Background: Misfolded protein aggregates are recruited to the aggresome by a protein complex consisting of histone deacetylase 6 (HDAC6).
Results: The ubiquitin-binding domain (ZnF-UBP) of HDAC6 binds to ubiquitin C termini generated by ataxin-3.
Conclusion: The exposure of ubiquitin C termini within protein aggregates enables HDAC6 recognition.
Significance: This study provides the role of HDAC6 in aggresome formation and suggests a novel ubiquitin-mediated signaling pathway.
Keywords: Deubiquitination, Protein Aggregation, Protein Structure, Protein-Protein Interactions, Ubiquitin, Aggresome, Ataxin-3
Abstract
The aggresome pathway is activated when proteasomal clearance of misfolded proteins is hindered. Misfolded polyubiquitinated protein aggregates are recruited and transported to the aggresome via the microtubule network by a protein complex consisting of histone deacetylase 6 (HDAC6) and the dynein motor complex. The current model suggests that HDAC6 recognizes protein aggregates by binding directly to polyubiquitinated proteins. Here, we show that there are substantial amounts of unanchored ubiquitin in protein aggregates with solvent-accessible C termini. The ubiquitin-binding domain (ZnF-UBP) of HDAC6 binds exclusively to the unanchored C-terminal diglycine motif of ubiquitin instead of conjugated polyubiquitin. The unanchored ubiquitin C termini in the aggregates are generated in situ by aggregate-associated deubiquitinase ataxin-3. These results provide structural and mechanistic bases for the role of HDAC6 in aggresome formation and further suggest a novel ubiquitin-mediated signaling pathway, where the exposure of ubiquitin C termini within protein aggregates enables HDAC6 recognition and transport to the aggresome.
Introduction
The ubiquitin proteasome pathway is the primary “quality control” mechanism that cells use to break down misfolded proteins, which are polyubiquitinated in preparation for degradation (1, 2). When proteasomal functions are disrupted or overloaded, misfolded proteins cannot be cleared and protein aggregation ensues (3). In such cases, the aggresome pathway is activated, and cytoplasmic protein aggregates are transported to the microtubule organizing center, where aggresomes are formed (3). Aggresomes are dynamic structures that can recruit various chaperones and proteasomes to aid in the disposal of aggregated proteins (4).
Aggresome formation is often considered a cytoprotective response, allowing for sequestration of potentially toxic misfolded proteins and promoting their clearance by autophagy (5). Accumulating evidence suggests an important role for HDAC66 in the transport and clearance of misfolded proteins by favoring the formation of aggresomes and activating autophagy (6, 7). HDAC6 modulates a wide range of cellular activities through distinct interactions with cellular components including tubulin, cortactin, HSP90, and other proteins and protein complexes (8, 9). In the aggresome pathway, HDAC6 plays the key role of gathering scattered polyubiquitinated protein aggregates and transporting them to aggresomes via microtubule tracts. HDAC6 has been shown to bind polyubiquitinated aggregates and dynactin/p150Glued, a component of the dynein motor complex, bridging the ubiquitinated proteins to the dynein motors and promoting transport of the cargo toward the microtubule organizing center to enable aggresome formation (10, 11).
HDAC6 is thought to interact with aggregated proteins through its ZnF-UBP domain (11), a domain typically found in deubiquitinases capable of binding the unanchored ubiquitin C terminus with high affinity and specificity (12, 13). However, in polyubiquitinated proteins, ubiquitin moieties are conjugated to target polypeptide chains through C-terminal diglycine motifs (14) and serve as molecular tags that are usually recognized by downstream proteins through its globular surface, often through a hydrophobic area around Ile-44 (15, 16). Therefore, it is difficult to reconcile how HDAC6 can potentially bind polyubiquitin chains of aggregated proteins through its ZnF-UBP domain.
Here we demonstrate the molecular mechanism of how scattered protein aggregates are recognized by HDAC6. First, we show that the HDAC6 ZnF-UBP domain interacts directly with protein aggregates using immunoprecipitation approaches. Then we solved the crystal structure of the ZnF-UBP domain of HDAC6 and its complexes with either full-length ubiquitin or a ubiquitin-C-terminal peptide. This structural and biochemical analysis revealed that an intact and unanchored (noncovalently modified) ubiquitin C-terminal diglycine motif is necessary and sufficient for binding to HDAC6 ZnF-UBP. Second, we show that such unanchored ubiquitin C termini are present abundantly in aggresomes. It is the solvent-accessible ubiquitin C termini that tag protein aggregates for HDAC6 recognition. These unanchored C termini are generated by the aggregate-associated deubiquitinase, ataxin-3, which cleaves (poly)ubiquitin moieties in polyubiquitinated proteins to expose their C-terminal diglycine motifs for HDAC6 binding and subsequent transport to the aggresome. Our work thus offers a critical piece of the mechanism underlying how HDAC6 recruits protein aggregates.
EXPERIMENTAL PROCEDURES
Materials
Purchased protein reagents used in various experiments included: HDAC6 (Biomol), ubiquitin (Boston Biochem), tetraubiquitin (Boston Biochem), and ubiquitin-AFC (7-amino-4-trifluoromethylcoumarin) (Boston Biochem). Peptide RLRGG was synthesized at the Tufts University Core Facility (Boston).
Antibodies used in various experiments included: anti-GFP antibody (product number A11122; Invitrogen), anti-ataxin3 antibody (product #MABN37; Millipore), anti-HDAC6 antibody (product number AB12173; Abcam), anti-C-terminal ubiquitin antibody (product number 04-454; Millipore), anti-N-terminal ubiquitin antibody (product number AP1228a; Abgent), anti-USP5 antibody (product number AP2134a; Abgent), anti-USP13 antibody (product number AB99421; Abcam), Alexa 555- or Alexa 647-conjugated secondary antibodies (Molecular Probes); and infrared dye-conjugated secondary antibodies, IR700 and IR800 (LI-COR Biosciences).
Cloning, Protein Expression, Purification, and Crystallization
ZnF-UBP domains of HDAC6 (1109–1215) and USP5 (171–290) were amplified by PCR from the Mammalian Gene Collection clone (with GenBankTM accession numbers BC013737 and BC004889, respectively) and subcloned into a modified pET28a-LIC vector. The mutants were prepared by the QuikChange (Stratagene) site-directed mutagenesis method. The recombinant proteins were overexpressed in LB medium as N-terminal His6-tagged proteins at 15 °C using Escherichia coli strain BL21 (DE3) codon plus RIL (Stratagene). For purification, the cells were lysed by passing through a Microfluidizer (Microfluidics Corporation) at 18,000 p.s.i. The purification procedure comprised of two steps: affinity chromatography on a Ni-NTA column (Qiagen) and gel filtration on a Superdex 75 column (26/60; GE Healthcare). The His tag was cleaved with thrombin after the affinity column step while dialyzing against 20 mm Tris, pH 8.0, 0.15 m NaCl, and 1 mm TCEP (Buffer A). Crystals of the HDAC6 ZnF-UBP domain were grown at 18 °C using the sitting drop method by mixing equal volumes of 3.5 m sodium formate, 100 mm Bis-Tris, pH 7.0, and 15 mg/ml protein. Crystals of the HDAC6 ZnF-UBP (15 mg/ml) and monoubiquitin (15 mg/ml) were grown at 28 °C using the sitting drop method by mixing with equal volumes of 15% PEG 3350, 0.1 m (NH4)2SO4, 0.1 m Bis-Tris, pH 5.6. Crystals of the HDAC6 ZnF-UBP domain and RLRGG complex were grown at 18 °C using the hanging drop method by mixing equal volumes of 20% PEG3350, 0.2 m CaCl2, 0.1 m Bis-Tris, pH 7.0, and 15 mg/ml protein solution with 3 mm peptide.
Data Collection and Structure Determination
The single wavelength anomalous diffraction data set of HDAC6 ZnF-UBP domain was collected at 100 K on BNL Beamline X29 with wavelength of 1.26 Å. The HDAC6 ZnF-UBP with peptide RLRGG data were collected on a Rigaku FR-E super bright rotating anode x-ray generator. Program HKL2000 was used for both data processing and scaling. The HDAC6 ZnF-UBP structure was determined using the anomalous signal from native-bound zinc atoms at 1.26Å. The program SOLVE was used to search the zinc sites, and RESOLVE was used to perform the site refinement and initial model building. ARPwARP was used for model building. The graphics program COOT (17) was used for manual model refinement and visualization. Refmac5 was also used to refine the model. The structure of HDAC6 with RLRGG peptide was determined using HDAC6 (3C5K) as a molecular replacement model. Diffraction data on HDAC6 with ubiquitin complex structure was collected on the APS beamline 23ID-B, and the structure was determined using the HDAC6 (3C5K) structure as a model. The program BUSTER (18) was used for structural refinement.
Pulldown Assay and Dynamic Light Scattering (DLS)
Proteins of interest (one of the interacting proteins was His-tagged) at 1 mg/ml in buffer A were incubated at room temperature for 30 min allowing for batch binding before either Ni-NTA (Qiagen) or Talon (Clontech) was added. The resin was washed extensively prior to elution with SDS-PAGE loading buffer. Eluted proteins were analyzed using NuPAGE 4–12% Bis-Tris gels (Invitrogen). For DLS experiments, equimolar amounts of HDAC6 Zn-UBP domain and ubiquitin or ubiquitin mutants were incubated together at room temperature for 30 min before centrifugation at 13,000 rpm for 10 min. The final protein concentration was determined using the Bradford protein assay (Bio-Rad). The samples were then transferred into 384-well plates for DLS analysis. DLS data were recorded using DynaPro Plate Reader with Dynamics software (Wyatt Technology).
Immunoprecipitation
293T cells were transfected with GFP-CFTRΔ508 using Lipofectamine 2000 (Invitrogen). At 48 h post-transfection, the cells were treated with either Me2SO or MG132 (EMD Biosciences). After 16 h of treatment, the cells were lysed, and proteins were quantified. Subsequently, 1 mg of total protein lysate/condition was incubated overnight with 50 μg of recombinant peptide at 4 °C before immunoprecipitation with 10 μg of GFP antibody/sample using protein A-agarose beads (Invitrogen). For ataxin-3 knockdown experiments, 293T cells were transfected with ataxin-3-specific siRNA (ON_TARGETplus SMARTpool ATXN3 L-012013-00-0005; Dharmacon RNAi Technologies) or scramble control siRNA (ON-TARGETplus nontargeting pool D-001810-10-05; Dharmacon RNAi Technologies), using Dharmafect transfection reagent according to the manufacturer's instructions. After 24 h post-transfection, these cells were transfected with GFP-CFTRΔ508, GFP-Atx1-82Q, or GFP and treated as described above.
Western Blot Analysis
Proteins were extracted from cells with a lysis buffer (50 mm Tris, pH 7.5, 150 mm KCl, 1 mm PMSF, 1% Triton, protease inhibitor mixture (Sigma)). Protein concentrations were measured using the Bradford protein assay (Bio-Rad). Sample lysates were resolved by SDS-PAGE and probed with anti-GFP 1:2000, anti-ataxin3 1:1000, anti-HDAC6 1:1000, anti-C-terminal ubiquitin 1:1000, and anti-N-terminal ubiquitin 1:1000. Western blot analysis was performed by incubating with the infrared dye-conjugated secondary antibodies, IR700 and IR800 (LI-COR Biosciences) for 1 h at room temperature; blots were imaged and processed on an Odyssey® infrared imaging system.
Immunofluorescence and Image Acquisition and Processing
Cultured A549 or 293T cells were transfected with respective plasmids and fixed post-treatment with PBS with 3.5% formaldehyde for 15 min and washed in PBS with 0.4% Triton X-100. The cells were blocked in 5% goat serum (in PBS) for 1 h before incubating in appropriate primary antibody mixtures with mild shaking at 4 °C overnight. Primary antibody dilutions used included anti-C-terminal ubiquitin antibody 1:250, anti-ataxin-3 antibody 1:250, anti-HDAC6 antibody 1:100, anti-USP5 antibody 1:250, anti-USP13 antibody 1:250, and secondary antibodies conjugated to Alexa 555 or Alexa 647 were used at 1:250. All of the antibody incubations were performed at 4 °C overnight in the presence of 5% normal goat serum. Fluorescence images were acquired on Olympus FV1000 confocal microscope and processed using FluoView10ASW (Olympus). The figure panels were assembled using Adobe Photoshop CS4.
RESULTS
HDAC6 ZnF-UBP Domain Interacts with Polyubiquitinated Protein Aggregates
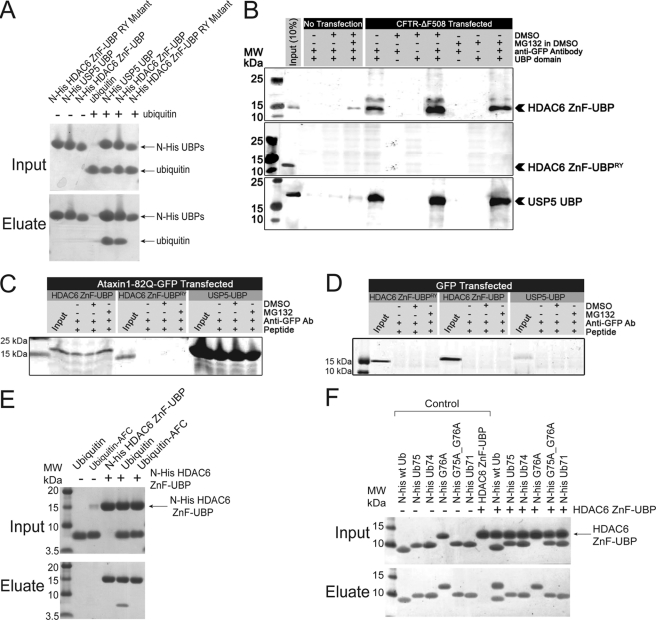
An earlier report showed that HDAC6 binds polyubiquitinated protein aggregates through its ZnF-UBP domain (11). However, because typical ZnF-UBP domains only bind unanchored ubiquitin C termini (19) and all the C termini of ubiquitin moieties in polyubiquitinated proteins are conjugated either to protein substrate or other ubiquitin moieties (20), it has been suggested that the ZnF-UBP domain may play a regulatory role rather than bind directly to the aggregates (12). To examine the direct interaction between HDAC6 ZnF-UBP domain and protein aggregates, we used the mutant cystic fibrosis transmembrane conductance regulator CFTR-ΔF508-induced protein aggregation as a model (3, 11). Cultured 293T cells were transfected with GFP-CFTR-ΔF508 and treated with a proteasome inhibitor, MG132, to induce aggresome formation. The interaction between HDAC6 and aggregates was tested by incubating cell lysates with recombinant HDAC6 ZnF-UBP domain. The binding activity of the recombinant HDAC6 ZnF-UBP domain to ubiquitin was first tested and confirmed by an in vitro pull down assay. As shown in Fig. 1A, when incubated with ubiquitin, His-tagged HDAC6 ZnF-UBP pulled down ubiquitin in a manner similar to the His-tagged UBP domain of USP5, a known ubiquitin-binding protein (13). We generated an active site mutant R1155A/Y1184A based on homologue analysis (12, 13). Although the HDAC6 ZnF-UBP RY mutant protein was properly folded, as confirmed by one-dimensional proton NMR (supplemental Fig. S1), the binding between mutant HDAC6 ZnF-UBP domain and ubiquitin was abolished (Fig. 1A). These results confirmed the specific interaction between HDAC6 ZnF-UBP domains and ubiquitin and the essential role of the predicted ubiquitin-binding site. We next tested the direct binding between HDAC6 ZnF-UBP domain and GFP-CFTR-ΔF508 aggregates using immunoprecipitation assays. As shown in Fig. 1B, when these ZnF-UBP domains were incubated with GFP-CFTR-ΔF508 aggregates, the wild-type HDAC6 ZnF-UBP co-immunoprecipitated with GFP-CFTR-ΔF508 (top panel), whereas the mutant R1155A/Y1184A (middle panel) failed to interact with the protein aggregates. We found that the well characterized homologue ZnF-UBP domain from USP5 also binds these aggregates (Fig. 1B, bottom panel). These observations are consistent with previous reports that the ZnF-UBP is responsible for protein aggregate recognition (11, 21, 22). Furthermore, when we used a different ubiquitinating substrate to form aggregates, i.e. human ataxin-1-82Q-GFP, we again observed interactions of USP5-UBP and HDAC6-UBP with these aggregates (Fig. 1C). Taken together, our results suggest that the ZnF-UBP domain alone is sufficient to bind protein aggregates, and the binding is dependent on an intact ubiquitin-binding site. These interactions were specific for GFP-CFTR-ΔF508-induced aggregation, because expression of GFP by itself failed to induce aggresome formation or allow ZnF-UBP binding to any aggregates (Fig. 1D).
FIGURE 1.
HDAC6 ZnF-UBP domain interacts with CFTR-ΔF508 aggregates and ubiquitin C terminus. A, protein-protein interaction assay of ubiquitin with either His-tagged wild-type HDAC6 ZnF-UBP domain, a R1155A/Y1184A mutant, or wild-type USP5 UBP domain. His-tagged proteins were incubated with or without ubiquitin at room temperature for 30 min before Ni-NTA was added. The resin was washed extensively prior to elution with SDS-PAGE loading buffer. The input and eluted proteins were resolved by SDS-PAGE and visualized by Coomassie staining. B, both HDAC6 ZnF-UBP and USP5 UBP, but not HDAC6 ZnF-UBP RY mutant, interact with GFP-CFTR-ΔF508 protein aggregates. Cultured 293T cells were transfected with GFP-CFTR-ΔF508 and treated with either MG132 or Me2SO, as indicated. The lysates were incubated with HDAC6 ZnF-UBP (top panel) or HDAC6 ZnF-UBP RY mutant (bottom panel). CFTR-ΔF508 aggregates were immunoprecipitated with an anti-GFP antibody, resolved by SDS-PAGE, and visualized by Coomassie staining. C, both HDAC6 ZnF-UBP and USP5-UBP interact with ataxin-1-82Q-GFP aggregates. Cultured 293T cells were transfected with human ataxin-1-82Q-GFP and treated with either MG132 or Me2SO, as indicated. The lysates were incubated with HDAC6 ZnF-UBP, HDAC6 ZnF-UBP RY mutant, or USP5-UBP. Ataxin-1-82Q-GFP aggregates were immunoprecipitated with an anti-GFP antibody, resolved by SDS-PAGE, and visualized by Coomassie staining. D, GFP overexpression did not cause aggresome formation. The cells were transfected with pLenti-GFP construct and treated with either Me2SO or MG132 in Me2SO. The cell lysates were collected 2 days after transfection; incubated with either USP5 UBP, HDAC6 ZnF-UBP, or HDAC6 ZnF-UBP-RY mutant proteins; and immunoprecipitated by anti-GFP antibody. Input and precipitated samples were resolved by SDS-PAGE and visualized by Coomassie staining. E, protein-protein interaction assays of His-tagged HDAC6 ZnF-UBP domain with ubiquitin and ubiquitin-AFC. The His-tagged HDAC6 ZnF-UBP domain was incubated with ubiquitin or ubiquitin-AFC before applied to a Talon (cobalt) column. The input and eluted proteins were resolved by SDS-PAGE and visualized by Coomassie staining. F, protein-protein interaction assays of N-terminal His-tagged ubiquitin and ubiquitin mutants with HDAC6 ZnF-UBP domain. N-terminal His-tagged ubiquitin and ubiquitin mutant proteins were incubated with HDAC6 ZnF-UBP domain before applied to a Talon (cobalt) column. The input and eluted proteins were resolved by SDS-PAGE and visualized by Coomassie staining. MW, molecular mass.
HDAC6 ZnF-UBP Domain Binds Specifically to Unanchored Ubiquitin C Terminus
The next question that needed to be addressed was how HDAC6 ZnF-UBP binds polyubiquitinated protein aggregates. There are two possibilities: the ZnF-UBP domain of HDAC6 either binds unanchored C-terminal ubiquitin tails similar to other characterized ZnF-UBP domains or binds polyubiquitinated protein in a mode different from known ZnF-UBPs. To differentiate these possibilities, we performed protein-protein interaction assays to examine the direct binding between HDAC6 and ubiquitin. As shown in Fig. 1E, His-tagged HDAC6 ZnF-UBP interacted only with unconjugated ubiquitin but not C-terminally capped ubiquitin-AFC (7-amino-4-trifluoromethylcoumarin). When the last one or two glycine residues of ubiquitin were mutated (G76A and G75A/G76A) or deleted (Ub75, one C-terminal glycine deletion; Ub74, two glycine deletions), the interaction with HDAC6 ZnF-UBP was abolished (Fig. 1F). The loss of interaction was further confirmed by gel filtration and DLS experiments (supplemental Fig. S2 and Table S1). These results showed that an intact and unanchored (noncovalently modified) ubiquitin C-terminal diglycine motif is necessary for binding to HDAC6 ZnF-UBP.
To reveal the structural details of this interaction, we solved the crystal structures of the ZnF-UBP domain of HDAC6 and its complexes with either full-length ubiquitin or a ubiquitin C-terminal peptide (Table 1). The crystal structure of the HDAC6 ZnF-UBP domain (residues 1109–1215), determined at 1.55 Å resolution using anomalous signals from natively bound Zn2+ ions, revealed a compact structure consisting of five anti-parallel β-strands, two α-helices, and three Zn2+ ions (Fig. 2A). A distinct pocket was visible on the surface, formed by a cluster of aromatic residues (Fig. 2A). The ZnF-UBP domain of HDAC6 has similar folds overall to those of USP5 (Protein Data Bank code 2G43) and USP16 (Protein Data Bank code 2I50) (12, 13) but with significant differences in structural details. For example, in contrast to HDAC6 ZnF-UBP, the USP5 UBP domain contains only one Zn2+ ion and has a longer loop between β2 and αA (L2A) and a shorter αB helix, which is rotated almost 90° (13). The USP16 UBP domain has a much longer N-terminal loop that contains a two-turn α-helical conformation in parallel to αB (12). The structure of the HDAC6 ZnF-UBP domain when in complex with full-length ubiquitin showed that the last three residues of ubiquitin C terminus were bound in an extended conformation within the predicted aromatic pocket. Importantly, and consistent with our previous data, there was no major interaction between the HDAC6 ZnF-UBP and the globular region of ubiquitin, which had weak or missing electron density in parts, presumably because of conformational heterogeneity within the crystal (Fig. 2B and Table 1). A higher resolution (1.7 Å) structure of HDAC6 ZnF-UBP with a bound ubiquitin C-terminal pentapeptide, RLRGG, confirmed such a binding mode (Fig. 2, B, C, and F). Interestingly, significant conformational changes were observed at Arg-1155, whose side chain ϵ-carbon atom (guanidino-C) swings 5.6 Å toward the binding pocket upon peptide binding, enabling hydrogen bond formation between the ω-nitrogen and the carbonyl group of ubiquitin Gly-75. Tyr-1156 was also rotated ∼20° toward the peptide, causing the ubiquitin C-terminal diglycine motif to be completely buried inside the ZnF-UBP domain. Thus, Arg-1155 and Tyr-1156 act as gatekeeper residues that move from an “open” to a “closed” conformation upon ubiquitin binding (Fig. 2D). The HDAC6 ZnF-UBP domain and ubiquitin interacted mainly through a hydrogen bond network (Fig. 2E), which was very similar to the one in the USP5 ZnF-UBP and ubiquitin complex (Protein Data Bank code 2G45). However, HDAC6 ZnF-UBP lacks the additional contacts that USP5 ZnF-UBP makes with the globular surface of ubiquitin (13). Our structural studies therefore demonstrate that the HDAC6 ZnF-UBP domain binds ubiquitin by recognizing the unanchored C-terminal tail, with the vast majority of the interactions made exclusively through the C-terminal diglycine motif. This domain makes no specific interaction with ubiquitin side chains beyond the C-terminal tail; indeed, the ubiquitin residues beyond the diglycine motif became increasingly “mobile” as indicated by their crystallographic B-factors (Fig. 2E).
TABLE 1.
Crystallographic data collection and phasing statistics
| HDAC6 | HDAC + peptide RLRGG | HDAC6 + ubiquitin | |
|---|---|---|---|
| Protein Data Bank code | 3C5K | 3GV4 | 3PHD |
| Space group | P 21 2121 | P 212121 | P43212 |
| Unit cell | a = 40.5 b = 45.1 c = 55.8 | a = 31.7 b = 36.1 c = 89.0 | a = b = 133.7, c = 118.8 |
| Beamline | BNL X29A | RIGAKU FR-E | APS,23ID-B |
| Wavelengtha | 1.26 | 1.54 | 1.03317 |
| Resolution | 50–1.55 | 50–1.72 | 50–3.0 |
| Unique reflections | 10,033 | 10,865 | 22,204 |
| Data redundancy | 10.8 (6.0) | 5.8 (3.1) | 16.2 (16.5) |
| Completeness (%) | 98.0 (80.5) | 94.9 (72.5) | 100 (100) |
| I/σ(I) | 43.7 (5.8) | 28.0 (4.3) | 45.8 (4.7) |
| Rsymb | 0.071 (0.20) | 0.073 (0.23) | 0.091 (0.767) |
| Refinement | |||
| Resolution | 35.07–1.55 | 33.45–1.72 | 32.7–3.0 |
| Reflections used | 9546 | 10,213 | 22,135 |
| All atoms (solvent) | 1000 (156) | 1010 (205) | 3331 (2) |
| Rwork/Rfree (%)c | 17.3/22.7 | 17.4/23.0 | 23.5 (26.5) |
| Root mean square deviation | |||
| Bond length | 0.008 | 0.010 | 0.01 |
| Bond angle | 1.15 | 1.19 | 1.12 |
| Mean B factor | 10.9 | 22.0 | 72.9 |
| Molprobidity Ramachandran plot | |||
| Favored (%) | 97.2 | 96.0 | 93.98 |
| Allowed (%) | 100 | 100 | 99.8 |
| Outliers (%) | 0 | 0 | 1 |
a Apo-structure was collected at peak zinc wavelength.
b Rsym = sum(|I − <I>|)/sum(<I>), where I is the observed intensity, and <I> is the average intensity from multiple observations of symmetry-related reflections.
c Rfree value was calculated with ∼5% of the data.
FIGURE 2.
The structures of HDAC6 ZnF-UBP domain and its complexes with ubiquitin and RLRGG peptide. A, ribbon diagram of HDAC6 ZnF-UBP domain. The residues in the ubiquitin-binding site are shown in cyan. B, overlay of structures of HDAC6 ZnF-UBP domain (shown in ribbon representation) in complex with ubiquitin (yellow sticks) and RLRGG peptide (cyan sticks). C, structures of HDAC6 ZnF-UBP domain and its complex with RLRGG peptide, showing the ubiquitin binding pocket of HDAC6 ZnF-UBP domain. The last three residues of the peptide (RGG) are shown as sticks. D, overlay of the ubiquitin-binding site of HDAC6 ZnF-UBP (green) and its complex with RLRGG peptide (cyan). The peptide is shown in gray. The different conformations of Arg-1155 and Tyr-1156 can be clearly observed. E, the complex structure of HDAC6-UBP and RLRGG peptide, showing the hydrogen bond network. The peptide is colored by B-factors. The water molecules are shown in red. F, overlay of structures of HDAC6 ZnF-UBP (cyan) and its complex with RLRGG peptide (green), showing that minimal change occurred in the overall protein fold upon peptide binding. The protein is shown in ribbon representation, and the peptide is shown as sticks (yellow).
Polyubiquitinated Protein Aggregates Contain Unanchored Ubiquitin C Termini
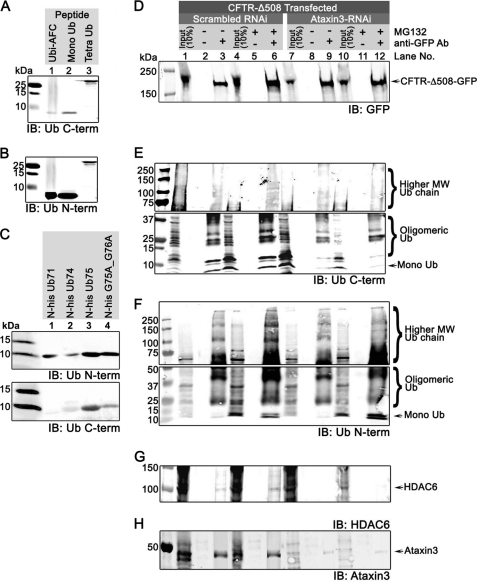
So far, our work has indicated that HDAC6 ZnF-UBP specifically recognizes ubiquitin C terminus and is capable of binding protein aggregates. This result further suggests that HDAC6 ZnF-UBP interacts with protein aggregates exclusively through unanchored ubiquitin C termini in the aggregates and not through binding polyubiquitinated proteins directly. To detect the presence of unanchored ubiquitin C termini in protein aggregates, we used a ubiquitin C terminus-specific antibody (anti-UbiC) to probe the immunoprecipitated aggregates resolved by SDS-PAGE. The specificity of the anti-UbiC antibody was confirmed by Western analysis (Fig. 3A) in which the antibody detected free ubiquitin C-terminal in pure mono- and tetraubiquitin peptides but only poorly detected the C-terminal AFC-conjugated ubiquitin. The low yet visible signal was likely due to contamination by a trace amount of unconjugated mono-ubiquitin, as seen from mass spectrometry analysis of the commercial ubiquitin-AFC sample (supplemental Fig. S3). AFC-conjugated ubiquitin appears as a peak at 8776 Da in the mass spectrum; however, we also detected a peak at 8565 Da that corresponds to unmodified ubiquitin with an unconjugated C terminus. This trace amount of unconjugated ubiquitin contributed to the low signal detected by the anti-UbiC antibody. When the same AFC-conjugated fraction was probed with an antibody specific for the ubiquitin N terminus (anti-UbiN), a robust signal was shown (Fig. 3B), suggesting that the anti-UbiC antibody recognized ubiquitin C-terminal with high preference. To further examine the specificity of anti-UbiC antibody, we probed various ubiquitin mutants with C-terminal truncations or point mutations with anti-UbiC and anti-UbiN antibody. As shown in Fig. 3C, anti-UbiN antibody could recognize all the peptides (Fig. 3C, upper panel); however, anti-UbiC antibody failed to recognize Ub71, where the last five amino acids were missing, and only poorly recognized the other truncated (Ub74) or mutant (G75A/G76A) ubiquitin peptides (Fig. 3C, lower panel). Collectively, these experiments demonstrated the specificity of the ubiquitin C-terminal (anti-UbiC) antibody.
FIGURE 3.
CFTR-ΔF508 protein aggregates contain unanchored ubiquitin generated by deubiquitinase ataxin-3. A–C, the specificity of the C-terminal ubiquitin anti-UbiC antibody. A and B, AFC-conjugated ubiquitin (lane 1), pure monoubiquitin (lane 2), and tetraubiquitin (lane 3) peptides were resolved by SDS-PAGE and detected in Western analysis by either anti-UbiC antibody (A) or anti-UbiN antibody (B). C, ubiquitin C-terminal truncation mutants Ub71 (lane 1), Ub74 (lane 2), and Ub75 (lane 3) and point mutant G75A/G76A were resolved by SDS-PAGE and detected in Western analysis by either anti-UbiN antibody (top panel) or anti-UbiC antibody (bottom panel). D–H, cultured 293T cells were transfected with GFP-CFTRΔF508 and either scrambled (lanes 1–6) or ataxin-3 siRNA (lanes 7–12) and treated with either Me2SO (lanes 1–3 and 7–9) or MG132 (lanes 4–6 and 10–12). Cell lysates were collected 2 days after transfection. GFP-CFTRΔF508 aggregates were immunoprecipitated with GFP antibody and resolved by SDS-PAGE and probed for GFP (D), ubiquitin C termini by anti-UbiC antibody (E), ubiquitin N termini by anti-UbiN antibody (F), HDAC6 (G), and ataxin-3 (H). IB, immunoblot; Ub, ubiquitin.
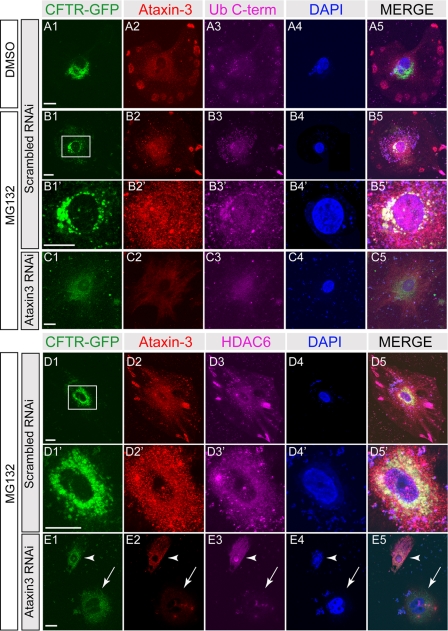
Next, we used the anti-UbiC antibody to detect the presence of unanchored ubiquitin in aggregates in cultured cells. When GFP-CFTR-ΔF508 mutant protein aggregates formed in cultured 293T cells were pulled down with an anti-GFP antibody, a significant amount of unanchored mono- and polyubiquitin (oligomeric-ubiquitin and higher molecular weight ubiquitins) were detected by anti-UbiC antibody (Fig. 3E, lanes 1–3), and the level of unanchored ubiquitin from mono- and polyubiquitin was enhanced with MG132 treatment (Fig. 3E, lanes 4–6), confirming the presence of unanchored ubiquitin C termini in protein aggregates. Note that the results pertaining to lanes 7–12 in Fig. 3E will be described in the following section. Next, we used the anti-UbiC antibody to examine the distribution of ubiquitin C termini in cells. To achieve better resolution, we used A549 cells for their larger cell morphology and compared their protein distribution with 293T cells. In untransfected A549 cells or 293T cells (supplemental Fig. S4), unconjugated ubiquitin C termini were evenly distributed throughout the cells without significant clustering (supplemental Fig. S4, A2 and B2). Expression of GFP-CFTR-ΔF508 caused protein aggregation (Fig. 4B, and supplemental Fig. S4, C and D). Unanchored ubiquitin C termini were detected in small clusters of GFP-CFTR-ΔF508 (supplemental Fig. S4, C2) and in large and perinuclear aggregates of GFP-CFTR-ΔF508 upon MG132 treatment, indicative of aggresome formation (Fig. 4, B and D, and supplemental Fig. S4, D2). Ubiquitin C termini were also detected in perinuclear clusters in MG132-treated cells that were not transfected with CFTR (supplemental Fig. S4, D2′, arrowheads), suggesting aggresome formation upon ubiquitin proteasome blockage in normal/untransfected cells. These results further confirm the presence of unanchored ubiquitin C termini in protein aggregates that can be accessed by the antibody.
