Abstract
Most solid tumors and their metastases experience periods of low oxygen or hypoxia, which is of major clinical significance as it promotes both tumor progression and resistance to therapy. Critical mediators of the hypoxic response are the hypoxia-inducible factors HIF-1α and HIF-2α. The HIFs are nonredundant and regulate both overlapping and unique downstream target genes. Here, we describe a novel mechanism for the switch between HIF-1α– and HIF-2α–dependent transcription during tumor hypoxia caused by the hypoxia associated factor (HAF). HAF is overexpressed in a variety of tumors and its levels are decreased during acute hypoxia, but increased following prolonged hypoxia. We have previously identified HAF as an E3 ubiquitin ligase that binds and ubiquitinates HIF-1α by an oxygen and pVHL-independent mechanism, thus targeting HIF-1α for proteasomal degradation. Here, we show that HAF also binds to HIF-2α, but at a different site than HIF-1α, and increases HIF-2α transactivation without causing its degradation. HAF, thus, switches the hypoxic response of the cancer cell from HIF-1α–dependent to HIF-2α–dependent transcription and activates genes involved in invasion such as MMP9, PAI-1, and the stem cell factor OCT-3/4. The switch to HIF-2α–dependent gene expression caused by HAF also promotes an enriched tumor stem cell population, resulting in highly aggressive tumors in vivo. Thus, HAF, by causing a switch from a HIF-1α– to HIF-2α–dependent response to hypoxia, provides a mechanism for more aggressive growth of tumors under prolonged hypoxia.
Introduction
Regions of low oxygen or hypoxia can be found in most solid tumors and their metastases. Tumor hypoxia is of major clinical significance as it promotes both tumor progression and resistance to therapy (1). This occurs as a result of a coordinated set of responses orchestrating the cellular adaptation to hypoxia. Hypoxia is also associated with the promotion and maintenance of cancer stem cells, a minority subpopulation within the tumor cell mass with the capacity of self-renewal and long-term proliferation believed to be responsible for tumor recurrence and resistance to chemotherapy (2, 3) and for the epithelial to mesenchymal transition (EMT), a process involved in tumor invasion and metastatic spread (4).
Critical mediators of the hypoxic response are the hypoxia-inducible factor (HIF), transcription factors that transactivate a large number of genes promoting angiogenesis, anaerobic metabolism, and resistance to apoptosis (5). HIFs are hetero-dimers comprising 1 of 2 major oxygen labile HIF-α subunits (HIF-1α and HIF-2α), and a stable HIF-1β subunit (6, 7). Under aerobic conditions, HIF-α is ubiquitinated by the von Hippel Lindau protein (pVHL) and targeted for proteasomal degradation (8). During hypoxia, pVHL binding is abrogated and HIF-α is stabilized and enters the nucleus, where it hetero-dimerizes with HIF-1β and binds to a conserved DNA sequence known as the hypoxia-responsive element (HRE), to transactivate a variety of hypoxia-responsive genes (9).
HIF-1α and HIF-2α have 48% amino acid identity and similar protein structures, and exhibit both common and unique patterns of downstream gene induction. HIF-1α preferentially induces glycolytic enzyme genes (10, 11), whereas HIF-2α induces genes involved in invasion such as the matrix metalloproteinases (MMP; ref. 12), PAI-1 (13), and the stem cell factor OCT-3/4 (14).
Tumor cells are subjected to a range of oxygen tensions and experience periods of acute/intermittent hypoxia (such as during blood vessel occlusion and reperfusion events), or chronic/prolonged hypoxia (such as in tumor regions distant from blood vessels). The variability in hypoxic intensity and duration necessitates distinct sets of cellular responses appropriate for each condition. In this respect, HIF-1α seems to have the dominant role in controlling responses to acute hypoxia, whereas HIF-2α drives the response to chronic hypoxia (15). The mechanism responsible for the selectivity is unclear but may include HIF-1α feedback regulation under chronic hypoxia (16).
Elevated levels of tumor HIF-1α have been associated with poor patient survival in multiple tumor types (7). Elevated HIF-2α has also been associated with poor patient survival and prognosis in specific tumor types such as renal cell carcinoma (RCC), neuroblastoma, astrocytoma, glioblastoma (GBM), and non–small cell lung cancer (15, 17–19). HIF-2α drives tumor progression in RCC in which there is a gradual shift from HIF-1α to HIF-2α expression with increasing tumor grade (20). HIF-2α (but not HIF-1α) has been shown to cooperate with a number of oncoproteins frequently deregulated in cancer such as c-Myc, epidermal growth factor receptor (EGFR), and K-Ras (14, 18, 21) and has been linked to increased tumor aggressiveness through the promotion of self-renewal and EMT (14, 21). However, despite increased interest in delineating the distinct roles of HIF-1α and HIF-2α in cancer, the mechanisms by which tumor cells regulate and switch from HIF-1α– to HIF-2α–dependent transcription remain unclear.
The hypoxia-associated factor (HAF, also known as SART1800 or squamous cell carcinoma antigen recognized by T cells) is overexpressed in a variety of tumor types (22–24). We previously identified HAF as an E3 ubiquitin ligase that binds to and ubiquitinates HIF-1α by an oxygen- and pVHL-independent mechanism, targeting HIF-1α for proteasomal degradation (25). HAF expression lowers HIF-1α levels and decreases HIF-1 transactivating activity. We now show that HAF also binds to HIF-2α but does not lead to its degradation but instead increases HIF-2 transactivating activity. Thus, HAF expression switches the hypoxia response of the cancer cell from HIF-1α– to the HIF-2α–dependent transcription of genes such as MMP9 and OCT-3/4. We show that this switch by HAF promotes the cancer stem cell phenotype and invasion, resulting in highly aggressive tumors in vivo.
Materials and Methods
Tissue culture
PANC-1, LN229, U87, RCC4, and 786-0 cells were from ATCC. The identities of all cell lines were authenticated by the Molecular Cytogenetics Facility at MDACC. U87 cells were maintained in Minimum Essential Medium (Invitrogen), whereas all others were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen), supplemented with 10% FBS and 2 μg/mL puromycin where appropriate. Neurobasal media was prepared as previously described (3). Hypoxic incubations (1% or 0.5% O2) were done by using the INVIVO2 Hypoxia Workstation (Biotrace International Inc.).
Plasmid construction
The pCMV14-3XFLAG-HAF was previously described (25) and used as a template for PCR, with primers containing EcoRI sites and ligated into EcoRI-digested pMX-IRES-GFP (26). Full-length and HAF truncations were ligated into pGEX-4T1 by using EcoR1 and Xho1 (GE Healthcare). HIF-2α and truncations in pcDNA3.2-V5/GW (Invitrogen) were generated by using Gateway recombination. HIF-2α (604–750) was ligated into pCMV/myc/nuc (Invitrogen) by using Sal1 and Not1.
Cell transfection
Stable pools of HAF overexpressing cells were generated by retroviral infection of pMX-HAF-IRES-GFP or pMX-IRES-GFP (26). Transient siRNA transfections were done by using Lipofectamine 2000 and Dharmacon SMARTpool siRNAs for HAF, HIF-1α, HIF-2α, or nontargeting pool #5 (Scr) or HIF-2α predesigned siRNA duplex for HIF-2α (D-004814-02). Knockdown efficacy was determined by using TaqMan qRT-PCR (quantitative reverse transcriptase PCR).
Microarray analysis
Global gene expression was measured by using Affymetrix Exon Arrays (Affymetrix). Briefly, LN229 cells were transfected with siHIF-1α, siHIF-2α, or nontargeting (Scr) siRNA for 48 hours, placed into hypoxia for 16 hours, after which RNA was harvested by using the RNEasy Kit (Qiagen). Samples were processed by using Affymetrix GeneChip Whole Transcript (WT) Sense Target Labeling Assay protocol (see Supplementary Experimental Procedures). Partek software was used to generate the difference between 2 groups of the samples at the gene level. Cut offs were set at fold change greater than 2; P < 0.05; GEO Accession number GSE27523.
Quantitative RT-PCR
Total RNA was isolated from cell lysates by using the RNEasy Kit with DNAse I step. TaqMan qRT-PCR was done by using the ABI 7300 System with One-Step RT-PCR Master Mix Kit and predesigned primer/probes (Life technologies) as previously described (25).
Western blotting and immunoprecipitation
Western blotting was done as previously described (27). Primary antibodies: HIF-1α (BD Biosciences), HIF-2α (NB100–122; Novus Biologicals), FLAG (Sigma-Aldrich), V5 (Invitrogen), actin (Santa Cruz Biotechnology Inc.), and HAF (25). Immunoprecipitation (IP) was done as described (25). Gelatin zymography was done by using precast 10% gelatin gels and buffers (Bio-Rad) according to standard protocols.
Luciferase reporter assay
The HIF luciferase reporter plasmid in the pGL3 vector backbone (Promega) was a gift from R. Gillies (Arizona Cancer Center, