Abstract
Cleft palate represents one of the most common congenital birth defects. Transforming growth factor β (TGFβ) signaling plays crucial functions in regulating craniofacial development, and loss of TGFβ receptor type II in cranial neural crest cells leads to craniofacial malformations, including cleft palate in mice (Tgfbr2fl/fl;Wnt1-Cre mice). Here we have identified candidate target genes of TGFβ signaling during palatal formation. These target genes were selected based on combining results from gene expression profiles of embryonic day 14.5 palates from Tgfbr2fl/fl;Wnt1-Cre mice and previously identified cleft palate phenotypes in genetically engineered mouse models. We found that fibroblast growth factor 9 (Fgf9) and transcription factor pituitary homeobox 2 (Pitx2) expressions are significantly down-regulated in the palate of Tgfbr2fl/fl;Wnt1-Cre mice, and Fgf9 and Pitx2 loss of function mutations result in cleft palate in mice. Pitx2 expression is down-regulated by siRNA knockdown of Fgf9, suggesting that Fgf9 is upstream of Pitx2. We detected decreased expression of both cyclins D1 and D3 in the palates of Tgfbr2fl/fl;Wnt1-Cre mice, consistent with the defect in cell proliferation. Significantly, exogenous FGF9 restores expression of cyclins D1 and D3 in a Pitx2-dependent manner and rescues the cell proliferation defect in the palatal mesenchyme of Tgfbr2fl/fl;Wnt1-Cre mice. Our study indicates that a TGFβ-FGF9-PITX2 signaling cascade regulates cranial neural crest cell proliferation during palate formation.
Keywords: Cell Proliferation, Craniofacial Development, Development, Molecular Cell Biology, Mouse Genetics
Introduction
Cleft palate represents one of the most frequent congenital birth defects in the human population (1). The causes of cleft palate remain largely unknown, but they appear to be complex, including genetic and environmental factors (2, 3). Cleft palate causes many clinical symptoms and complications, such as difficulties with suckling and eating, dysfunction of tongue and oral muscles, dental abnormalities, and speech and language delay (4). Therefore, prevention of cleft palate is the ultimate objective, and a prerequisite of this aim is to elucidate the mechanisms of healthy palate development and the causes of cleft palate.
The majority of cells in the craniofacial region are derived from cranial neural crest cells (CNCC),2 which produce the facial skeleton, smooth muscle, and sensory nerve. Transforming growth factor β (TGFβ) signaling plays crucial functions in craniofacial development, including palate formation by mediating cell proliferation, differentiation, and extracellular matrix formation via regulation of downstream target genes (5–7). TGFβ ligands activate the membrane receptor serine/threonine kinase complex composed of TGFβ receptor type II (TβRII) and TGFβ receptor type I (TβRI/ALK5). The ligand-receptor complex phosphorylates SMAD2 and SMAD3, which form a transcriptional complex with SMAD4, and then the transcriptional complex translocates into the nucleus to control the expression of downstream target genes (8–10). TβRII is expressed in the CNCC-derived palatal mesenchyme (11, 12), and targeted null mutation of Tgfbr2 in CNCC leads to craniofacial malformations, including small mandible, dysmorphic calvaria, and cleft palate (13–18). In humans, mutations in TGFBR2 cause the multisystemic Loeys-Dietz syndrome (OMIM number 609192), which includes craniofacial malformations, such as cleft palate.
Our previous study showed that conditional gene ablation of Tgfbr2 in CNCC (Tgfbr2fl/fl;Wnt1-Cre mice) results in cleft palate with 100% phenotype penetrance, down-regulated cyclin D1 expression, and decreased CNCC proliferation in the palatal mesenchyme at embryonic day 14.5 (E14.5) (13). TGFβ signaling interacts with other growth factors, such as bone morphogenetic protein, Wnt, and fibroblast growth factor (FGF) (19). Although this interplay of pathways appears to be critical for the regulation of palate formation, little is known regarding the developmental molecular pathways that regulate cell proliferation during palatogenesis. Here we show that TGFβ regulates cell proliferation via the FGF9-PITX2 signaling cascade during palate formation.
EXPERIMENTAL PROCEDURES
Animals
Mating Tgfbr2fl/+;Wnt1-Cre with Tgfbr2fl/fl mice generated Tgfbr2fl/fl;Wnt1-Cre conditional null alleles that were genotyped using PCR primers as described previously (13). All mouse embryos used in this study were maintained in a C57BL/6J background. Animal usage was approved by the Institutional Animal Care and Use Committee at the University of Southern California.
Immunological Analysis
Immunoblots were performed as described previously (17). Antibodies used for immunoblotting were as follows: rabbit polyclonal antibodies against cyclin D1, cyclin D3, PITX2, and FGF9 (Cell Signaling Technology) and mouse monoclonal antibody against GAPDH (Chemicon).
Histological Examination
BrdU staining was performed as described previously (13, 14, 17). Immunohistochemical staining was performed as described previously (20, 21). Rabbit polyclonal antibodies used for immunohistochemistry were anti-cyclin D1 and anti-cyclin D3 (Cell Signaling Technology).
RNA Preparation and Quantitative RT-PCR
As described in a different analysis of these same samples,3 total RNA was isolated from mouse embryonic palate dissected at the indicated developmental stage or from primary mouse embryonic palatal mesenchymal (MEPM) cells. First-strand cDNA was synthesized from 2 μg of total RNA using an oligo(dT)20 primer and SuperScript III reverse transcriptase (Invitrogen). RNA was quantified and tested for quality by photometric measurement. Quantitative PCR was performed in triplicate by SYBR Green (Bio-Rad) in an iCycler (Bio-Rad) as previously described (17). A melting curve was obtained for each PCR product after each run to confirm that the SYBR Green signal corresponded to a unique and specific amplicon. The relative abundance of each transcript was calculated based on PCR efficiency and cycle number at which the fluorescence crosses a threshold for the GAPDH internal reference, and the gene was tested using iCycler iQ optical system software (Bio-Rad). PCR primers are available upon request.
Microarray Analysis
As described for these same samples,3 total RNA samples (1 μg/sample) were converted into biotin-labeled cRNA using the GeneChip® IVT labeling kit and standard protocols recommended by Affymetrix (Santa Clara, CA). Fragmented cDNA was applied to GeneChip® mouse genome 430 2.0 arrays (Affymetrix) that contain probe sets designed to detect over 39,000 transcripts. Microarrays were hybridized, processed, and scanned, as described previously using the manufacturer's recommended conditions (22). WebArray software was used to generate scaled log2 transformed gene expression values using the RMA algorithm (23, 24). Probes sets showing >1.5-fold differential expression with a <5% false discovery rate (FDR) were identified through LIMMA (linear models for microarray data)-based linear model statistical analysis, and FDR calculations were made using the SPLOSH (spacings LOESS histogram) method. All scaled gene expression scores, and .cel files are available at the National Center for Biotechnology Information GEO (Gene Expression Omnibus) repository under series accession number GSE22989.
