Abstract
The insulin-like growth factor (IGF) system plays an important role in mammary gland biology as well as in the etiology of breast cancer. The IGF-I receptor (IGF-IR), which mediates the biological actions of IGF-I and IGF-II, has emerged in recent years as a promising therapeutic target. The IGF and estrogen signaling pathways act in a synergistic manner in breast epithelial cells. The present study was aimed at investigating 1) the putative translocation of IGF-IR and the related insulin receptor (IR) to the nucleus in breast cancer cells, 2) the impact of IGF-IR and IR levels on IGF-IR biosynthesis in estrogen receptor (ER)-positive and ER-depleted breast cancer cells, and 3) the potential transcription factor role of IGF-IR in the specific context of IGF-IR gene regulation. We describe here a novel mechanism of autoregulation of IGF-IR gene expression by cellular IGF-IR, which is seemingly dependent on ER status. Regulation of the IGF-IR gene by IGF-IR protein is mediated at the level of transcription, as demonstrated by 1) binding assays (DNA affinity chromatography and ChIP) showing specific IGF-IR binding to IGF-IR promoter DNA and 2) transient transfection assays showing transactivation of the IGF-IR promoter by exogenous IGF-IR. The IR is also capable of translocating to the nucleus and binding the IGF-IR promoter in ER-depleted, but not in ER-positive, cells. However, transcription factors IGF-IR and IR display diametrically opposite activities in the context of IGF-IR gene regulation. Thus, whereas IGF-IR stimulated IGF-IR gene expression, IR inhibited IGF-IR promoter activity. In summary, we have identified a novel mechanism of IGF-IR gene autoregulation in breast cancer cells. The clinical implications of these findings and, in particular, the impact of IGF-IR/IR nuclear localization on targeted therapy require further investigation.
Keywords: Breast Cancer, Estrogen Receptor, Insulin, Insulin-like Growth Factor (IGF), Transcription Regulation, IGF Receptor, Promoter Activity
Introduction
The insulin-like growth factors (IGFs)2 play an important role in normal mammary gland development as well as in the biology of breast cancer (1–4). Most of the biological actions of IGF-I and IGF-II are mediated by the IGF-I receptor (IGF-IR), a tyrosine kinase-containing, membrane-bound heterotetramer with potent antiapoptotic and cell survival activities (5–8). The presence of the IGF-IR is a fundamental prerequisite for the acquisition of a neoplastic phenotype, and cells with a targeted disruption of the IGF-IR gene, with a few exceptions, do not undergo transformation when exposed to oncogenic agents (9). Furthermore, IGF-IR overexpression is a typical feature of most primary breast cancers (10, 11), albeit the biochemical and molecular mechanisms responsible for enhanced IGF-IR expression have been only partly dissected (12, 13). Likewise, the role of the IGF axis in the progression of breast tumors from early to advanced stages has been a controversial issue. Thus, whereas a number of studies showed down-regulation of the IGF-IR at advanced tumor stages, other studies showed sustained IGF-IR up-regulation at metastatic stages of the disease (14–17).
Control of IGF-IR expression is mainly attained at the level of transcription (13). The regulatory region of the IGF-IR gene lacks canonical TATA and CAAT sequences, two promoter elements that are generally required for accurate transcription initiation (18–20). Transcription of the IGF-IR gene, however, starts from a unique “initiator” motif, a promoter element able to direct initiation in the absence of a TATA box (13, 21). We have recently conducted a proteomic study based on DNA affinity chromatography followed by mass spectroscopic analyses aimed at identifying IGF-IR promoter-binding proteins in estrogen receptor (ER)-positive and ER-depleted breast cancer cell lines (22). These analyses identified a series of nuclear proteins that are potentially involved in the differential expression of the IGF-IR in ER-positive and ER-negative breast cancers.
The interplay between the IGF and estrogen signaling pathways has been the focus of significant basic and translational interest (23–26). The IGFs and estrogen act in a synergistic fashion in breast epithelial cells (27), and both the MAPK and Akt pathways were shown to mediate the activation of ERα by IGF-I (28, 29). On the other hand, estrogens regulate IGF-I signaling and the expression of key members of the IGF axis (30). Furthermore, estrogens were shown to induce a physical interaction between ERα and IGF-IR, leading to activation and phosphorylation of IGF-IR and downstream signaling molecules (31).
To fully understand IGF-IR function, the role of a number of protein modifications has been investigated. Recent studies have shown that the IGF-IR can be modified by the small ubiquitin-like modifier (SUMO) proteins, SUMO-1, -2, and/or -3, with ensuing translocation to the nucleus (32–34). Of interest, the ability of IGF-IR to translocate to the nucleus in human tumor cells allowed the receptor to interact with chromatin and to function as a transcriptional regulator (34). The functional significance of IGF-IR SUMOylation in the specific context of breast cancer is yet to be elucidated. Likewise, the potential impact of IGF-IR status (i.e. levels, cellular localization, activation, etc.) on IGF-IR gene expression has not yet been addressed in a systematic fashion. The aim of this study was to investigate the putative nuclear localization of IGF-IR and autoregulation of IGF-IR gene expression in ER-positive and ER-depleted breast cancer cells and to identify the pathways involved and biological significance of this novel mechanism. In addition, and in view of the structural homology between IGF-IR and the insulin receptor (IR) and given the overlapping signaling pathways downstream of the receptors, some of the effects of IGF-IR were compared with those of IR. The results of cell fractionation and confocal microscopy analyses demonstrated that both IGF-IR and IR are present in the nucleus in a SUMOylated form. Higher IGF-IR levels were seen in total, nuclear, and cytosolic fractions of ER-positive in comparison with ER-depleted cells. In contrast, higher IR levels were seen in ER-depleted in comparison with ER-positive cells. DNA affinity chromatography and chromatin immunoprecipitation (ChIP) assays revealed that both IGF-IR and IR can bind to IGF-IR promoter DNA in ER-depleted but not ER-expressing cells. However, whereas exogenous IGF-IR enhanced IGF-IR promoter activity, IR expression had a diametrically opposite effect, leading to IGF-IR promoter inhibition. These results identify a novel ER-dependent mechanism of IGF-IR gene autoregulation in breast cancer cells. The clinical relevance of this autoregulatory loop is yet to be investigated.
EXPERIMENTAL PROCEDURES
Cell Cultures
Human breast cancer-derived MCF7 cells (ER-positive; American Type Culture Collection, Manassas, VA) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), glutamine, and antibiotics. C4.12.5 (ER-depleted) cells were derived by clonal selection of MCF7 cells grown in the absence of estrogen for 9 months (23). C4.12.5 cells were maintained in phenol red-free DMEM with 10% charcoal/dextran-treated FBS, glutamine, and antibiotics. C4.12.5 cells were kindly provided by Dr. Wade V. Welshons (University of Missouri, Columbia, MO). Cells were incubated at 37 °C in a humidified atmosphere containing 5% CO2.
Treatments
Cells were serum-starved for 24 h, after which they were treated with increasing concentrations of IGF-I (PeproTech Ltd., Rocky Hill, NJ) or β-estradiol (Sigma-Aldrich) for 24 h. All experiments were carried out at least twice.
Cell Proliferation Studies
Cells were seeded in 24-well plates (105 cells/well) in triplicate and cultured for 24 h. For dose-response assays, IGF-I (1–50 ng/ml) or estradiol (1–100 nm) was added to the cells for 72 h. Cells maintained in starvation medium served as controls. After the incubation, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Sigma-Aldrich) was added to each well at a concentration of 50 μg/ml for 1 h, the medium was then removed, and DMSO was added to the cells. The colorimetric reaction was determined at 570 nm/630 nm using a 24-well plate reader (MRX, Dynex Technologies, Chantilly, VA).
Cell Fractionation
Whole cell extracts were prepared by lysing cells in a buffer solution containing protease inhibitors, as described previously (22). To prepare cytosolic and nuclear extracts, cells were washed with ice-cold phosphate-buffered saline (PBS) and centrifuged, and the pellet was resuspended in a buffer solution containing 10 mm Hepes (pH 7.9), 1.5 mm MgCl2, 10 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA, 1 mm dithiothreitol, and protease inhibitors and incubated for 30 min on ice. IGEPAL (Sigma-Aldrich) was then added to a final concentration of 3%, vortexed, and centrifuged, after which the supernatant containing the cytosolic fraction was saved at −70 °C. A second buffer (20 mm Hepes, 420 mm NaCl, 1 mm EDTA, 1 mm EGTA, and protease inhibitors) was added to the nuclei-containing pellet, incubated for 30 min at room temperature, and spun for 5 min. The supernatant containing the nuclear extract was frozen at −70 °C. Protein concentrations were determined with the Bradford reagent (Bio-Rad) using bovine serum albumin (BSA) as a standard. Given that total protein concentration was higher in the cytosol than in the nucleus, the cytosolic fraction was 3.3-fold more diluted than the nuclear fraction in order to obtain equal protein amounts in adjacent lanes in Western immunoblots.
Western Immunoblots
Samples were electrophoresed through 10% SDS-PAGE, followed by blotting of the proteins onto nitrocellulose membranes. After blocking with skim milk, the blots were incubated overnight with the antibodies listed below, washed, and incubated with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibody. Antibodies against IGF-IR β-subunit (C-20), ERα (MC-20), and lamin A/C (H110) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). An antibody against tubulin (B-5-1-2) was purchased from Sigma-Aldrich, and anti-actin (clone C4) was purchased from ICN Biomedicals Inc. (Aurora, OH). Anti-phospho-IGF-IR (catalog no. 3024), anti-IR β-subunit (catalog no. 3025), and anti-SUMO-1 (catalog no. 2A12) were obtained from Cell Signaling Technology, Inc. (Beverly, MA). The secondary antibodies were HRP-conjugated bovine (1:500) or donkey (1:500) anti-goat IgG (Santa Cruz Biotechnology, Inc.), goat anti-rabbit IgG (1:50,000), and donkey anti-mouse IgG (1:25,000; Jackson ImmunoResearch Laboratories, West Grove, PA). Proteins were detected using the SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Immunoprecipitation (IP) Assays
Nuclear extracts (35 μg) were diluted 1:2 with IP dilution buffer (0.5% Triton X-100, 0.5% deoxycholic acid, 150 mm NaCl, 20 mm Tris-HCl (pH 7.5), 30 mm sodium pyrophosphate, and 20 mm N-ethylmaleimide) and were immunoprecipitated by incubating overnight at 4 °C with anti-IGF-IR β-subunit (1:40) or anti-IR β-subunit (1:50). Protein A/G-agarose beads (SC-20003; Santa Cruz Biotechnology, Inc.) were added to the samples and incubated for 2 h. Samples were then washed with PBS, mixed with sample buffer, boiled for 10 min at 95 °C, and electrophoresed through 10% SDS-PAGE. Finally, membranes were blotted with anti-SUMO-1, anti-IGF-IR β-subunit, or anti-IR β-subunit, as described above.
Quantitative RT-PCR
To measure IGF-IR mRNA levels in MCF7 and C4.12.5 cells, quantitative real-time reverse transcription-PCR analysis was performed using TaqMan® gene expression assay kits (Applied Biosystems, Carlsbad, CA). The PCR amplification was done using an ABI Prism 7900HT sequence detection system under the following thermal cycler conditions: 2 min at 50 °C and 10 min at 95 °C for 40 cycles (30 s at 95 °C and 1 min at 60 °C). The data were analyzed with SDS software (Applied Biosystems) and the RQ (relative quantity) Manager software and were calculated based on the comparative threshold cycle (Ct) method. IGF-IR mRNA expression was normalized to GAPDH mRNA levels.
DNA Affinity Chromatography
For DNA affinity chromatography of the IGF-IR promoter, a 511-bp human proximal promoter biotinylated fragment (extending from nt −458 in the 5′-flanking region to nt +53 in the 5′-untranslated region; nt +1 corresponds to the transcription start site) was bound to streptavidin magnetic beads (Dynabeads® M-270 streptavidin; Dynak Biotech ASA, Oslo, Norway) and incubated with nuclear extracts of MCF7 or C4.12.5 cells (35). IGF-IR promoter-binding proteins were eluted with a high salt-containing buffer and analyzed by Western blots, as described previously (22).
ChIP Assays
ChIP assays were performed as described previously (22, 30). Briefly, MCF7 and C4.12.5 cells were cross-linked with 1% formaldehyde at 37 °C for 10 min. Cells were then rinsed with ice-cold PBS and centrifuged. The pellets were resuspended in lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl, pH 8.1, and protease inhibitor mixture) and sonicated for 3 min. The supernatant fraction was immunoprecipitated with anti-IGF-IR, anti-IR, anti-ERα, or normal rabbit serum (NRS) for 18 h at 4 °C. The next day, Protein A/G Plus-agarose beads were added for 1 h at 4 °C. For PCR analysis of antibody-immunoprecipitated chromatin, a set of primers encompassing the IGF-IR proximal promoter region (from nt −458 to +53) and ThermolaceTM DNA polymerase reagent (Invitrogen) were used.
Transient Transfection Assays
For transient cotransfection experiments, an expression vector encoding the full-length IGF-IR cDNA fused to a green fluorescent protein (GFP) marker (in pcDNA3) was used, along with an IGF-IR promoter luciferase reporter construct (p(−188/+640)LUC) (30). This plasmid includes 188 nucleotides of the 5′-flanking region and 640 bp of the 5′-untranslated region of the rat IGF-IR gene. The IGF-IR expression plasmid was kindly provided by Dr. Rosemary O'Connor (Department of Biochemistry, University of Cork, Cork, Ireland). Cells were seeded in 6-well plates the day before transfection and cotransfected with 2 μg of the IGF-IR promoter reporter construct along with 1 μg of the GFP-IGF-IR expression plasmid (or empty pcDNA3 vector) and 1.2 μg of a β-galactosidase expression plasmid (pCMVβ, Clontech, Palo Alto, CA) using the Jet-PEI transfection reagent (Polyplus, Illkirch, France). Cells were harvested after 48 h, and luciferase activity was measured as described previously (20). Promoter activities were expressed as luciferase values normalized for β-galactosidase activity.
IR Coexpression Studies
An IR recombinant adenovirus was constructed using the pAd/CMV/V5-DEST vector containing the CMV promoter and C-terminal V5 epitope (ViraPower Adenoviral Expression System, Invitrogen). A pGEM containing the wild type IR cDNA (a generous gift of Dr. Domenico Accili, Columbia University, New York) was digested with SalI, inserted into the pENTR vector (Invitrogen), and then recombined with the pAD/CMV/V5-DEST vector according to the manufacturer's protocol. The generated recombinant adenovirus was amplified and purified from lysed 293 cells and utilized for infection of cells. MCF7 and C4.12.5 cells were infected with the IR adenovirus for 24 h, after which the virus-containing medium was removed, and cells were transfected with the IGF-IR promoter luciferase construct, as described above.
Confocal Microscopy Analyses
MCF7 and C4.12.5 cells were cultured for 36 h, harvested, and washed twice with PBS. Cells were resuspended in 0.5 ml of PBS, and 20–30 μl of the cell suspension were spread on microscope slides (poly-l-lysine, Superfrost/Plus, Electron Microscopy Sciences, Hatfield, PA). Slides were air-dried and permeabilized with 0.5% Triton X-100 for 20 min, washed with PBS, and blocked with 4% BSA, 0.1% Triton X-100 in PBS. After 1 h, the slides were incubated with polyclonal antibodies against IGF-IR or IR (or NRS; 1:100 in 1% BSA, 0.1% Triton X-100) overnight at 4 °C. The next day, the slides were rinsed three times with 0.1% Triton X-100 in PBS and further incubated with 45 μl of a 1:50 dilution of fluorescent goat anti-rabbit IgG (Bio-Yeda, Rehovot, Israel) for 1 h. The cells were then washed with PBS, propidium iodide (PI; 0.01 mg/ml) was added for 30 min, cells were washed again, and then cells were mounted on Vectashield mounting medium (Vector Laboratories Inc., Burlingame, CA). Staining was assessed using a laser-scanning confocal microscope driven by Scanware software (Zeiss LSM image browser). Images were collected using the same settings for similar detections and were processed by setting identical thresholds using Powerpoint, in order to facilitate visual comparison.
Bioinformatic Analyses
The IGF-IR sequence was analyzed for functionality by domain analysis and DNA binding prediction. Various domains (including nuclear localization signals) are shown as described in the UniProt database (UniProt: P08069 (IGF1R_HUMAN)) (36). Four DNA binding prediction sites were summarized to a consensus sequence (37–40). IGF-IR sequence conservation was obtained using the ConSurf server (supplemental Fig. 1) (available on the World Wide Web) (41). Twenty-four homologues were obtained from the Pfam data base (PF01030; receptor L-domain family), and incomplete sequences lacking the kinase or receptor domains were omitted. Sequence alignment was performed by MAFFT (42).
RESULTS
Analysis of Subcellular Distribution of IGF-IR and IR in ER-positive and ER-depleted Breast Cancer Cells
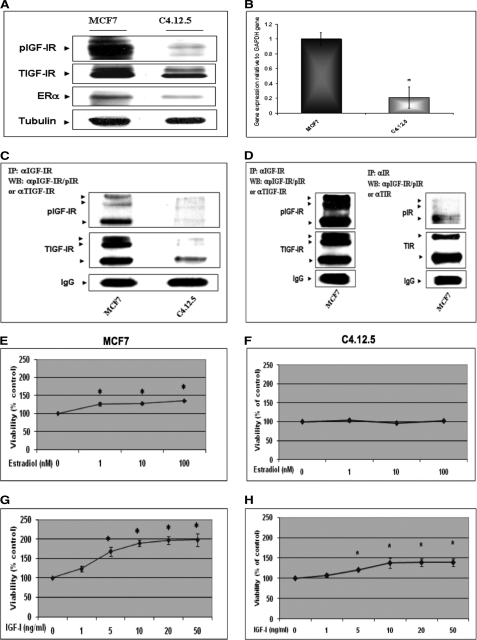
The stimulatory effect of ER action on IGF-IR levels has been well established (30). In addition, recent studies have shown that IGF-IR can translocate to the nucleus, although the biological significance of this event remains undefined (32–34). To evaluate the impact of ER status on the nuclear translocation of IGF-IR, the human breast cancer-derived MCF7 (ER-positive) and MCF7-derived C4.12.5 (ER-depleted) cell lines were used. C4.12.5 cells were generated by clonal selection of MCF7 cells that were maintained in estrogen-free conditions for 9 months (23). These cells constitute a validated model that allows the analysis of ER effects on cellular and biochemical variables in an otherwise identical genetic background. Furthermore, these cells reflect early (MCF7) and advanced (C4.12.5) stages of the disease. Western blots showed that both total IGF-IR and phospho-IGF-IR (pIGF-IR) levels were largely reduced in C4.12.5 in comparison with MCF7 cells (76% for IGF-IR and 30% for phospho-IGF-IR) (Fig. 1A). These results were previously reported (22, 30). To assess whether these values were correlated with corresponding changes in IGF-IR mRNA, quantitative RT-PCR was performed. Results obtained indicate that IGF-IR mRNA levels in C4.12.5 cells were ∼20 ± 14% of those measured in MCF7 cells (Fig. 1B).
FIGURE 1.
IGF-IR gene expression in ER-positive and ER-depleted breast cancer cells. A, Western blot analysis of IGF-IR levels. MCF7 and C4.12.5 cell lines were lysed in the presence of protease inhibitors, and equal amounts of protein (100 μg) were separated by 10% SDS-PAGE. After electrophoresis, proteins were transferred onto nitrocellulose membranes and blotted with antibodies against total and phosphorylated IGF-IR (pIGF-IR) and ERα, followed by incubation with an HRP-conjugated secondary antibody. Membranes were reprobed with a tubulin antibody. The figure shows the results of a typical experiment, repeated multiple times with similar results. B, quantitative real-time PCR of IGF-IR mRNA levels. Total RNA was prepared from MCF7 and C4.12.5 cells, and IGF-IR mRNA and GAPDH mRNA values were measured using the TaqMan® real-time PCR system. Analysis of the data was performed as described under “Experimental Procedures.” *, p < 0.01 versus MCF7 cells. C and D, IP analysis of phospho-IGF-IR and phospho-IR abundance in MCF7 and C4.12.5 cells. Total cell extracts (650 μg) were immunoprecipitated with antibodies against total IGF-IR or total-IR, electrophoresed, and immunoblotted with antibodies against phospho-IGF-IR/IR, total IGF-IR, or total-IR. Exposure time for the autoradiogram shown in C was 1 min, whereas the autoradiogram shown in D was exposed for 1 h. E–H, mitogenic effects of estradiol and IGF-I in MCF7 and C4.12.5 cells. MCF7 and C4.12.5 cells were starved overnight and then incubated with increasing doses of estradiol (1, 10, and 100 nm) or IGF-I (1, 5, 10, 20, and 50 ng/ml) for 72 h in serum-free medium. Cellular proliferation was assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assays. p < 0.05 versus control cells. A value of 100% was given to the number of cells in control (unstimulated) cultures. Error bars, S.E.
To corroborate the results of Western blots and in view of the fact that the antibody used can recognize the activated forms of both IGF-IR and IR, IP studies were performed. To this end, MCF7 and C4.12.5 total cell extracts were immunoprecipitated with antibodies against total-IGF-IR or total-IR, electrophoresed, and immunoblotted with anti-phospho-IGF-IR/phospho-IR, anti-total IGF-IR, or anti-total IR. Comparative analyses confirmed that phospho-IGF-IR levels were ∼10-fold higher in MCF7 than in C4.12.5 cells (Fig. 1C). Furthermore, phospho-IGF-IR levels in MCF7 cells were ∼7.5-fold higher than phospho-IR levels (Fig. 1D).
Proliferation in response to estradiol or IGF-I treatments was determined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide cell viability assay. Consistent with their reduced ER levels, C4.12.5 cells do not proliferate in response to estradiol stimulation with doses of up to 100 nm for 72 h. MCF7 cells, on the other hand, displayed an increase in proliferation in response to estrogen stimulation, with an almost maximal effect (127 ± 4.5% of control) seen at a dose of 1 nm estradiol (Fig. 1, compare E and F). In addition, as expected from the lower IGF-IR levels, the mitogenic response of C4.12.5 cells to IGF-I (1–50 ng/ml) stimulation was significantly blunted in comparison with MCF7 cells (139 ± 3% versus 198 ± 16.5% increase at 50 ng/ml; Fig. 1, compare G and H).