FIGURE 4.
Unanchored ubiquitin was co-localized with ataxin-3 in CFTR-ΔF508 aggregates. A549 cells expressing GFP-CFTR-ΔF508 were transfected with either scrambled RNAi (A, B, and D) or ataxin-3 RNAi (C and E) and treated with either Me2SO (DMSO, A) or MG132 (B–E). The cells were labeled for GFP (green), ataxin-3 (red), C-terminal ubiquitin (Ub C-term, magenta in A–C), HDAC6 (magenta in D and E), and DAPI (blue). Perinuclear areas in B1 and D1 (white boxes) are shown in B′ and D′, respectively. The ataxin-3 siRNA-mediated knockdown efficiency varied among cells, because we observed that some cells have efficient knockdown (arrow in E), whereas other cells have significant residual levels of ataxin-3 (arrowhead in E). Scale bars, 10 μm.
Unanchored Ubiquitin C Termini Are Exposed by Deubiquitinase Ataxin-3
It has been reported that HDAC6 is involved in aggresome formation of only polyubiquitinated proteins, but not nonubiquitinated proteins, such as GFP-250 aggregates (11). Therefore, it is likely that unanchored ubiquitin tails are generated in situ from polyubiquitinated proteins by a deubiquitinase associated with aggregate particles. Among known deubiquitinases, ataxin-3 has been shown to interact with polyubiquitinated proteins (23) and to be involved in aggresome formation without a known role (24, 25). We therefore examined the presence and localization of ataxin-3 during aggresome formation using an ataxin-3-specific antibody (26–28). As shown in Fig. 3H, ataxin-3 was co-immunoprecipitated with GFP-CFTR-ΔF508 aggregates upon MG132 treatment, confirming that ataxin-3 is associated with aggregates (Fig. 3H, lanes 3 and 6). In unstressed cells, ataxin-3 was evenly distributed (Fig. 4, A2, and supplemental Fig. S4, A3) as previously reported (29). Upon MG132 treatment, ataxin-3 co-localized with GFP-CFTR-ΔF508 clusters and unanchored ubiquitin C termini (Fig. 4B and supplemental Fig. S4, D2 and D2′). To test whether ataxin-3 is required for producing unanchored ubiquitin C termini in protein aggregates, we performed ataxin-3 siRNA knockdown experiments. Using siRNA transfection-mediated knockdown, we were able to reduce the cellular ataxin-3 level to less than 27% of the normal level (supplemental Fig. S5). Consequently, the level of ataxin-3 associated with aggregates was reduced (Fig. 3H, lanes 9 and 12). In ataxin-3 knockdown cells, GFP-CFTR-ΔF508 showed a diffuse signal, indicating that aggresome formation was hindered (Fig. 4E and supplemental Fig. S4F). Although unanchored ubiquitin C termini were detected in knockdown cells, their co-localization with GFP-CFTR-ΔF508 was not observed, suggesting a lack of unanchored ubiquitin C termini in GFP-CFTR-ΔF508 aggregate particles. The importance of ataxin-3 in aggresome formation is further shown in Fig. 4E, where ataxin-3 knockdown in one of the cells was not efficient, leaving a significant amount of ataxin-3 to enable CFTR-ΔF508 aggresome formation (Fig. 4E, arrowhead). In contrast, a neighboring cell with efficient ataxin-3 knockdown failed to form aggresomes.
The reduced amount of unanchored ubiquitin C-terminal was further demonstrated by the Western analysis, in which the level of ubiquitin C termini associated with GFP-CFTR-ΔF508 aggregates in ataxin-3 siRNA-treated cells was reduced (Fig. 3E, lanes 9 and 12). Specifically, the amount of low molecular weight oligomeric ubiquitin and mono-ubiquitin with free C termini pulled down with aggresomes was significantly reduced (Fig. 3E, compare lanes 3 and 6 with lanes 9 and 12). However, the signal for high molecular weight ubiquitin chains was indistinguishable among lanes 3, 6, 9, and 12 because the anti-UbiC antibody did not recognize conjugated C termini present in the higher molecular weight ubiquitinated protein aggregates. To rule out the possibility that ataxin-3 siRNA treatment affected the expression of GFP-CFTR-ΔF508, we determined the protein level of GFP-CFTR-ΔF508 in siRNA-treated cells, and as shown in supplemental Fig. S6, the total level of GFP-CFTR-ΔF508 expressed in siRNA-treated cells was not affected by ataxin-3 knockdown. Furthermore, the amount of immunoprecipitated GFP-CFTR-ΔF508 was not affected by siRNA treatment, because similar amounts of GFP-CFTR-ΔF508 were precipitated with or without ataxin-3 knockdown (Fig. 3D). Interestingly, the amount of HDAC6 associated with GFP-CFTR-ΔF508 aggregates was reduced in ataxin-3 siRNA-treated cells (Fig. 3G, lanes 9 and 12), indicating that the interaction between HDAC6 and protein aggregates is dependent on the presence of exposed ubiquitin C termini generated by ataxin-3. To further confirm that the effect of ataxin-3 was restricted to generating unconjugated free C termini in the aggregates, we probed the aggregates pulled down with or without ataxin-3 knockdown with an antibody specific for ubiquitin N termini (anti-UbiN). As shown in Fig. 3B, anti-UbiN antibody robustly recognizes the ubi-AFC, where the C termini are capped by AFC conjugate. With anti-UbiN antibody, we show that altering ataxin-3 levels did not affect the levels of ubiquitin moieties expressed in cells (Fig. 3F, comparing input lanes 1 and 7, and lanes 4 and 10) or pulled down in the aggregates (Fig. 3F, comparing lanes 3 and 9 and lanes 6 and 12), suggesting that ataxin-3 knockdown did not reduce the total level of ubiquitin expressed in cells or the ubiquitin associated with protein aggregates but only specifically reduced the amount of unanchored ubiquitin C termini in protein aggregates. We further confirmed these observations using a different ubiquitinated protein substrate, human ataxin-1-82Q-GFP, which is known to form aggregates in cells when treated with MG132 (30). We transfected 293 cells with ataxin-1-82Q-GFP, treated with MG132 to induce aggregate formation, and siRNA to knock down ataxin-3. Similar to the results with GFP-CFTR-ΔF508 aggregates, we found that in ataxin-3 siRNA-treated cells, ataxin-3 levels were significantly reduced (supplemental Fig. S7E, lanes 7–12), there was a reduction of ubiquitin C termini (supplemental Fig. S7B), and ataxin-1-82Q-GFP failed to recruit HDAC6 (supplemental Fig. S7D, lanes 9 and 12). Collectively, these results support the notion that HDAC6 requires the unanchored ubiquitin C termini generated by ataxin-3 to interact with protein aggregates.