Comparative Analysis of Transcription Factor Binding Sites
We used the University of California Santa Cruz genome browser (available on the University of California Santa Cruz Web site) to obtain the genomic sequences of the human FGF9 (RefSeq accession NM_002010.2) and PITX2, transcript variant 1 (RefSeq accession NM_153427.2) genes, including 5 kb upstream and 5 kb downstream of the respective transcription start sites, based on human genome Build 19. These sequences were then mapped to seven additional mammalian genomes (chimpanzee (Build 2.1.3), orangutan (Build 2.0.2), rhesus macaque (Build 1.0), mouse (Build 37), rat (Build 3.4), dog (Build 2), and horse (Build equCab2)) through the Blat tool (available on the University of Santa Cruz Web site). We obtained multiple alignments for these sequences using the ClustalW2 tool (available on the EMBL-EBI Web site) with default parameters and settings (25). To account for uncertainty in the quality of the horse and dog draft genome sequences, we performed sequence alignments with and without information from these species. We searched for transcription factor binding motifs relevant to SMAD and p38 MAPK pathway elements within or proximal to these genes (i.e. 5 kb upstream and 5 kb downstream of the human transcription start site for each gene) that are conserved across diverse mammals. We focused our analysis of canonical TGFβ signaling on the 5′-GTCT-3′, 5′-AGAC-3′, and 5′-GTCTAGAC-3′ SMAD recognition sites (26, 27). For our analysis of non-canonical TGFβ signaling, we focused on the well defined recognition sequences of ATF family members (5′-TGACGTCA-3′, as reported in Ref. 28) as well as that of ATF2 CRE-binding heterodimers, which recognize the 8-base sequence 5′-TGACGTAG-3′ (29) and related sequence variants 5′-TGACG(Y/M/R)-3′ (30, 31).
In Situ Hybridizations
Fgf9 and Pitx2 expression patterns in the palate were examined by in situ hybridization performed using methods described previously (14, 17). Pitx2 and Fgf9 probes were gifts from Drs. Liu and Thesleff, respectively (32, 33). Several negative controls (e.g. sense probe and no probe) were run in parallel with the experimental reaction. Cryosections were pretreated with proteinase K (Sigma), refixed in fresh 4% PFA, 0.2% glutaraldehyde in phosphate-buffered saline (PBS), and then prehybridized for 5 h at 55 °C in a hybridization buffer including 50% formamide. After hybridization, tissues were washed in high stringency conditions and preblocked in antibody blocking solution and then incubated with preabsorbed antibody. Color development was performed in nitro blue tetrazolium/5-bromo-4-chloro-3′-indolylphosphate color development solution (Sigma). Following visualization, the tissues were postfixed and cleared in 50% glycerol before photography.
Small Interfering RNA (siRNA) Transfection
MEPM cells (2 × 106 cells) were plated in a 6-well cell culture plate until the cells reached 60–80% confluence. siRNA duplex and reagents were purchased from Invitrogen and Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), respectively. The siRNA mixture in transfection medium was incubated with cells for 6 h at 37 °C in a CO2 incubator.
Organ Culture of Palate and FGF9 Bead Implantation
Timed pregnant mice were sacrificed at E13.5. Genotyping was carried out as described above. The palatal shelves were microdissected and cultured in serum-free chemically defined medium as described previously (13). To remove the epithelium, palatal shelves were treated with 0.05% trypsin, EDTA for 10 min prior to careful dissection. After 24 h in culture with p38 mitogen-activated protein kinase (MAPK) inhibitor SB203580 (100 μm), palates were harvested, fixed in 4% paraformaldehyde, 0.1 m phosphate buffer (pH 7.4), and processed. Heparin beads (Sigma) were used for delivery of FGF9 protein. The beads were washed in PBS and then incubated for 1 h at room temperature in 100 μg/ml FGF9 (R&D Systems). The FGF9-containing beads were delivered into palatal explants. Control beads were incubated with 0.1% bovine serum albumin (BSA). FGF9- or BSA-containing beads were placed into palatal explants.
Primary Cultured Cells Derived from Mouse Embryonic Palatal Mesenchyme
Primary MEPM cells were obtained from 13.5-day-old embryos. Palates were dissected at E13.5 and trypsinized for 30 min at 37 °C in a CO2 incubator. After pipetting thoroughly, cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum supplemented with penicillin, streptomycin, l-glutamate, sodium pyruvate, and non-essential amino acids. MEPM cells were treated with or without p38 MAPK inhibitor SB203580 at 10 μm for 24 h.
Statistical Analysis
Two-tailed Student's t test was applied for statistical analysis. For all graphs, data are represented as mean ± S.D. A p value of <0.05 was considered statistically significant.
RESULTS
Selection of TGFβ Downstream Signaling Molecules Crucial for Palatogenesis
To investigate the cause of cleft palate resulting from TGFBR2 mutations in humans, we examined mice with CNCC-specific conditional inactivation of Tgfbr2 (Tgfbr2fl/fl;Wnt1-Cre). As will be described in detail,4 we obtained gene expression profiles of E14.5 palate from Tgfbr2fl/fl;Wnt1-Cre and Tgfbr2fl/fl control mice using Affymetrix U430 2.0 GeneChips (during palatal fusion, n = 5 per genotype). We chose this stage because it is the time at which normal mouse palatal fusion takes place. Overall, we found that a total of 293 transcripts were differentially expressed (149 were up-regulated, and 144 were down-regulated; ≥1.5-fold, <5% FDR) in Tgfbr2fl/fl;Wnt1-Cre mice relative to the controls (Fig. 1A). Although TGFβ signaling interacts with other growth factors, such as bone morphogenetic protein, Wnt, and FGF family members, Fgf9 was the only member of these growth factor families that was differentially expressed in E14.5 Tgfbr2fl/fl;Wnt1-Cre palates relative to controls (1.52-fold down-regulated, FDR = 0.011).
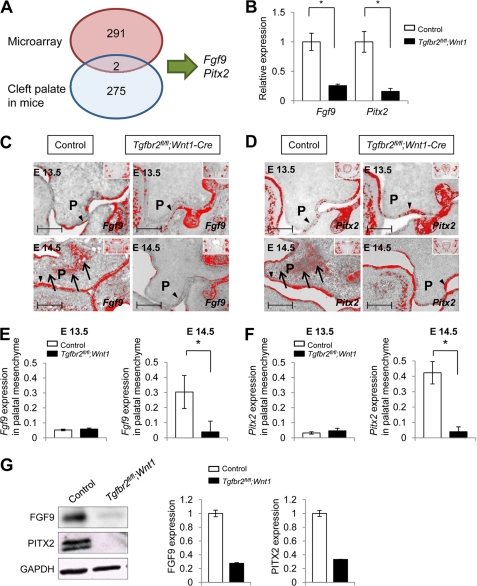
FIGURE 1.
Fgf9 and Pitx2 are down-regulated in Tgfbr2fl/fl;Wnt1-Cre mice at E14.5. A, diagram depicts the strategy used to identify Fgf9 and Pitx2 genes in our approach using the combination of E14.5 palate microarray analysis and known mutations in mice that result in cleft palate phenotypes. B, quantitative RT-PCR analysis of Fgf9 and Pitx2 in Tgfbr2fl/fl;Wnt1-Cre (closed columns) mice compared with Tgfbr2fl/fl control mice (open columns) in the palate at E14.5. *, p < 0.05. C and D, in situ hybridization of Fgf9 (C) and Pitx2 (D) in Tgfbr2fl/fl;Wnt1-Cre and Tgfbr2fl/fl mice at E13.5 and E14.5. The arrows indicate expression of Fgf9 and Pitx2 in the palatal mesenchyme of Tgfbr2fl/fl control mice. Arrowheads, Fgf9 and Pitx2 expression in the palatal epithelium of Tgfbr2fl/fl control and Tgfbr2fl/fl;Wnt1-Cre mice. Insets show low magnification of craniofacial regions to indicate global expression pattern of Fgf9 and Pitx2. P, palatal shelf. Bar, 50 μm. E and F, quantitative RT-PCR analysis of Fgf9 (E) and Pitx2 (F) in the palatal mesenchyme of E13.5 and E14.5 Tgfbr2fl/fl control (open columns) and Tgfbr2fl/fl;Wnt1-Cre (closed columns) mice. *, p < 0.05. G, immunoblotting analysis of FGF9 and PITX2 in E14.5 Tgfbr2fl/fl control and Tgfbr2fl/fl;Wnt1-Cre palates. The graph shows quantitative densitometry analysis of the immunoblotting data. Error bars, S.D.