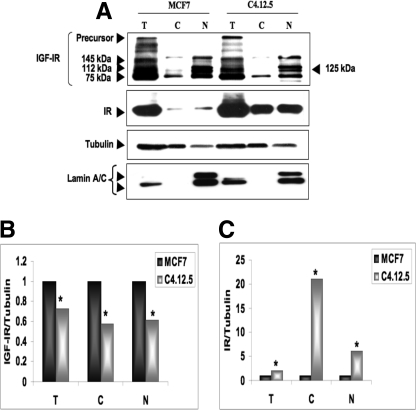
To address the subcellular distribution of IGF-IR and IR in the MCF7 and C4.12.5 cell lines, cells were fractionated as described under “Experimental Procedures,” and IGF-IR and IR expressions were measured by Western blots of cytosolic and nuclear fractions (Fig. 2A). In addition to the precursor and mature IGF-IR forms, a number of bands were visualized in total lysates, which may correspond to partially processed or SUMOylated molecules (see below). Hence, at least four molecular species were visualized in the nuclear fractions of both cells, and only one band was visualized in the cytosolic fractions. Higher IGF-IR levels were seen in total, cytosolic, and nuclear fractions of MCF7 in comparison with C4.12.5 cells (Fig. 2B). Of interest, IR displayed an opposite pattern of expression, with higher IR levels seen in total and subcellular fractions of C4.12.5 in comparison with MCF7 cells (Fig. 2C).
FIGURE 2.
Subcellular distribution of IGF-IR in MCF7 and C4.12.5 cells. A, confluent MCF7 and C4.12.5 cells were lysed and fractionated into cytosolic and nuclear fractions, as described under “Experimental Procedures.” Total lysates (T; 80 μg) and cytosolic (C; 20 μg) and nuclear fractions (N; 20 μg) were resolved on 10% SDS-PAGE and blotted with anti-IGF-IR, anti-IR, anti-tubulin, and anti-lamin A/C (as a control for contamination of cytosolic fractions). Quantitative analysis of IGF-IR (B) and IR (C) abundance in MCF7 and C4.12.5 cells was done by scanning densitometry of the corresponding bands. The bars represent the mean ± S.E. of three independent experiments. p < 0.01 versus MCF7 cells.
Analysis of IGF-IR and IR SUMOylation
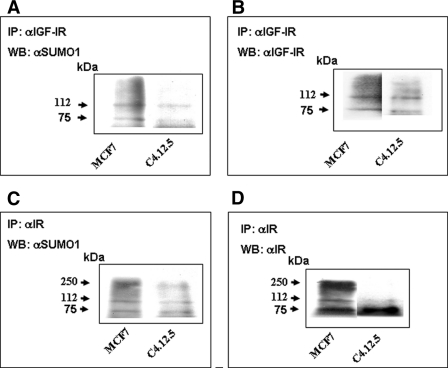
Previous studies have suggested that SUMOylation of the receptor takes place in a ligand-dependent fashion and is required for its nuclear translocation (32, 33). To determine whether IGF-IR and IR are SUMOylated in breast cancer cells with different ER backgrounds, nuclear extracts of MCF7 and C4.12.5 cells were immunoprecipitated with anti-IGF-IR β-subunit or anti-IR β-subunit, electrophoresed through 10% SDS-PAGE, and immunoblotted with anti-SUMO-1. The results of IP assays revealed that SUMO-1 was conjugated to IGF-IR and IR in both cell lines, as indicated by the presence of ∼75 and ∼112 kDa bands (Fig. 3, A–D), which reflect the modification of IGF-1R and IR by the SUMO-1 molecule. In addition, the IR precursor (∼250 kDa) was also detected with anti-SUMO-1 (Fig. 3C).
FIGURE 3.
Coimmunoprecipitation analysis of IGF-IR and IR SUMOylation. Nuclear extracts of MCF7 and C4.12.5 (35 μg) were immunoprecipitated (IP) with anti-IGF-IR or anti-IR, electrophoresed through 10% SDS-polyacrylamide gels, and immunoblotted (WB) with anti-SUMO-1 (A and C) as described under “Experimental Procedures.” Membranes were reprobed with IGF-IR (B) or IR (D) antibodies.
Confocal Microscopy Analysis of Nuclear IGF-IR and IR Localization
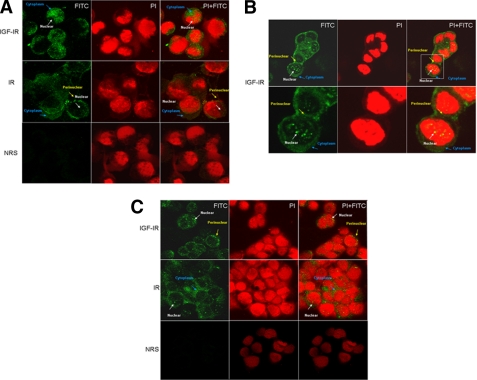
To corroborate the results of cell fractionation experiments showing a nuclear localization of IGF-IR and IR in MCF7 and C4.12.5 cells, we conducted microscopic analyses using a confocal system. The cells were stained for IGF-IR and IR with FITC (green) and for DNA with PI (red). The merged pictures (PI + FITC) show that IGF-IR and IR stainings are predominantly cytoplasmic but are also detectable in the nuclei and perinuclear areas of both cells (Fig. 4). The nuclear speckled pattern may correspond to a nucleolar localization, as reported previously for insulin receptor substrate-1 (43). The co-localization of IGF-IR or IR and DNA is evidenced by the yellow grains in the nuclei. Visual inspection of confocal images confirmed Western blot data, indicating that nuclear IGF-IR levels were higher in MCF7 (Fig. 4, A and B) than in C4.12.5 (Fig. 4C) cells, whereas, in contrast, IR expression was higher in C4.12.5 cells. No staining was detected with NRS in either cell line, and it was used as a negative control.
FIGURE 4.
Confocal microscopy analysis of IGF-IR nuclear localization. A, fluorescence microscope imaging of IGF-IR and IR-expressing MCF7 cells by confocal immunofluorescence microscopy. B, amplified fluorescence imaging of IGF-IR-expressing MCF7 cells. C, fluorescence microscope imaging of IGF-IR- and IR-expressing C4.12.5 cells. Fixed cells were stained for DNA with PI (red) and for IGF-IR and IR with fluorescent goat anti-rabbit IgG (FITC) (green). Merging of pictures (FITC + PI) gives a yellow color with yellow grains in the nucleus. NRS was used as a negative control (A and C).