USP5 and USP13 are two known deubiquitinases with a specialized ZnF-UBP domain that recognizes the C-terminal diglycine motif in unattached chains (13, 31, 32). We examined the cellular distribution of these two well characterized deubiquitinases and found that neither of them co-localized with CFTR-ΔF508 aggresomes (supplemental Fig. S8), suggesting that they are unlikely to be involved in aggresome formation. This observation supports a specific role for deubiquitinase ataxin-3 in aggresome formation. Collectively, our results suggest a novel mechanism of ataxin-3-mediated HDAC6-dependent aggresome formation, where the aggregate-specific deubiquitinase, ataxin-3, cleaves (poly)ubiquitin moieties in polyubiquitinated proteins to expose their C-terminal diglycine motifs, providing a handle for HDAC6 binding and subsequent transport to the aggresome.
DISCUSSION
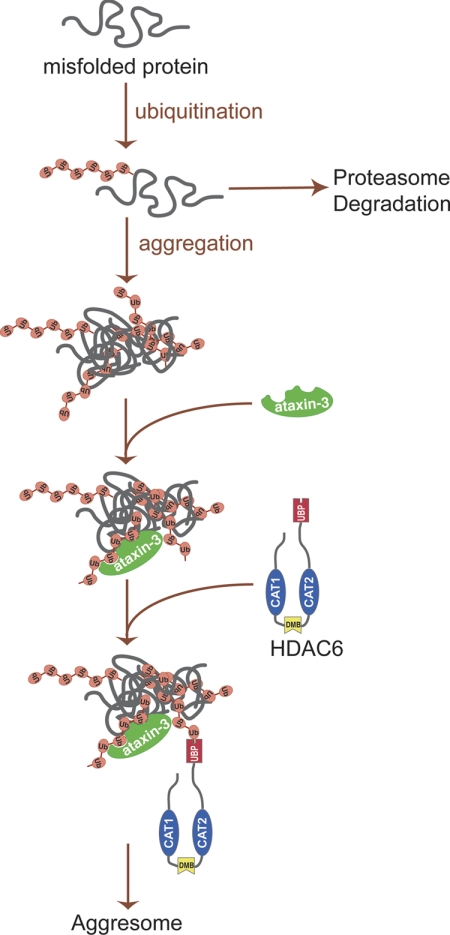
Our x-ray crystallography and biochemical results indicated that the HDAC6 ZnF-BUZ domain binds unconjugated diglycine motif at the C terminus of ubiquitin to promote aggresome formation. Using cellular analyses, we further showed that ataxin-3 is the deubiquitinase that generates the unanchored ubiquitin C termini in protein aggregates. The exposed ubiquitin C-terminal tails serve as “tags” for HDAC6 recognition and subsequent transport to the aggresome. Our work provides a critical piece of the mechanism underlying HDAC6-mediated aggresome formation (Fig. 5), where aggregate-associated ataxin-3 deubiquitinase cleaves polyubiquitin chains and exposes unanchored ubiquitin C termini for HDAC6 recognition of aggregate particles and their subsequent transport to form aggresomes.
FIGURE 5.
A schematic representation of aggresome formation. Under normal conditions, polyubiquitinated misfolded proteins are efficiently degraded by ubiquitin proteasome. When ubiquitin proteasome is disrupted or overwhelmed, polyubiquitinated misfolded proteins form aggregates. Deubiquitinase ataxin-3 interacts with polyubiquitinated protein aggregates, generating unanchored ubiquitin or ubiquitin chains. HDAC6 recognizes and binds these unanchored ubiquitin C-terminal tails in protein aggregates and recruits them to dynein motor complexes that subsequently transport the aggregated cargo to the aggresomes.
Previous work has suggested that HDAC6 binds protein aggregates via direct interactions with the polyubiquitin moiety through its ZnF-UBP domain (11). Our work indicates that such direct binding of HDAC6 to polyubiquitinated proteins is not feasible, because ubiquitin moieties are conjugated to protein substrates through C-terminal diglycine motifs (14). All known ZnF-UBP domains have been shown to bind unanchored ubiquitin C termini exclusively, and this structural property would preclude the possibility of HDAC6 to interact with polyubiquitinated misfolded proteins via its ZnF-UBP domain. Because protein aggregates consist primarily of polyubiquitinated proteins, an alternative mechanism has been proposed that HDAC6 ZnF-UBP may play a regulatory role rather than directly binding to protein aggregates and further suggested that there is probably another binding site on HDAC6 that interacts with polyubiquitinated proteins (12). Our work here demonstrated that HDAC6 ZnF-UBP alone interacts with protein aggregates, and such an interaction is dependent on intact diglycine-binding sites. This result is consistent with the earlier notion that the HDAC6 ZnF-UBP domain is responsible for binding protein aggregates (11). The fact that such an interaction is dependent on the intact diglycine-binding site points to a previously unknown possibility that protein aggregates contain unanchored ubiquitin or ubiquitin chains.
Protein aggregates need to be sequestered by aggresomes to avoid cytotoxicity. It is believed that protein aggregates are recruited to aggresomes by HDAC6. However, it has remained mysterious how scattered protein aggregates are recognized by HDAC6. Our biochemical, cellular, and imaging evidence unambiguously showed that the unconjugated ubiquitins are integrated parts of protein aggregates. These unconjugated ubiquitin tails are solvent-accessible on the surface of the aggregates, which can interact with HDAC6 ZnF-UBP domain or anti-UbiC antibody (Fig. 3). We provided evidence that unanchored ubiquitin chains with various lengths are presented in protein aggregates (Figs. 3 and 4), serving as “tags” for HDAC6 to recognize and bind. When the proteasome function is disrupted, ubiquitin is no longer “recycled” efficiently, which may deplete the free ubiquitin pool in the cytoplasm (33). The exposed unanchored ubiquitin C termini on protein aggregates thus act as “signals” for HDAC6 recognition. However, it remains unclear how the unanchored mono- or polyubiquitin chains are retained within protein aggregates without diffusing away.
An intriguing question is how the unanchored ubiquitin tails are exposed in aggregates. It has been shown that the ubiquitinase ataxin-3 is required for aggresome formation (24, 25), but it was unclear why the deubiquitination activity is essential for this process. Although the exact deubiquitination mechanism of ataxin-3 has yet to be fully elucidated, several reports have suggested that ataxin-3 is a polyubiquitin-editing enzyme that could cut within ubiquitin chains in polyubiquitinated proteins (23, 34). Our results show that ataxin-3 is one of the deubiquitinases responsible for exposing unanchored C termini on the aggregates. RNAi-mediated knockdown of ataxin-3 significantly reduces the amount of unanchored ubiquitin chains, which in turn eliminates HDAC6 binding (Fig. 3E). Therefore, our model defines an essential role for ataxin-3 deubiquination activity in aggresome formation.
In summary, our structural, biochemical, and cellular analyses demonstrate that the ZnF-UBP domain of HDAC6 binds unconjugated C-terminal diglycine motifs of ubiquitin and that this interaction is important for the binding and transport of polyubiquitinated protein aggregates. Aggresome formation is also dependent on ataxin-3, which is known to cleave polyubiquitin chains and to likely expose unanchored ubiquitin C termini for HDAC6 recognition of aggregate particles. Although proteins targeted for proteasome degradation are covalently tagged by ubiquitin through isopeptide linkages, our findings suggest that protein aggregates targeted for aggresomal clearance are tagged noncovalently by unanchored ubiquitin C termini.