Next, we created a list of various genetically engineered mouse models known to exhibit the cleft palate phenotype during embryogenesis, as originally compiled by Mouse Genome Informatics (MGI) and modified by us based on the literature. Currently, this includes 277 different genetically engineered mice with cleft palate (Fig. 1A and supplemental Table 1). To screen for candidate genes related to cleft palate and TGFβ signaling, we compared the list of known mutant mouse models exhibiting cleft palate and the microarray analysis performed on E14.5 Tgfbr2fl/fl;Wnt1-Cre palates. We identified two overlapping genes (Fgf9 and Pitx2) in these lists (Fig. 1A and supplemental Table 1), which were both predicted to be less highly expressed in the palates of Tgfbr2fl/fl;Wnt1-Cre mice relative to those of controls (Fgf9, 1.52-fold reduced, FDR = 0.011; Pitx2, 1.74-fold reduced, FDR = 0.006). Fgf9-null and Pitx2-null mice have been reported to show cleft palate (34, 35). We confirmed the reduction of Fgf9 and Pitx2 gene expression in Tgfbr2fl/fl;Wnt1-Cre mice by quantitative RT-PCR using palates of Tgfbr2fl/fl;Wnt1-Cre and Tgfbr2fl/fl control mice. Gene expression of both Fgf9 and Pitx2 was significantly down-regulated in Tgfbr2fl/fl;Wnt1-Cre mice relative to control mice (Fig. 1B).
We performed in situ hybridization to investigate the expression patterns of Fgf9 and Pitx2 during palate formation. The expression of Fgf9 was localized in the palatal epithelium in both Tgfbr2fl/fl;Wnt1-Cre and Tgfbr2fl/fl control mice at E13.5 (Fig. 1C). At E14.5, Fgf9 gene expression was detectable in both the palatal epithelium and mesenchyme in Tgfbr2fl/fl control mice; however, Fgf9 gene expression was not detectable in the palatal mesenchyme of E14.5 Tgfbr2fl/fl;Wnt1-Cre mice, although it remained detectable in the palatal epithelium (Fig. 1, C and E, and supplemental Fig. 1). Pitx2 gene expression was detectable in the palatal epithelium of both Tgfbr2fl/fl;Wnt1-Cre and Tgfbr2fl/fl control mice at E13.5 and E14.5. In Tgfbr2fl/fl control mice, Pitx2 expression was also detectable in the palatal mesenchyme at E14.5 (Fig. 1, D and F, and supplemental Fig. 1). In E14.5 Tgfbr2fl/fl;Wnt1-Cre mice, Pitx2 expression was not detectable in the palatal mesenchyme (Fig. 1, D and F, and supplemental Fig. 1). The expression patterns were essentially the same for both Fgf9 and Pitx2. We also analyzed FGF9 and PITX2 protein expression in the palates of Tgfbr2fl/fl;Wnt1-Cre and Tgfbr2fl/fl control mice at E14.5 and found that they were both reduced in Tgfbr2fl/fl;Wnt1-Cre palates (Fig. 1G and supplemental Fig. 2).
Fgf9 Regulates Pitx2 Gene Expression via SMAD-independent TGFβ Signaling during Palate Formation
Here, we investigated the means by which TGFβ regulated Fgf9 and Pitx2 expressions. There are at least two possibilities; one is through the non-canonical TGFβ signaling (such as p38 MAPK activation), and another is through the canonical SMAD-dependent pathway. Our recent study showed that loss of Tgfbr2 in CNCC resulted in an activation of an alternative TGFβ signaling cascade that involved the activation of p38 MAPK.3 We hypothesized that blocking p38 MAPK activation will restore the gene expression of Fgf9 and Pitx2 in the palatal mesenchyme of Tgfbr2fl/fl;Wnt1-Cre mice. To test this, we analyzed Fgf9 and Pitx2 gene expression after treatment with p38 MAPK inhibitor SB203580 using primary MEPM cells derived from the palates of Tgfbr2fl/fl;Wnt1-Cre and Tgfbr2fl/fl control mice. We found that Fgf9 and Pitx2 expression increased in Tgfbr2fl/fl;Wnt1-Cre MEPM cells treated with p38 MAPK inhibitor to a level comparable with that of Tgfbr2fl/fl control MEPM cells, consistent with Fgf9 and Pitx2 down-regulation by p38 MAPK activation in the absence of Tgfbr2 (Fig. 2A). Moreover, we added p38 MAPK inhibitor to an ex vivo palate organ culture system and confirmed that gene expression of Fgf9 and Pitx2 in Tgfbr2fl/fl;Wnt1-Cre palates increased after treatment (Fig. 2B and supplemental Fig. 3).
FIGURE 2.
Fgf9 regulates the gene expression of Pitx2. A, quantitative RT-PCR analysis of Fgf9 and Pitx2 after treatment (+) or no treatment (−) with p38 MAPK inhibitor of primary MEPM cells from Tgfbr2fl/fl control (open columns) and Tgfbr2fl/fl;Wnt1-Cre (closed columns) mice. *, p < 0.05. B, quantitative RT-PCR analysis of Fgf9 and Pitx2 after treatment (+) or no treatment (−) with p38 MAPK inhibitor in organ culture of E14.5 palatal mesenchyme from Tgfbr2fl/fl control (open columns) and Tgfbr2fl/fl;Wnt1-Cre (closed columns) mice. *, p < 0.05. C and D, gene expressions of Fgf9 (C) and Pitx2 (D) after siRNA knockdown of Fgf9 in primary MEPM cells from Tgfbr2fl/fl mice. *, p < 0.05. Antisense siRNA treatment was used as control (Ctrl). *, p < 0.05. E and F, gene expression of Fgf9 (E) and Pitx2 (F) after siRNA knockdown for Pitx2 in primary MEPM cells from Tgfbr2fl/fl mice. *, p < 0.05. Antisense siRNA treatment was used as control (Ctrl). Error bars, S.D.
Next, to explore the mechanism by which Fgf9 and Pitx2 interact with each other during palate development, we reduced the gene expression of Fgf9 or Pitx2 in primary MEPM cells using a siRNA knockdown approach (Fig. 2, C–F). Gene expression of Fgf9 and Pitx2 was successfully suppressed by the siRNA treatment by 73 and 54%, respectively (Fig. 2, C and E). We found that gene expression of Pitx2 was significantly decreased 3.5-fold after Fgf9 siRNA treatment, but Pitx2 siRNA treatment had no effect on gene expression of Fgf9 (Fig. 2, D and F). These data suggest that Fgf9 may regulate the gene expression of Pitx2 in the palatal mesenchyme during palatogenesis.