Analysis of IGF-IR Binding to IGF-IR Gene Promoter
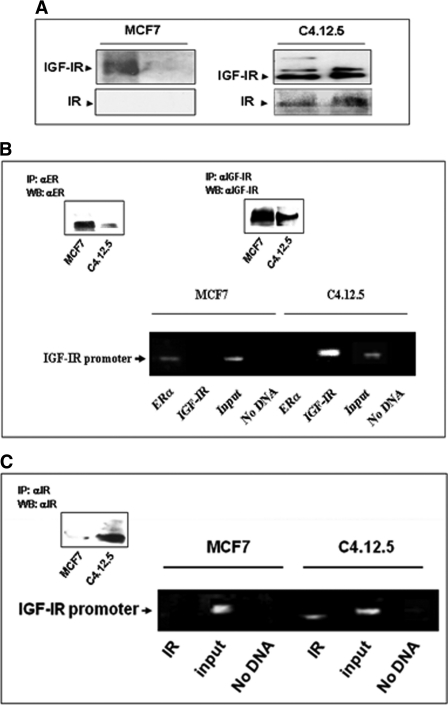
To evaluate the hypothesis that IGF-IR can control IGF-IR gene transcription, we assessed initially the capacity of the receptor protein to bind to the IGF-IR promoter region. To this end, we used a proteomic approach based on DNA affinity chromatography followed by Western blot analyses. A 511-bp human proximal IGF-IR promoter fragment extending from nucleotide −458 to +53 was labeled using a 5′-biotinylated antisense primer, as described (22). This fragment includes most of the proximal 5′-flanking region and comprises the “initiator” motif from which transcription starts in vivo. The rationale for using this genomic fragment was the fact that previous studies have shown that this specific region is responsible for the largest part of IGF-IR promoter activity (20). Nuclear extracts of MCF7 and C4.12.5 cells were incubated with the PCR-amplified, biotin-labeled, streptavidin beads-attached IGF-IR proximal promoter DNA probe. Bound proteins were eluted with a high salt buffer. Western blot analysis identified both the IGF-IR and IR in eluates of IGF-IR promoter bound material in C4.12.5 but not in MCF7 cells (Fig. 5A).
FIGURE 5.
Binding of IGF-IR and IR to IGF-IR promoter DNA. A, DNA affinity chromatography. Nuclear extracts of MCF7 and C4.12.5 cells were incubated with a PCR-amplified, biotin-labeled IGF-IR proximal promoter DNA probe extending from nt −458 to +53, after which DNA-protein complexes were adsorbed to streptavidin beads. Bound proteins were eluted with a high salt buffer, electrophoresed through 10% SDS-PAGE, and blotted with antibodies against IGF-IR or IR β-subunits. The left lanes in each gel correspond to nuclear extracts, and the right lanes represent the DNA affinity chromatography eluates. B and C, chromatin immunoprecipitation. MCF7 and C4.12.5 cells were cross-linked with formaldehyde, lysed, sonicated, and immunoprecipitated with IGF-IR (B), IR (C), or ERα (B) antibodies, followed by PCR amplification of precipitated chromatin using primers encompassing the IGF-IR promoter. The position of the 510-bp amplified fragments is indicated. The input bands represent the amplified PCR product in the absence of antibodies. Immunoprecipitated (IP) IGF-IR, IR, and ER were detected by Western blots (WB) using specific antibodies (insets).
To corroborate the results of DNA affinity chromatography, ChIP analyses were performed using antibodies against IGF-IR, IR, and ERα, followed by PCR amplification of the immunoprecipitated DNA using a set of primers against the proximal IGF-IR promoter region, encompassing the transcription start site. The results of ChIP assays confirmed that IGF-IR (Fig. 5B) and IR (Fig. 5C) bound in vivo directly to the IGF-IR promoter in ER-depleted C4.12.5 cells but not in ER-positive MCF7 cells. Conversely, ERα bound to the IGF-IR promoter in MCF7 but not in C4.12.5 cells (Fig. 5B). The input bands represent the amplified PCR product in the absence of antibodies.
Effect of IGF-IR and IR Levels on IGF-IR Promoter Activity
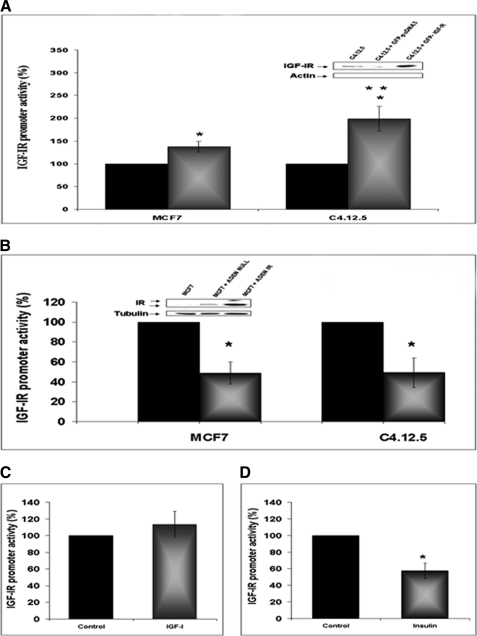
The functional relevance of DNA binding data was assessed by cotransfection experiments using IGF-IR or IR expression vectors along with an IGF-IR promoter luciferase reporter. To this end, MCF7 and C4.12.5 cells were cotransfected with an expression vector encoding the full-length IGF-IR cDNA along with the p(−188/+640)LUC IGF-IR promoter luciferase reporter plasmid (Fig. 6A). The results of transient cotransfections indicate that IGF-IR induced an increase in IGF-IR promoter activity in both cells lines, although significant differences were seen between cells. Thus, IGF-IR enhanced IGF-IR promoter activity by 199 ± 27% in C4.12.5 cells (exhibiting IGF-IR binding to the IGF-IR promoter) and only by 137.5 ± 11.5% in MCF7 cells (lacking IGF-IR binding) (p < 0.05).
FIGURE 6.
Effect of IGF-IR and IR levels on IGF-IR promoter activity. A, MCF7 and C4.12.5 cells were cotransfected with the p(−188/+640)LUC IGF-IR reporter construct, along with a full-length IGF-IR cDNA expression vector (GFP-IGF-IR) (or empty pcDNA3) and a β-galactosidase vector. Forty-eight hours after transfection, cells were harvested and luciferase and β-galactosidase activities were measured. Promoter activities are expressed as luciferase values normalized to β-gal values. A value of 100% was given to the promoter activity generated by the reporter plasmid in empty vector-transfected MCF7 or C4.12.5 cells. Bars, mean ± S.E. (error bars) of three independent experiments in duplicate wells. *, p < 0.05 versus control cells; **, p < 0.05 versus MCF7 cells transfected with GFP-IGF-IR. The inset shows a Western blot (WB) of C4.12.5 cells transfected with GFP-IGF-IR or GFP-pcDNA in comparison with endogenous IGF-IR expression. B, MCF7 and C4.12.5 cells were infected with an IR-containing adenoviral vector (or empty virus). After 24 h, the medium was changed, and cells were transfected with the p(−188/+640)LUC IGF-IR reporter construct for 48 h. Bars, mean ± S.E. of three independent experiments in duplicate wells. *, p < 0.01 versus control cells. The inset shows a Western blot using a specific IR antibody of MCF7 cells infected with the IR viral vector or empty vector in comparison with endogenous IR expression. C and D, MCF7 cells were transfected with the p(−188/+640)LUC IGF-IR promoter plasmid along with the full-length IGF-IR (C) or IR (D) vectors described above, in serum-containing medium. After 24 h, medium was replaced with serum-free medium, and cells were incubated for an additional 24 h in the presence of 50 ng/ml of IGF-I (C) or insulin (D) (or left untreated). After an additional 24 h, cells were harvested, and luciferase and β-galactosidase activities were measured as described above. A value of 100% was given to the promoter activities of expression vector-transfected cells in the absence of exogenous ligand treatments; *, p < 0.05 versus control cells.
To evaluate the functional significance of IR binding to the IGF-IR promoter, we next analyzed the effect of IR protein on IGF-IR promoter activity. For this purpose, MCF7 and C4.12.5 cells were infected with an IR-encoding adenovirus for 24 h, following which the cells were transfected with the p(−188/+640)LUC IGF-IR promoter luciferase plasmid, as described above. After an additional 48 h, cells were harvested, and luciferase and β-galactosidase activities were measured. Results obtained indicate that IR coexpression significantly decreased IGF-IR promoter activity in both cell lines (∼50%) (Fig. 6B).
To examine the ligand dependence of these effects, MCF7 cells were transfected with the p(−188/+640)LUC IGF-IR promoter plasmid along with the full-length IGF-IR or IR vectors described above, in serum-containing medium. After 24 h, medium was replaced with serum-free medium, and cells were incubated for an additional 24 h in the presence of 50 ng/ml IGF-I or insulin (or left untreated, control). Luciferase assays indicate that IGF-I treatment slightly (∼114 ± 16%) increased IGF-IR promoter activity compared with untreated cells (Fig. 6C). On the other hand, insulin reduced IGF-IR promoter activity by ∼43 ± 9.2% (Fig. 6D).
Bioinformatic Analyses
Three functional SUMOylation sites in the human IGF-IR sequence were recently published (33). These sites are highlighted in the IGF-IR scheme shown in supplemental Fig. 1. Lysines 1055 and 1130 are conserved only in the IGF-IR and IR, whereas lysine 1150 is also conserved in the epidermal growth factor receptor and ErbB protein family (see supplemental Fig. 2 for sequence conservation). A consensus of four different DNA binding predictions (as presented under “Experimental Procedures”) is also provided (conserved sites are colored in blue). These predictions support the hypothesis that IGF-IR may function as a transcription factor by binding DNA in various locations and by including SUMOylation sites.
DISCUSSION
The role of the IGF system in mammary gland biology has been well established (44). Similarly well established is the role of the IGF-IR as the main mediator of the proliferative and antiapoptotic activities of IGF-I and IGF-II in breast cancer (7, 27, 45). Although the significance of IGF-IR expression levels in terms of prognosis and clinical correlates is still a controversial issue (11, 12), the IGF-IR emerged in recent years as a promising molecular target in biological therapy protocols (46–48). Targeting efforts, however, are hampered by the extremely complex, overlapping activities of the IGF-IR and IR in many organs. In addition to mediating the effects of insulin, IR was shown to mediate the mitogenic activities of IGF-II in breast cancer cells, mainly via activation of the IR-A isoform (8, 49). Therefore, the physical and functional interactions between the IGF-IR and IR signaling pathways are of major clinical relevance. An additional major player in breast cancer development and progression is estrogen. The finely tuned bidirectional interactions between the IGF-IR/IR and ER signaling pathways, however, are still ill defined. The present study was aimed at investigating the impact of IGF-IR levels on IGF-IR biosynthesis in ER-positive and ER-depleted breast cancer cells and, furthermore, to address the putative transcription factor role of IGF-IR. Our cellular model employed MCF7-derived C4.12.5 cells, which were generated by clonal selection of MCF7 cells that were maintained in estrogen-free culture medium for a long period of time. Similarly to hormone-independent breast cancer, C4.12.5 cells express reduced levels of ERα and IGF-IR. Our study has identified a novel mechanism of autoregulation of IGF-IR gene expression by cellular IGF-IR, which is tightly dependent on ER status. Regulation of the IGF-IR gene by IGF-IR protein is mediated at the level of transcription, as indicated by 1) binding assays (DNA affinity chromatography and ChIP) showing specific IGF-IR binding to IGF-IR promoter DNA and 2) transient transfection assays showing transactivation of the IGF-IR promoter by exogenous IGF-IR. Similarly to IGF-IR, the IR is also capable of translocating to the nucleus and binding the IGF-IR promoter in ER-depleted C4.12.5 cells but not in ER-positive MCF7 cells. However, transcription factors IGF-IR and IR display diametrically opposite activities in the context of IGF-IR gene regulation. Thus, whereas IGF-IR stimulated IGF-IR gene expression, IR inhibited IGF-IR promoter activity. The ability of IR to inhibit IGF-IR promoter activity in cells with both high and low IR binding to the IGF-IR promoter is most probably explained by the fact that these regulatory mechanisms involve multiprotein complexes (22), and it is often difficult to appreciate the effect of individual transcription factors. Consistent with these results, the addition of exogenous insulin (in starvation medium) had an inhibitory effect on IGF-IR promoter activity. IGF-I, on the other hand, had only a slight stimulatory effect, which can be explained by endogenous growth factor production by MCF7 cells (22). Finally, we cannot discard the possibility that increased IGF-IR or IR expression may affect IGF-IR promoter activity via other mechanisms that do not involve direct binding to the IGF-IR promoter region.
Most early studies have shown down-regulation of IGF-IR by its cognate ligand. One of us has previously demonstrated that IGF-I reduced IGF-IR mRNA levels and IGF-IR promoter activity in muscle and neuroblastoma cell lines (50). The present study evaluated the regulation of IGF-IR gene expression by IGF-IR or IR levels, regardless of their activation status. The finding that nuclear IGF-IR, but not IR, can enhance IGF-IR promoter activity in ER-depleted cells may constitute an important mechanism responsible for cell cycle progression at advanced (ER-independent) stages of the disease.
Recent proteomic analyses led to the identification of a series of nuclear proteins that are probably responsible for the regulation of IGF-IR gene expression in ER-positive and ER-negative breast cancers (22). In addition, we have previously established that the IGF-IR gene is under inhibitory control by a number of tumor suppressors (e.g. p53 and BRCA1) with important roles in the etiology of the disease (20, 51–53). Mutant forms of p53 and BRCA1 are impaired in their ability to repress IGF-IR promoter activity and may lead to enhanced IGF-IR levels, associated with a more aggressive disease (17). Likewise, we have shown that ERα enhances IGF-IR promoter activity via interaction with zinc finger protein Sp1, a potent GC box-binding transactivator of the IGF-IR gene (30). Our results are consistent with competition between IGF-IR or IR and ERα proteins for Sp1 binding to IGF-IR promoter sequences. Specifically, transcription factors IGF-IR and IR are able to bind to the IGF-IR promoter only in cells with reduced ER levels and, consequently, diminished ER binding to IGF-IR promoter DNA (most probably via GC box-binding Sp1). Using electrophoretic mobility shift assays, Sehat et al. (33) have recently shown binding of IGF-IR to randomly synthesized double-stranded DNA probes. To the best of our knowledge, no previous studies have shown IGF-IR and IR binding to the IGF-IR promoter.
Our data confirm recently published results showing that IGF-IR nuclear translocation is mediated by SUMOylation (33). The SUMOylation sites on lysine residues within the tyrosine kinase domain were found to be conserved among a variety of homologues from different organisms, and their mutagenesis abolished the ability to translocate to the nucleus and activate transcription. Nuclear localization of the IGF-IR close homologue, epidermal growth factor receptor, and its potential role as a transcription factor was shown previously (54). IGF-IR and epidermal growth factor receptor show 20.5% identity (32.7% similarity, by global pairwise alignment), both belonging to the receptor-L-domain protein family (PF01030). However, when comparing the protein-kinase domain in these two proteins, 33% identity (50% similarity) is found. Such similarity is thought to suggest common functions proposed for both epidermal growth factor receptor and IGF-IR. More specifically, the faculty of IGF-IR to translocate to the nucleus in human tumor cells, to interact with chromatin, and, consequently, to function as a transcriptional regulator was predicted to be localized at the tyrosine kinase domain (34). We show here that predictions for DNA binding residues are mainly at the tyrosine kinase domain. These DNA binding sites are rather conserved (supplemental Fig. 2). Few other binding sites along the protein are also found.
