI] [PubMed] [Google Scholar]
- 27.Mori K, et al. Endocytic delivery of lipocalin-siderophore-iron complex rescues the kidney from ischemia-reperfusion injury. J Clin Invest. 2005;115:610–621. doi: 10.1172/JCI23056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu SY, Pauksen K, Venge P. Serum measurements of human neutrophil lipocalin (HNL) discriminate between acute bacterial and viral infections. Scand J Clin Lab Invest. 1995;55:125–131. doi: 10.3109/00365519509089604. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.