TGFβ Signaling Regulates Expression of Cyclins D1 and D3 via SMAD-independent Pathway during Palate Formation
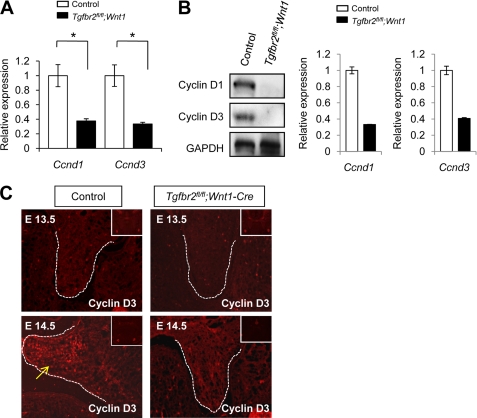
Our previous work demonstrated that cyclin D1 expression is down-regulated in the palatal mesenchyme in Tgfbr2fl/fl;Wnt1-Cre mice at E14.5 (13). Therefore, we investigated the gene expression of cyclin D isoforms at E14.5 and found that both cyclins D1 and D3 were significantly down-regulated in palates from E14.5 Tgfbr2fl/fl;Wnt1-Cre mice (Fig. 3A). Cyclins D1 and D3 protein levels were also down-regulated in Tgfbr2fl/fl;Wnt1-Cre palates at E14.5 (Fig. 3B). Furthermore, we detected cyclin D3 in the palatal mesenchyme of E14.5 Tgfbr2fl/fl control mice, but we failed to detect them in the palates of Tgfbr2fl/fl;Wnt1-Cre mice at E14.5 (Fig. 3C). Significantly, decreased gene expression of Ccnd1 and Ccnd3 in Tgfbr2fl/fl;Wnt1-Cre MEPM cells was restored by treatment with p38 MAPK inhibitor SB203580, suggesting that the SMAD-independent p38 MAPK pathway regulates gene expression of Ccnd1 and Ccnd3 in Tgfbr2fl/fl;Wnt1-Cre mice (Fig. 4A).
FIGURE 3.
Expression of cyclins D1 and D3 is decreased in Tgfbr2fl/fl;Wnt1-Cre mice during palatal development. A, quantitative RT-PCR analysis of cyclins D1 and D3 (Ccnd1 and Ccnd3) in the palate of Tgfbr2fl/fl control (open columns) and Tgfbr2fl/fl;Wnt1-Cre (closed columns) mice. *, p < 0.05. B, Western blot of cyclins D1 and D3 (Ccnd1 and Ccnd3) in the palate of Tgfbr2fl/fl control and Tgfbr2fl/fl;Wnt1-Cre mice at E14.5. The graph shows quantitative densitometry analysis of the immunoblotting data. C, immunohistochemical analysis of cyclin D3 in the palate of Tgfbr2fl/fl control and Tgfbr2fl/fl;Wnt1-Cre mice at E13.5 and E14.5. The arrows indicate expression of cyclin D3 in the palatal mesenchyme of Tgfbr2fl/fl mice at E14.5. Dotted lines, palate. Insets, low magnification of craniofacial regions to indicate global expression pattern of cyclin D3. Error bars, S.D.
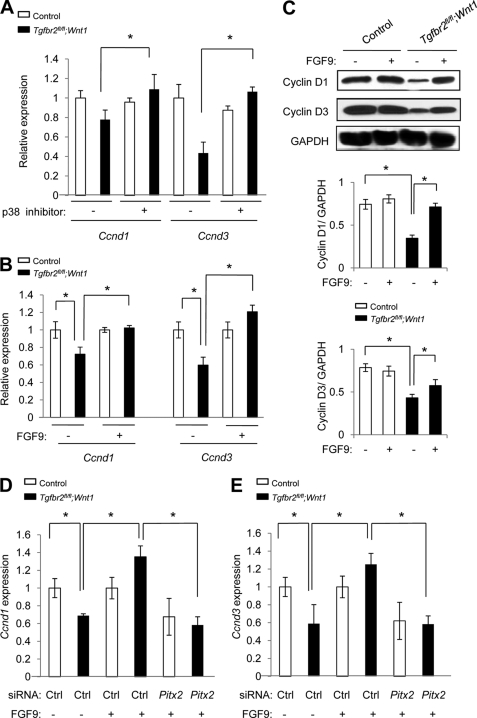
FIGURE 4.
TGFβ-mediated FGF9-PITX2 signaling regulates gene expression of Ccnd1 and Ccnd3 in palatal mesenchymal cells. A, quantitative RT-PCR analysis of cyclins D1 (Ccnd1) and D3 (Ccnd3) in MEPM cells of Tgfbr2fl/fl control (open columns) and Tgfbr2fl/fl;Wnt1-Cre (closed columns) mice with (+) or without (−) p38 MAPK inhibitor. *, p < 0.05. B, quantitative RT-PCR analysis of cyclins D1 and D3 in primary MEPM cells of Tgfbr2fl/fl control (open columns) and Tgfbr2fl/fl;Wnt1-Cre (closed columns) mice after treatment (+) or no treatment (−) with FGF9 protein. *, p < 0.05. C, Western blots of cyclins D1 and D3 after treatment (+) or no treatment (−) with FGF9 protein. The graphs (below) show quantitative densitometry analysis of the immunoblotting data. *, p < 0.05. D and E, quantitative RT-PCR analysis of cyclins D1 (D; Ccnd1) and D3 (E; Ccnd3) in MEPM cells of Tgfbr2fl/fl control (open columns) and Tgfbr2fl/fl;Wnt1-Cre (closed columns) mice after mock treatment (Ctrl), treatment with FGF9 protein (+), or treatment with a combination of FGF9 protein and siRNA for Pitx2 for 24 h. *, p < 0.05. Error bars, S.D.
To investigate the role of FGF9 in TGFβ-mediated cell proliferation during palate formation, we analyzed the gene expression of cyclins D1 and D3 after treatment with FGF9 protein using the ex vivo palate organ culture system. Decreased gene expression of cyclins D1 and D3 in Tgfbr2fl/fl;Wnt1-Cre palate was restored to control levels after FGF9 treatment (Fig. 4B). Protein levels of cyclins D1 and D3 were also restored after FGF9 treatment in Tgfbr2fl/fl;Wnt1-Cre palates (Fig. 4C).
Next, we investigated the possible requirement for Pitx2 during FGF9 induction of Ccnd1 and Ccnd3 expression. As described above, gene expression of Ccnd1 and Ccnd3 was induced by FGF9 treatment in Tgfbr2fl/fl;Wnt1-Cre MEPM cells; however, Ccnd1 and Ccnd3 expression was not induced by FGF9 treatment following siRNA knockdown of Pitx2 (Fig. 4, D and E). These data again indicate that Pitx2 is a downstream target for FGF9 and show that Pitx2 is required for FGF9-mediated induction of Ccnd1 and Ccnd3 expression. Taken together, our results demonstrate that expression of cyclins D1 and D3 is regulated by a TGFβ-FGF9-PITX2 signaling pathway.
FGF9 Rescues the Cell Proliferation Defect in Tgfbr2fl/fl;Wnt1-Cre Mice via Pitx2
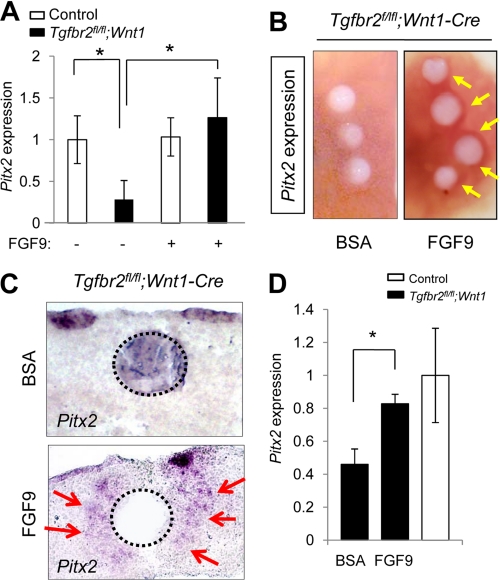
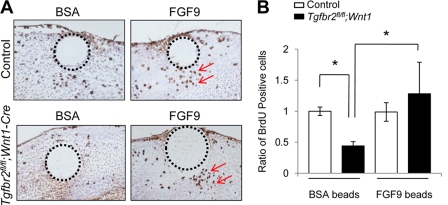
We investigated whether the cell proliferation defect in Tgfbr2fl/fl;Wnt1-Cre mice was rescued by the addition of FGF9. First, we treated MEPM cells with exogenous FGF9 and found that gene expression of Pitx2 in Tgfbr2fl/fl;Wnt1-Cre MEPM cells was increased to that of control Tgfbr2fl/fl MEPM cells (Fig. 5A). We also implanted FGF9 or BSA beads in palates from Tgfbr2fl/fl;Wnt1-Cre mice. FGF9-beads induced gene expression of Pitx2 in Tgfbr2fl/fl;Wnt1-Cre palates (Fig. 5, B–D). Thus, FGF9 can induce expression of Pitx2 both in vitro and ex vivo in Tgfbr2fl/fl;Wnt1-Cre mice. Furthermore, FGF9 treatment resulted in restored cell proliferation in the palate of Tgfbr2fl/fl;Wnt1-Cre mice (Fig. 6, A and B). These data demonstrate that FGF9 can induce Pitx2 expression and cell proliferation in the absence of TβRII. Thus, our results suggest that the FGF9-PITX2 pathway mediates TGFβ signaling to contribute to mesenchymal cell proliferation during palatal formation (supplemental Fig. 4).
FIGURE 5.
Exogenous FGF9 rescues Pitx2 expression in Tgfbr2fl/fl;Wnt1-Cre mice during palatal development. A. Quantitative RT-PCR analysis of Pitx2 in primary MEPM cells of Tgfbr2fl/fl control (open columns) and Tgfbr2fl/fl;Wnt1-Cre (closed columns) mice after treatment (+) or no treatment (−) with FGF9 protein. *, p < 0.05. B, whole mount in situ hybridization after implantation of FGF9 or BSA control beads in the palate of Tgfbr2fl/fl;Wnt1-Cre mice. The arrows indicate the increased expression of Pitx2 around FGF9 beads. C, section in situ hybridization for Pitx2 after implantation of FGF9 or BSA control beads in the palate of the Tgfbr2fl/fl;Wnt1-Cre mice. The arrows indicate the increased expression of Pitx2 around FGF9 beads. Dotted lines, perimeter of the beads. D, quantitative RT-PCR analysis of Pitx2 in the mesenchyme of Tgfbr2fl/fl;Wnt1-Cre (closed columns) and Tgfbr2fl/fl control (open column) mice after FGF9 or BSA treatment. *, p < 0.05. Error bars, S.D.
FIGURE 6.
Exogenous FGF9 rescues the cell proliferation defect of Tgfbr2fl/fl;Wnt1-Cre mice during palatal development. A, BrdU staining after treatment with FGF9 or BSA beads in the palate of Tgfbr2fl/fl control and Tgfbr2fl/fl;Wnt1-Cre mice. The arrows indicate BrdU-positive cells. Dotted lines, perimeter of the beads. B, quantitative analysis of BrdU-positive cells in A. BrdU-positive cells were significantly increased in the palate of Tgfbr2fl/fl;Wnt1-Cre mice after the treatment with FGF9-containing beads. *, p < 0.05. Error bars, S.D.
Phylogenetically Conserved SMAD and ATF2 Recognition Sequences Exist Proximal to or within the Human FGF9 and PITX2 Genes
We analyzed sequences of the human FGF9 and PITX2 genes (5-kb upstream and 5-kb downstream of their transcription start sites, respectively) and found that the FGF9 genomic region has 69 potential SMAD recognition sites (15 of which were conserved in at least six mammals) and one potential ATF2 recognition site that was conserved in all eight mammalian genomes investigated (supplemental Tables 2 and 3). We focused on the ATF2 transcription factor because it is known to be regulated by the p38 MAPK pathway (36). The interrogated human PITX2 genomic region had 65 potential SMAD recognition sites (seven of which were conserved in at least six mammals) and no ATF2 recognition sites (supplemental Tables 4 and 5). However, we found one ATF2 recognition site that was 5,032 bp downstream of the human PITX2 transcription start site that was conserved in all eight mammalian genomes investigated. Although this focused analysis does not preclude the existence of other potential transcription factor binding sites relevant to non-canonical TGFβ signaling, it is conceivable that canonical SMAD-mediated TGFβ signaling may promote Fgf9 expression, whereas non-canonical p38 MAPK-mediated signaling inhibits Fgf9 expression.
DISCUSSION
TGFβ Signaling Is Crucial in Regulating Craniofacial Development
Heterozygous mutations in TGFBR1 or TGFBR2 are associated with Loeys-Dietz syndrome, which can manifest with craniofacial malformations, such as cleft palate, craniosynostosis, and hypertelorism; skeletal defects, such as scoliosis, arachnodactaly, and joint laxity; and vascular problems, including arterial tortuosity with the potential for aneurysms and dissections (37–39). In addition, heterozygous mutations in FBN-1, which encodes an elastic extracellular matrix protein called fibrillin-1, lead to excessive TGFβ signaling and cause Marfan syndrome, which exhibits clinical phenotypes similar to Loeys-Dietz syndrome (40–42). Furthermore, DiGeorge syndrome, which results from a variably sized deletion on chromosome 22 (del22q11), including about 30 genes, such as the transcription factor Tbx1 and the signal adaptor protein CrkL, exhibits altered TGFβ signaling in ∼80% of patients who have cleft palate and other craniofacial malformations (43–45). Thus, TGFβ signaling is crucial in regulating palate formation during embryonic development, and altered TGFβ signaling can lead to multiple human syndromes.
The utility of animal models with cleft palate is impressive, because multiple studies have begun to elucidate the cellular and molecular mechanisms of palate formation. In some cases, the human gene deficiency was identified first and replicated in an animal model, but in other cases, animal models provided leads to understanding the molecular mechanism of clinical syndromes. Now, over 200 genetically mutated mice exhibit cleft palate, and ∼400 known human syndromes exhibit cleft palate as one of their clinical symptoms. Human gene linkage studies have shown that point mutations in TGFBR1 or TGFBR2 cause craniofacial deformities, such as cleft palate (37–39). Our strategy of identifying TGFβ downstream signaling molecules that are also crucial for palatogenesis will advance our understanding of the signaling network involved in regulating palate development in mice and humans.
TGFβ Signaling Regulates Fgf9 and Pitx2 Gene Expression
The FGF family includes 18 receptor-binding ligands and four FGF receptors (46). Among the FGF ligands, FGF9 can induce osteoblast proliferation and new bone formation (47, 48). Fgf9-null mice die at birth due to lung hypoplasia, and ∼40% of Fgf9−/− embryos exhibit cleft palate (34). Interestingly, FGF9 can induce Pitx2 expression in the mesenchyme derived from small intestine and lung (49, 50). The loss of TGFβ2 disrupts the expression of the causative genes for developmental glaucoma, Foxc1 and Pitx2 (51). Patients with TGFBR2 or TGFBR1 mutations show craniofacial defects and signs of elevated TGFβ signaling (38, 52, 53). Similarly, mutations in TGFβ receptor gene family members cause craniofacial deformities in mice (13, 54). However, it was unknown whether TGFβ ligands may still signal in Tgfbr2 mutant mice. Our recent study showed that loss of Tgfbr2 in CNCC results in elevated TGFβ2 and TβRIII expression, activation of a TβRI/TβRIII-mediated, SMAD-independent signaling pathway, and a cell proliferation defect in the palatal mesenchyme.3 Strikingly, Tgfb2, Tgfbr1/Alk5, or Tak1 haploinsufficiency disrupts TβRI/TβRIII-mediated signaling and rescues craniofacial deformities in Tgfbr2 mutant mice, indicating that the activation of this non-canonical TGFβ signaling pathway is responsible for craniofacial malformations in Tgfbr2 mutant mice.3 In this study, we treated MEPM cells and palatal mesenchyme with a p38 MAPK inhibitor, and we found that gene expression of Fgf9 was restored after the treatment with p38 MAPK inhibitor. These results indicate that non-canonical TGFβ-mediated p38 MAPK signaling regulates gene expression of Fgf9 during palate formation. Furthermore, the presence of highly conserved candidate SMAD and ATF2 recognition sites proximal to or within the human FGF9 and PITX2 genes (supplemental Tables 2–5) provides preliminary evidence suggesting that a TGFβ2-FGF9-PITX2 signaling cascade could be a well conserved mechanism in regulating mammalian organogenesis.