In summary, our studies demonstrate that IGF-IR and IR are localized in the nuclear and perinuclear areas of breast cancer cells. However, whereas nuclear IGF-IR acts as a transcriptional activator of its own promoter, nuclear IR functions as a negative regulator of IGF-IR promoter activity. The clinical implications of these findings and, in particular, the impact of IGF-IR/IR nuclear localization on targeted therapy require further investigation.
Supplementary Material
Acknowledgments
We thank W. V. Welshons, D. Accilli, and R. O'Connor for providing cell lines and reagents.

This article contains supplemental Figs. 1 and 2.
- IGF
- insulin-like growth factor
- IGF-IR
- insulin-like growth factor-I receptor
- IR
- insulin receptor
- ER
- estrogen receptor
- SUMO
- small ubiquitin-like modifier
- IP
- immunoprecipitation
- nt
- nucleotide
- PI
- propidium iodide
- NRS
- normal rabbit serum.
REFERENCES
- 1. Rowzee A. M., Lazzarino D. A., Rota L., Sun Z., Wood T. L. (2008) IGF ligand and receptor regulation of mammary development. J. Mammary Gland Biol. Neoplasia 13, 361–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Werner H., Le Roith D. (2000) New concepts in regulation and function of the insulin-like growth factors. Implications for understanding normal growth and neoplasia. Cell Mol. Life Sci. 57, 932–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Surmacz E. (2000) Function of the IGF-I receptor in breast cancer. J. Mammary Gland Biol. Neoplasia 5, 95–105 [DOI] [PubMed] [Google Scholar]
- 4. Chitnis M. M., Yuen J. S., Protheroe A. S., Pollak M., Macaulay V. M. (2008) The type 1 insulin-like growth factor receptor pathway. Clin. Cancer Res. 14, 6364–6370 [DOI] [PubMed] [Google Scholar]
- 5. Werner H., Bruchim I. (2009) The insulin-like growth factor-I receptor as an oncogene. Arch. Physiol. Biochem. 115, 58–71 [DOI] [PubMed] [Google Scholar]
- 6. Samani A. A., Yakar S., LeRoith D., Brodt P. (2007) The role of the IGF system in cancer growth and metastasis. Overview and recent insights. Endocr. Rev. 28, 20–47 [DOI] [PubMed] [Google Scholar]
- 7. Pollak M. (2008) Insulin and insulin-like growth factor signaling in neoplasia. Nat. Rev. Cancer 8, 915–928 [DOI] [PubMed] [Google Scholar]
- 8. Belfiore A., Frasca F. (2008) IGF and insulin receptor signaling in breast cancer. J. Mammary Gland Biol. Neoplasia 13, 381–406 [DOI] [PubMed] [Google Scholar]
- 9. Sell C., Rubini M., Rubin R., Liu J. P., Efstratiadis A., Baserga R. (1993) Simian virus 40 large tumor antigen is unable to transform mouse embryonic fibroblasts lacking type 1 insulin-like growth factor receptor. Proc. Natl. Acad. Sci. U.S.A. 90, 11217–11221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Happerfield L. C., Miles D. W., Barnes D. M., Thomsen L. L., Smith P., Hanby A. (1997) The localization of the insulin-like growth factor receptor 1 (IGFR-1) in benign and malignant breast tissue. J. Pathol. 183, 412–417 [DOI] [PubMed] [Google Scholar]
- 11. Yerushalmi R., Gelmon K. A., Leung S., Gao D., Cheang M., Pollak M., Turashvili G., Gilks B. C., Kennecke H. (2011) Insulin-like growth factor receptor (IGF-1R) in breast cancer subtypes. Breast Cancer Res. Treat. [DOI] [PubMed] [Google Scholar]
- 12. Werner H. (2009) For debate. The pathophysiological significance of IGF-I receptor overexpression. New insights. Pediatr. Endocrinol. Rev. 7, 2–5 [PubMed] [Google Scholar]
- 13. Werner H., Maor S. (2006) The insulin-like growth factor-I receptor gene. A downstream target for oncogene and tumor suppressor action. Trends Endocrinol. Metab. 17, 236–242 [DOI] [PubMed] [Google Scholar]
- 14. Peyrat J. P., Bonneterre J. (1992) Type 1 IGF receptor in human breast diseases. Breast Cancer Res. Treat. 22, 59–67 [DOI] [PubMed] [Google Scholar]
- 15. Jones R. A., Campbell C. I., Gunther E. J., Chodosh L. A., Petrik J. J., Khokha R., Moorehead R. A. (2007) Transgenic overexpression of IGF-IR disrupts mammary ductal morphogenesis and induces tumor formation. Oncogene 26, 1636–1644 [DOI] [PubMed] [Google Scholar]
- 16. Schnarr B., Strunz K., Ohsam J., Benner A., Wacker J., Mayer D. (2000) Down-regulation of insulin-like growth factor-I receptor and insulin receptor substrate-1 expression in advanced human breast cancer. Int. J. Cancer 89, 506–513 [DOI] [PubMed] [Google Scholar]
- 17. Maor S., Yosepovich A., Papa M. Z., Yarden R. I., Mayer D., Friedman E., Werner H. (2007) Elevated insulin-like growth factor-I receptor (IGF-IR) levels in primary breast tumors associated with BRCA1 mutations. Cancer Lett. 257, 236–243 [DOI] [PubMed] [Google Scholar]
- 18. Werner H., Bach M. A., Stannard B., Roberts C. T., Jr., LeRoith D. (1992) Structural and functional analysis of the insulin-like growth factor I receptor gene promoter. Mol. Endocrinol. 6, 1545–1558 [DOI] [PubMed] [Google Scholar]
- 19. Cooke D. W., Bankert L. A., Roberts C. T., Jr., LeRoith D., Casella S. J. (1991) Analysis of the human type I insulin-like growth factor receptor promoter region. Biochem. Biophys. Res. Commun. 177, 1113–1120 [DOI] [PubMed] [Google Scholar]
- 20. Idelman G., Glaser T., Roberts C. T., Jr., Werner H. (2003) WT1-p53 interactions in insulin-like growth factor-I receptor gene regulation. J. Biol. Chem. 278, 3474–3482 [DOI] [PubMed] [Google Scholar]
- 21. Smale S. T., Baltimore D. (1989) The “initiator” as a transcription control element. Cell 57, 103–113 [DOI] [PubMed] [Google Scholar]
- 22. Sarfstein R., Belfiore A., Werner H. (2010) Identification of insulin-like growth factor-I receptor (IGF-IR) gene promoter-binding proteins in estrogen receptor (ER)-positive and ER-depleted breast cancer cells. Cancers 2, 233–261 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23. Oesterreich S., Zhang P., Guler R. L., Sun X., Curran E. M., Welshons W. V., Osborne C. K., Lee A. V. (2001) Re-expression of estrogen receptor α in estrogen receptor α-negative MCF-7 cells restores both estrogen and insulin-like growth factor-mediated signaling and growth. Cancer Res. 61, 5771–5777 [PubMed] [Google Scholar]
- 24. Becker M. A., Ibrahim Y. H., Cui X., Lee A. V., Yee D. (2011) The IGF pathway regulates ERα through a S6K1-dependent mechanism in breast cancer cells. Mol. Endocrinol. 25, 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Casa A. J., Potter A. S., Malik S., Lazard Z., Kuiatse I., Kim H. T., Tsimelzon A., Creighton C. J., Hilsenbeck S. G., Brown P. H., Oesterreich S., Lee A. V. (2011) Estrogen and insulin-like growth factor-I (IGF-I) independently down-regulate critical repressors of breast cancer growth. Breast Cancer Res. Treat., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martin M. B., Stoica A. (2002) Insulin-like growth factor-I and estrogen interactions in breast cancer. J. Nutr. 132, 3799S–3801S [DOI] [PubMed] [Google Scholar]