The aggresome pathway has emerged as a potential therapeutic target for cancer treatment (35, 36). Our studies suggest the ZnF-UBP domain of HDAC6 as a site of interest. The deep and hydrophobic C-terminal ubiquitin-binding site of HDAC6 represents an attractive site for the development of small molecule antagonists.
Supplementary Material
Acknowledgments
We thank Drs. James E. Bradner, Brian Raught, Aled Edwards, Dalia Barsyte, and Brant Watson for critically reviewing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R01NS064269 (to R. G. Z.). This work was also supported by Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust.

This article contains supplemental Table S1 and Figs. S1–S8.
The atomic coordinates and structure factors (codes 3C5K, 3GV4, and 3PHD) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- HDAC
- histone deacetylase
- Ni-NTA
- nickel-nitrilotriacetic acid
- DLS
- dynamic light scattering.
REFERENCES
- 1. Goldberg A. L. (2003) Nature 426, 895–899 [DOI] [PubMed] [Google Scholar]
- 2. Tai H. C., Schuman E. M. (2008) Nat. Rev. Neurosci. 9, 826–838 [DOI] [PubMed] [Google Scholar]
- 3. Johnston J. A., Ward C. L., Kopito R. R. (1998) J. Cell Biol. 143, 1883–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garcia-Mata R., Gao Y. S., Sztul E. (2002) Traffic 3, 388–396 [DOI] [PubMed] [Google Scholar]
- 5. Chin L. S., Olzmann J. A., Li L. (2010) Biochem. Soc. Trans 38, 144–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee J. Y., Koga H., Kawaguchi Y., Tang W., Wong E., Gao Y. S., Pandey U. B., Kaushik S., Tresse E., Lu J., Taylor J. P., Cuervo A. M., Yao T. P. (2010) EMBO J. 29, 969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pandey U. B., Nie Z., Batlevi Y., McCray B. A., Ritson G. P., Nedelsky N. B., Schwartz S. L., DiProspero N. A., Knight M. A., Schuldiner O., Padmanabhan R., Hild M., Berry D. L., Garza D., Hubbert C. C., Yao T. P., Baehrecke E. H., Taylor J. P. (2007) Nature 447, 859–863 [DOI] [PubMed] [Google Scholar]
- 8. Matthias P., Yoshida M., Khochbin S. (2008) Cell Cycle 7, 7–10 [DOI] [PubMed] [Google Scholar]
- 9. Valenzuela-Fernández A., Cabrero J. R., Serrador J. M., Sánchez-Madrid F. (2008) Trends Cell Biol. 18, 291–297 [DOI] [PubMed] [Google Scholar]
- 10. Dompierre J. P., Godin J. D., Charrin B. C., Cordelières F. P., King S. J., Humbert S., Saudou F. (2007) J. Neurosci 27, 3571–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kawaguchi Y., Kovacs J. J., McLaurin A., Vance J. M., Ito A., Yao T. P. (2003) Cell 115, 727–738 [DOI] [PubMed] [Google Scholar]
- 12. Pai M. T., Tzeng S. R., Kovacs J. J., Keaton M. A., Li S. S., Yao T. P., Zhou P. (2007) J. Mol. Biol. 370, 290–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reyes-Turcu F. E., Horton J. R., Mullally J. E., Heroux A., Cheng X., Wilkinson K. D. (2006) Cell 124, 1197–1208 [DOI] [PubMed] [Google Scholar]
- 14. Finley D., Chau V. (1991) Annu. Rev. Cell Biol. 7, 25–69 [DOI] [PubMed] [Google Scholar]
- 15. Hicke L., Schubert H. L., Hill C. P. (2005) Nat. Rev. Mol. Cell Biol. 6, 610–621 [DOI] [PubMed] [Google Scholar]
- 16. Raiborg C., Slagsvold T., Stenmark H. (2006) Trends Biochem. Sci. 31, 541–544 [DOI] [PubMed] [Google Scholar]
- 17. Emsley P., Cowtan K. (2004) Acta Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 18. Bricogne G. (1993) Acta Crystallogr. 49, 37–60 [DOI] [PubMed] [Google Scholar]
- 19. Bonnet J., Romier C., Tora L., Devys D. (2008) Trends Biochem. Sci. 33, 369–375 [DOI] [PubMed] [Google Scholar]
- 20. Hochstrasser M. (1996) Annu. Rev. Genet. 30, 405–439 [DOI] [PubMed] [Google Scholar]
- 21. Boyault C., Gilquin B., Zhang Y., Rybin V., Garman E., Meyer-Klaucke W., Matthias P., Müller C. W., Khochbin S. (2006) EMBO J. 25, 3357–3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boyault C., Sadoul K., Pabion M., Khochbin S. (2007) Oncogene 26, 5468–5476 [DOI] [PubMed] [Google Scholar]
- 23. Burnett B., Li F., Pittman R. N. (2003) Hum. Mol. Genet. 12, 3195–3205 [DOI] [PubMed] [Google Scholar]
- 24. Burnett B. G., Pittman R. N. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4330–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Donaldson K. M., Li W., Ching K. A., Batalov S., Tsai C. C., Joazeiro C. A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8892–8897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Menzies F. M., Huebener J., Renna M., Bonin M., Riess O., Rubinsztein D. C. (2010) Brain 133, 93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Paulson H. L., Perez M. K., Trottier Y., Trojanowski J. Q., Subramony S. H., Das S. S., Vig P., Mandel J. L., Fischbeck K. H., Pittman R. N. (1997) Neuron 19, 333–344 [DOI] [PubMed] [Google Scholar]
- 28. Trottier Y., Cancel G., An-Gourfinkel I., Lutz Y., Weber C., Brice A., Hirsch E., Mandel J. L. (1998) Neurobiol. Dis. 5, 335–347 [DOI] [PubMed] [Google Scholar]
- 29. Pozzi C., Valtorta M., Tedeschi G., Galbusera E., Pastori V., Bigi A., Nonnis S., Grassi E., Fusi P. (2008) Neurobiol. Dis. 30, 190–200 [DOI] [PubMed] [Google Scholar]
- 30. Zhai R. G., Zhang F., Hiesinger P. R., Cao Y., Haueter C. M., Bellen H. J. (2008) Nature 452, 887–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Amerik A., Swaminathan S., Krantz B. A., Wilkinson K. D., Hochstrasser M. (1997) EMBO J. 16, 4826–4838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Komander D., Clague M. J., Urbé S. (2009) Nat. Rev. Mol. Cell Biol. 10, 550–563 [DOI] [PubMed] [Google Scholar]
- 33. Mimnaugh E. G., Chen H. Y., Davie J. R., Celis J. E., Neckers L. (1997) Biochemistry 36, 14418–14429 [DOI] [PubMed] [Google Scholar]
- 34. Mao Y., Senic-Matuglia F., Di Fiore P. P., Polo S., Hodsdon M. E., De Camilli P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12700–12705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Olzmann J. A., Li L., Chin L. S. (2008) Curr. Med. Chem. 15, 47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rodriguez-Gonzalez A., Lin T., Ikeda A. K., Simms-Waldrip T., Fu C., Sakamoto K. M. (2008) Cancer Res. 68, 2557–2560 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.