Multiple studies have demonstrated that PITX2 has diverse roles in cell proliferation, differentiation, hematopoiesis, and organogenesis (32, 55–60). During early embryogenesis, PITX2 is a key regulator in the establishment of embryonic left-right asymmetry (61). At a later embryonic stage, PITX2 regulates cell type-specific cell proliferation in response to growth factors during the development of the cardiac outflow tract (58). PITX2 is required for the differentiation of neural crest cells (51). Previous studies have also shown that PITX2 cooperates with β-catenin and LEF/TCF to regulate cell proliferation by directly activating transcription of cyclin Ds and c-Myc (51, 62). Our data indicate that PITX2 is downstream of TGFβ and FGF signaling and may also regulate cell proliferation by directly activating cyclin D1 and D3 gene expression. Pitx2-deficient mice die at an embryonic stage and show severe defects in heart, eye, pituitary gland, palate, and tooth development (35, 58). In addition, PITX2 mutations have been identified in several human disorders, such as Axenfeld-Rieger syndrome, iridoniodysgenesis syndrome, and sporadic Peter syndrome (63–65). Some patients with these syndromes exhibit cleft palate among their defects.
We have previously reported that TGFβ signaling regulates FGF signaling in frontal bone development, and FGF2-containing beads rescue a defect in cell proliferation in frontal bone primordium in Tgfbr2fl/fl;Wnt1-Cre mice (14). In the current study, we rescued a defect in cell proliferation in Tgfbr2fl/fl;Wnt1-Cre palates by the addition of FGF9 ex vivo during palate development. Thus, TGFβ-mediated FGF signaling may be an important mechanism to regulate cell proliferation during embryogenesis.
Mice with neural crest cell-specific deletion of Tgfbr2 (Tgfbr2fl/fl;Wnt1-Cre) are useful for the investigation of craniofacial anomalies, such as cleft palate, because Tgfbr2fl/fl;Wnt1-Cre mice exhibit craniofacial deformities, including cleft palate, at 100% penetrance (13, 17, 18, 66, 67). In addition, the defects in Tgfbr2fl/fl;Wnt1-Cre embryos mimic defects in patients with Loeys-Dietz and DiGeorge syndromes (13, 45). Therefore, identification of downstream targets of TGFβ signaling, such as Fgf9 and Pitx2, may be informative for future prevention, early diagnosis, and treatment of congenital birth defects.
Supplementary Material
Acknowledgments
We thank Julie Mayo for critical reading of the manuscript. We thank Dr. Harold Moses for Tgfbr2fl/fl mice, Dr. Harold Slavkin for discussion, and the Microarray Core Facility of Children's Hospital Los Angeles for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health, NIDCR, Grants DE012711, DE014078, and DE017007 (to Y. C.).

This article contains supplemental Tables 1–5 and Figs. 1–4.
J. Iwata, J. G. Hacia, A. Suzuki, P. A. Sanchez-Lara, M. Urata, and Y. Chai, submitted for publication.
J. Iwata, R. Pelikan, Y. Chai, and J. G. Hacia, manuscript in preparation.
- CNCC
- cranial neural crest cell(s)
- FDR
- false discovery rate
- MEPM
- mouse embryonic palatal mesenchymal
- TβRI
- TβRII, and TβRIII, TGFβ receptor type I, II, and III, respectively
- E13.5 and E14.5
- embryonic day 13.5 and 14.5, respectively.
REFERENCES
- 1. Tolarová M. M., Cervenka J. (1998) Classification and birth prevalence of orofacial clefts. Am. J. Med. Genet. 75, 126–137 [PubMed] [Google Scholar]
- 2. Mossey P. A., Little J., Munger R. G., Dixon M. J., Shaw W. C. (2009) Cleft lip and palate. Lancet 374, 1773–1785 [DOI] [PubMed] [Google Scholar]
- 3. Yu W., Serrano M., Miguel S. S., Ruest L. B., Svoboda K. K. (2009) Cleft lip and palate genetics and application in early embryological development. Indian J. Plast. Surg. 42, (suppl.) S35–S50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ciminello F. S., Morin R. J., Nguyen T. J., Wolfe S. A. (2009) Cleft lip and palate. Review. Compr. Ther. 35, 37–43 [PubMed] [Google Scholar]
- 5. Shah N. M., Groves A. K., Anderson D. J. (1996) Alternative neural crest cell fates are instructively promoted by TGFβ superfamily members. Cell. 85, 331–343 [DOI] [PubMed] [Google Scholar]
- 6. Dorsky R. I., Moon R. T., Raible D. W. (2000) Environmental signals and cell fate specification in premigratory neural crest. BioEssays. 22, 708–716 [DOI] [PubMed] [Google Scholar]
- 7. Nomura M., Li E. (1998) Smad2 role in mesoderm formation, left-right patterning, and craniofacial development. Nature. 393, 786–790 [DOI] [PubMed] [Google Scholar]
- 8. Heldin C. H., Miyazono K., ten Dijke P. (1997) TGF-β signaling from cell membrane to nucleus through SMAD proteins. Nature. 390, 465–471 [DOI] [PubMed] [Google Scholar]
- 9. Massagué J. (1998) TGF-β signal transduction. Annu. Rev. Biochem. 67, 753–791 [DOI] [PubMed] [Google Scholar]
- 10. Pelton R. W., Hogan B. L., Miller D. A., Moses H. L. (1990) Differential expression of genes encoding TGFs β 1, β 2, and β 3 during murine palate formation. Dev. Biol. 141, 456–460 [DOI] [PubMed] [Google Scholar]
- 11. Cui X. M., Warburton D., Zhao J., Crowe D. L., Shuler C. F. (1998) Immunohistochemical localization of TGF-β type II receptor and TGF-β3 during palatogenesis in vivo and in vitro. Int. J. Dev. Biol. 42, 817–820 [PubMed] [Google Scholar]
- 12. Wang Y. Q., Sizeland A., Wang X. F., Sassoon D. (1995) Restricted expression of type-II TGF β receptor in murine embryonic development suggests a central role in tissue modeling and CNS patterning. Mech. Dev. 52, 275–289 [DOI] [PubMed] [Google Scholar]
- 13. Ito Y., Yeo J. Y., Chytil A., Han J., Bringas P., Jr., Nakajima A., Shuler C. F., Moses H. L., Chai Y. (2003) Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development 130, 5269–5280 [DOI] [PubMed] [Google Scholar]
- 14. Sasaki T., Ito Y., Bringas P., Jr., Chou S., Urata M. M., Slavkin H., Chai Y. (2006) TGFβ-mediated FGF signaling is crucial for regulating cranial neural crest cell proliferation during frontal bone development. Development 133, 371–381 [DOI] [PubMed] [Google Scholar]