- 27. Yee D., Lee A. V. (2000) Cross-talk between the insulin-like growth factors and estrogens in breast cancer. J. Mammary Gland Biol. Neoplasia 5, 107–115 [DOI] [PubMed] [Google Scholar]
- 28. Kato S., Endoh H., Masuhiro Y., Kitamoto T., Uchiyama S., Sasaki H., Masushige S., Gotoh Y., Nishida E., Kawashima H., Metzger D., Chambon P. (1995) Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science 270, 1491–1494 [DOI] [PubMed] [Google Scholar]
- 29. Martin M. B., Franke T. F., Stoica G. E., Chambon P., Katzenellenbogen B. S., Stoica B. A., McLemore M. S., Olivo S. E., Stoica A. (2000) A role for Akt in mediating the estrogenic functions of epidermal growth factor and insulin-like growth factor I. Endocrinology 141, 4503–4511 [DOI] [PubMed] [Google Scholar]
- 30. Maor S., Mayer D., Yarden R. I., Lee A. V., Sarfstein R., Werner H., Papa M. Z. (2006) Estrogen receptor regulates insulin-like growth factor-I receptor gene expression in breast tumor cells. Involvement of transcription factor Sp1. J. Endocrinol. 191, 605–612 [DOI] [PubMed] [Google Scholar]
- 31. Kahlert S., Nuedling S., van Eickels M., Vetter H., Meyer R., Grohe C. (2000) Estrogen receptor α rapidly activates the IGF-1 receptor pathway. J. Biol. Chem. 275, 18447–18453 [DOI] [PubMed] [Google Scholar]
- 32. Deng H., Lin Y., Badin M., Vasilcanu D., Strömberg T., Jernberg-Wiklund H., Sehat B., Larsson O. (2011) Overaccumulation of nuclear IGF-1 receptor in tumor cells requires elevated expression of the receptor and the SUMO-conjugating enzyme Ubc9. Biochem. Biophys. Res. Commun. 404, 667–671 [DOI] [PubMed] [Google Scholar]
- 33. Sehat B., Tofigh A., Lin Y., Trocmé E., Liljedahl U., Lagergren J., Larsson O. (2010) SUMOylation mediates the nuclear translocation and signaling of the IGF-1 receptor. Sci. Signal. 3, ra10. [DOI] [PubMed] [Google Scholar]
- 34. Aleksic T., Chitnis M. M., Perestenko O. V., Gao S., Thomas P. H., Turner G. D., Protheroe A. S., Howarth M., Macaulay V. M. (2010) Type 1 insulin-like growth factor receptor translocates to the nucleus of human tumor cells. Cancer Res. 70, 6412–6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Werner H., Rauscher F. J., 3rd, Sukhatme V. P., Drummond I. A., Roberts C. T., Jr., LeRoith D. (1994) Transcriptional repression of the insulin-like growth factor I receptor (IGF-I-R) gene by the tumor suppressor WT1 involves binding to sequences both upstream and downstream of the IGF-I-R gene transcription start site. J. Biol. Chem. 269, 12577–12582 [PubMed] [Google Scholar]
- 36. Boutet E., Lieberherr D., Tognolli M., Schneider M., Bairoch A. (2007) UniProtKB/Swiss-Prot. Methods Mol. Biol. 406, 89–112 [DOI] [PubMed] [Google Scholar]
- 37. Wang L., Brown S. J. (2006) BindN. A web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res. 34, W243–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ahmad S., Gromiha M. M., Sarai A. (2004) Analysis and prediction of DNA-binding proteins and their binding residues based on composition, sequence, and structural information. Bioinformatics 20, 477–486 [DOI] [PubMed] [Google Scholar]
- 39. Hwang S., Gou Z., Kuznetsov I. B. (2007) DP-Bind. A web server for sequence-based prediction of DNA-binding residues in DNA-binding proteins. Bioinformatics 23, 634–636 [DOI] [PubMed] [Google Scholar]
- 40. Ofran Y., Mysore V., Rost B. (2007) Prediction of DNA-binding residues from sequence. Bioinformatics 23, i347–i353 [DOI] [PubMed] [Google Scholar]
- 41. Ashkenazi H., Erez E., Martz E., Pupko T., Ben-Tal N. (2010) ConSurf 2010. Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Katoh K., Kuma K., Toh H., Miyata T. (2005) MAFFT version 5. Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sun H., Tu X., Prisco M., Wu A., Casiburi I., Baserga R. (2003) Insulin-like growth factor I receptor signaling and nuclear translocation of insulin receptor substrates 1 and 2. Mol. Endocrinol. 17, 472–486 [DOI] [PubMed] [Google Scholar]
- 44. Cannata D., Lann D., Wu Y., Elis S., Sun H., Yakar S., Lazzarino D. A., Wood T. L., Leroith D. (2010) Elevated circulating IGF-I promotes mammary gland development and proliferation. Endocrinology 151, 5751–5761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Creighton C. J., Casa A., Lazard Z., Huang S., Tsimelzon A., Hilsenbeck S. G., Osborne C. K., Lee A. V. (2008) Insulin-like growth factor-I activates gene transcription programs strongly associated with poor breast cancer prognosis. J. Clin. Oncol. 26, 4078–4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Scotlandi K., Picci P. (2008) Targeting insulin-like growth factor 1 receptor in sarcomas. Curr. Opin. Oncol. 20, 419–427 [DOI] [PubMed] [Google Scholar]
- 47. Yuen J. S., Macaulay V. M. (2008) Targeting the type 1 insulin-like growth factor receptor as a treatment for cancer. Expert Opin. Ther. Targets 12, 589–603 [DOI] [PubMed] [Google Scholar]
- 48. Bruchim I., Attias Z., Werner H. (2009) Targeting the IGF1 axis in cancer proliferation. Expert Opin. Ther. Targets 13, 1179–1192 [DOI] [PubMed] [Google Scholar]
- 49. Vigneri R., Squatrito S., Sciacca L. (2010) Insulin and its analogs. Actions via insulin and IGF receptors. Acta Diabetol. 47, 271–278 [DOI] [PubMed] [Google Scholar]
- 50. Hernández-Sánchez C., Werner H., Roberts C. T., Jr., Woo E. J., Hum D. W., Rosenthal S. M., LeRoith D. (1997) Differential regulation of insulin-like growth factor-I (IGF-I) receptor gene expression by IGF-I and basic fibroblastic growth factor. J. Biol. Chem. 272, 4663–4670 [DOI] [PubMed] [Google Scholar]
- 51. Werner H., Karnieli E., Rauscher F. J., LeRoith D. (1996) Wild-type and mutant p53 differentially regulate transcription of the insulin-like growth factor I receptor gene. Proc. Natl. Acad. Sci. U.S.A. 93, 8318–8323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Abramovitch S., Werner H. (2003) Functional and physical interactions between BRCA1 and p53 in transcriptional regulation of the IGF-IR gene. Horm. Metab. Res. 35, 758–762 [DOI] [PubMed] [Google Scholar]
- 53. Maor S. B., Abramovitch S., Erdos M. R., Brody L. C., Werner H. (2000) BRCA1 suppresses insulin-like growth factor-I receptor promoter activity. Potential interaction between BRCA1 and Sp1. Mol. Genet. Metab. 69, 130–136 [DOI] [PubMed] [Google Scholar]
- 54. Lin S. Y., Makino K., Xia W., Matin A., Wen Y., Kwong K. Y., Bourguignon L., Hung M. C. (2001) Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell Biol. 3, 802–808 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.