- 15. Oka S., Oka K., Xu X., Sasaki T., Bringas P., Jr., Chai Y. (2007) Cell autonomous requirement for TGF-β signaling during odontoblast differentiation and dentin matrix formation. Mech. Dev. 124, 409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oka K., Oka S., Sasaki T., Ito Y., Bringas P., Jr., Nonaka K., Chai Y. (2007) The role of TGF-β signaling in regulating chondrogenesis and osteogenesis during mandibular development. Dev. Biol. 303, 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iwata J., Hosokawa R., Sanchez-Lara P. A., Urata M., Slavkin H., Chai Y. (2010) Transforming growth factor-β regulates basal transcriptional regulatory machinery to control cell proliferation and differentiation in cranial neural crest-derived osteoprogenitor cells. J. Biol. Chem. 285, 4975–4982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hosokawa R., Oka K., Yamaza T., Iwata J., Urata M., Xu X., Bringas P., Jr., Nonaka K., Chai Y. (2010) TGF-β mediated FGF10 signaling in cranial neural crest cells controls development of myogenic progenitor cells through tissue-tissue interactions during tongue morphogenesis. Dev. Biol. 341, 186–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Massagué J. (2000) How cells read TGF-β signals. Nat. Rev. Mol. Cell Biol. 1, 169–178 [DOI] [PubMed] [Google Scholar]
- 20. Iwata J., Ezaki J., Komatsu M., Yokota S., Ueno T., Tanida I., Chiba T., Tanaka K., Kominami E. (2006) Excess peroxisomes are degraded by autophagic machinery in mammals. J. Biol. Chem. 281, 4035–4041 [DOI] [PubMed] [Google Scholar]
- 21. Sou Y. S., Waguri S., Iwata J., Ueno T., Fujimura T., Hara T., Sawada N., Yamada A., Mizushima N., Uchiyama Y., Kominami E., Tanaka K., Komatsu M. (2008) The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol. Biol. Cell. 19, 4762–4775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karaman M. W., Houck M. L., Chemnick L. G., Nagpal S., Chawannakul D., Sudano D., Pike B. L., Ho V. V., Ryder O. A., Hacia J. G. (2003) Comparative analysis of gene-expression patterns in human and African great ape cultured fibroblasts. Genome Res. 13, 1619–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xia X., McClelland M., Wang Y. (2005) WebArray. An online platform for microarray data analysis. BMC Bioinformatics 6, 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Y., McClelland M., Xia X. Q. (2009) Analyzing microarray data using WebArray. Cold Spring Harb. Protoc. 2009, pdb.prot5260 [DOI] [PubMed] [Google Scholar]
- 25. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) ClustalW and ClustalX version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 26. Denissova N. G., Pouponnot C., Long J., He D., Liu F. (2000) Transforming growth factor β-inducible independent binding of SMAD to the Smad7 promoter. Proc. Natl. Acad. Sci. U.S.A. 97, 6397–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zawel L., Dai J. L., Buckhaults P., Zhou S., Kinzler K. W., Vogelstein B., Kern S. E. (1998) Human Smad3 and Smad4 are sequence-specific transcription activators. Mol. Cell 1, 611–617 [DOI] [PubMed] [Google Scholar]
- 28. Hai T., Curran T. (1991) Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc. Natl. Acad. Sci. U.S.A. 88, 3720–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kimura N., Takamatsu N., Yaoita Y., Osamura R. Y. (2008) Identification of transcriptional regulatory elements in the human somatostatin receptor sst2 promoter and regions including estrogen response element half-site for estrogen activation. J. Mol. Endocrinol. 40, 75–91 [DOI] [PubMed] [Google Scholar]
- 30. Nakamura T., Okuyama S., Okamoto S., Nakajima T., Sekiya S., Oda K. (1995) Down-regulation of the cyclin A promoter in differentiating human embryonal carcinoma cells is mediated by depletion of ATF-1 and ATF-2 in the complex at the ATF/CRE site. Exp. Cell Res. 216, 422–430 [DOI] [PubMed] [Google Scholar]
- 31. Hai T. W., Liu F., Coukos W. J., Green M. R. (1989) Transcription factor ATF cDNA clones. An extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 3, 2083–2090 [DOI] [PubMed] [Google Scholar]
- 32. Liu C., Liu W., Palie J., Lu M. F., Brown N. A., Martin J. F. (2002) Pitx2c patterns anterior myocardium and aortic arch vessels and is required for local cell movement into atrioventricular cushions. Development 129, 5081–5091 [DOI] [PubMed] [Google Scholar]
- 33. Kettunen P., Karavanova I., Thesleff I. (1998) Responsiveness of developing dental tissues to fibroblast growth factors. Expression of splicing alternatives of FGFR1, -2, -3, and of FGFR4 and stimulation of cell proliferation by FGF-2, -4, -8, and -9. Dev. Genet. 22, 374–385 [DOI] [PubMed] [Google Scholar]
- 34. Colvin J. S., White A. C., Pratt S. J., Ornitz D. M. (2001) Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme. Development 128, 2095–2106 [DOI] [PubMed] [Google Scholar]
- 35. Lu M. F., Pressman C., Dyer R., Johnson R. L., Martin J. F. (1999) Function of Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature 401, 276–278 [DOI] [PubMed] [Google Scholar]
- 36. Zhu T., Lobie P. E. (2000) Janus kinase 2-dependent activation of p38 mitogen-activated protein kinase by growth hormone. Resultant transcriptional activation of ATF-2 and CHOP, cytoskeletal reorganization, and mitogenesis. J. Biol. Chem. 275, 2103–2114 [DOI] [PubMed] [Google Scholar]
- 37. Mizuguchi T., Collod-Beroud G., Akiyama T., Abifadel M., Harada N., Morisaki T., Allard D., Varret M., Claustres M., Morisaki H., Ihara M., Kinoshita A., Yoshiura K., Junien C., Kajii T., Jondeau G., Ohta T., Kishino T., Furukawa Y., Nakamura Y., Niikawa N., Boileau C., Matsumoto N. (2004) Heterozygous TGFBR2 mutations in Marfan syndrome. Nat. Genet. 36, 855–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Loeys B. L., Chen J., Neptune E. R., Judge D. P., Podowski M., Holm T., Meyers J., Leitch C. C., Katsanis N., Sharifi N., Xu F. L., Myers L. A., Spevak P. J., Cameron D. E., De Backer J., Hellemans J., Chen Y., Davis E. C., Webb C. L., Kress W., Coucke P., Rifkin D. B., De Paepe A. M., Dietz H. C. (2005) A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat. Genet. 37, 275–281 [DOI] [PubMed] [Google Scholar]
- 39. Loeys B. L., Schwarze U., Holm T., Callewaert B. L., Thomas G. H., Pannu H., De Backer J. F., Oswald G. L., Symoens S., Manouvrier S., Roberts A. E., Faravelli F., Greco M. A., Pyeritz R. E., Milewicz D. M., Coucke P. J., Cameron D. E., Braverman A. C., Byers P. H., De Paepe A. M., Dietz H. C. (2006) Aneurysm syndromes caused by mutations in the TGF-β receptor. N. Engl. J. Med. 355, 788–798 [DOI] [PubMed] [Google Scholar]
- 40. Brooke B. S., Habashi J. P., Judge D. P., Patel N., Loeys B., Dietz H. C., 3rd (2008) Angiotensin II blockade and aortic-root dilation in Marfan's syndrome. N. Engl. J. Med. 358, 2787–2795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Habashi J. P., Judge D. P., Holm T. M., Cohn R. D., Loeys B. L., Cooper T. K., Myers L., Klein E. C., Liu G., Calvi C., Podowski M., Neptune E. R., Halushka M. K., Bedja D., Gabrielson K., Rifkin D. B., Carta L., Ramirez F., Huso D. L., Dietz H. C. (2006) Losartan, an AT1 antagonist, prevents aortic aneurysm in a mouse model of Marfan syndrome. Science. 312, 117–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kalluri R., Han Y. (2008) Targeting TGF-β and the extracellular matrix in Marfan's syndrome. Dev Cell. 15, 1–2 [DOI] [PubMed] [Google Scholar]
- 43. Lindsay E. A. (2001) Chromosomal microdeletions. Dissecting del22q11 syndrome. Nat. Rev. Genet. 2, 858–868 [DOI] [PubMed] [Google Scholar]
- 44. Vitelli F., Morishima M., Taddei I., Lindsay E. A., Baldini A. (2002) Tbx1 mutation causes multiple cardiovascular defects and disrupts neural crest and cranial nerve migratory pathways. Hum. Mol. Genet. 11, 915–922 [DOI] [PubMed] [Google Scholar]
- 45. Wurdak H., Ittner L. M., Lang K. S., Leveen P., Suter U., Fischer J. A., Karlsson S., Born W., Sommer L. (2005) Inactivation of TGFβ signaling in neural crest stem cells leads to multiple defects reminiscent of DiGeorge syndrome. Genes Dev. 19, 530–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McKeehan W. L., Wang F., Kan M. (1998) The heparan sulfate-fibroblast growth factor family. Diversity of structure and function. Prog. Nucleic Acid Res. Mol. Biol. 59, 135–176 [DOI] [PubMed] [Google Scholar]
- 47. Riley B. M., Mansilla M. A., Ma J., Daack-Hirsch S., Maher B. S., Raffensperger L. M., Russo E. T., Vieira A. R., Dodé C., Mohammadi M., Marazita M. L., Murray J. C. (2007) Impaired FGF signaling contributes to cleft lip and palate. Proc. Natl. Acad. Sci. U.S.A. 104, 4512–4517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Govindarajan V., Overbeek P. A. (2006) FGF9 can induce endochondral ossification in cranial mesenchyme. BMC Dev. Biol. 6, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Geske M. J., Zhang X., Patel K. K., Ornitz D. M., Stappenbeck T. S. (2008) Fgf9 signaling regulates small intestinal elongation and mesenchymal development. Development 135, 2959–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bellusci S. (2008) Lung stem cells in the balance. Nat. Genet. 40, 822–824 [DOI] [PubMed] [Google Scholar]
- 51. Iwao K., Inatani M., Matsumoto Y., Ogata-Iwao M., Takihara Y., Irie F., Yamaguchi Y., Okinami S., Tanihara H. (2009) Heparan sulfate deficiency leads to Peters anomaly in mice by disturbing neural crest TGF-β2 signaling. J. Clin. Invest. 119, 1997–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Holm T. M., Habashi J. P., Doyle J. J., Bedja D., Chen Y., van Erp C., Lindsay M. E., Kim D., Schoenhoff F., Cohn R. D., Loeys B. L., Thomas C. J., Patnaik S., Marugan J. J., Judge D. P., Dietz H. C. (2011) Noncanonical TGFβ signaling contributes to aortic aneurysm progression in Marfan syndrome mice. Science 332, 358–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Habashi J. P., Doyle J. J., Holm T. M., Aziz H., Schoenhoff F., Bedja D., Chen Y., Modiri A. N., Judge D. P., Dietz H. C. (2011) Angiotensin II type 2 receptor signaling attenuates aortic aneurysm in mice through ERK antagonism. Science 332, 361–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dudas M., Kim J., Li W. Y., Nagy A., Larsson J., Karlsson S., Chai Y., Kaartinen V. (2006) Epithelial and ectomesenchymal role of the type I TGF-β receptor ALK5 during facial morphogenesis and palatal fusion. Dev. Biol. 296, 298–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lin C. R., Kioussi C., O'Connell S., Briata P., Szeto D., Liu F., Izpisúa-Belmonte J. C., Rosenfeld M. G. (1999) Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature 401, 279–282 [DOI] [PubMed] [Google Scholar]
- 56. Liu C., Liu W., Lu M. F., Brown N. A., Martin J. F. (2001) Regulation of left-right asymmetry by thresholds of Pitx2c activity. Development 128, 2039–2048 [DOI] [PubMed] [Google Scholar]
- 57. Liu W., Selever J., Lu M. F., Martin J. F. (2003) Genetic dissection of Pitx2 in craniofacial development uncovers new functions in branchial arch morphogenesis, late aspects of tooth morphogenesis and cell migration. Development 130, 6375–6385 [DOI] [PubMed] [Google Scholar]
- 58. Kioussi C., Briata P., Baek S. H., Rose D. W., Hamblet N. S., Herman T., Ohgi K. A., Lin C., Gleiberman A., Wang J., Brault V., Ruiz-Lozano P., Nguyen H. D., Kemler R., Glass C. K., Wynshaw-Boris A., Rosenfeld M. G. (2002) Identification of a Wnt/Dvl/beta-Catenin → Pitx2 pathway mediating cell type-specific proliferation during development. Cell. 111, 673–685 [DOI] [PubMed] [Google Scholar]
- 59. Hayashi M., Maeda S., Aburatani H., Kitamura K., Miyoshi H., Miyazono K., Imamura T. (2008) Pitx2 prevents osteoblastic transdifferentiation of myoblasts by bone morphogenetic proteins. J. Biol. Chem. 283, 565–571 [DOI] [PubMed] [Google Scholar]
- 60. Ai D., Liu W., Ma L., Dong F., Lu M. F., Wang D., Verzi M. P., Cai C., Gage P. J., Evans S., Black B. L., Brown N. A., Martin J. F. (2006) Pitx2 regulates cardiac left-right asymmetry by patterning second cardiac lineage-derived myocardium. Dev. Biol. 296, 437–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yoshioka H., Meno C., Koshiba K., Sugihara M., Itoh H., Ishimaru Y., Inoue T., Ohuchi H., Semina E. V., Murray J. C., Hamada H., Noji S. (1998) Pitx2, a bicoid-type homeobox gene, is involved in a lefty-signaling pathway in determination of left-right asymmetry. Cell 94, 299–305 [DOI] [PubMed] [Google Scholar]
- 62. Koziczak M., Holbro T., Hynes N. E. (2004) Blocking of FGFR signaling inhibits breast cancer cell proliferation through downregulation of D-type cyclins. Oncogene 23, 3501–3508 [DOI] [PubMed] [Google Scholar]
- 63. Semina E. V., Reiter R., Leysens N. J., Alward W. L., Small K. W., Datson N. A., Siegel-Bartelt J., Bierke-Nelson D., Bitoun P., Zabel B. U., Carey J. C., Murray J. C. (1996) Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat. Genet. 14, 392–399 [DOI] [PubMed] [Google Scholar]
- 64. Martin D. M., Probst F. J., Fox S. E., Schimmenti L. A., Semina E. V., Hefner M. A., Belmont J. W., Camper S. A. (2002) Exclusion of PITX2 mutations as a major cause of CHARGE association. Am. J. Med. Genet. 111, 27–30 [DOI] [PubMed] [Google Scholar]
- 65. Espinoza H. M., Cox C. J., Semina E. V., Amendt B. A. (2002) A molecular basis for differential developmental anomalies in Axenfeld-Rieger syndrome. Hum. Mol. Genet. 11, 743–753 [DOI] [PubMed] [Google Scholar]
- 66. Chai Y., Ito Y., Han J. (2003) TGF-β signaling and its functional significance in regulating the fate of cranial neural crest cells. Crit. Rev. Oral. Biol. Med. 14, 78–88 [DOI] [PubMed] [Google Scholar]
- 67. Zhao H., Oka K., Bringas P., Kaartinen V., Chai Y. (2008) TGF-β type I receptor Alk5 regulates tooth initiation and mandible patterning in a type II receptor-independent manner. Dev. Biol. 320, 19–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.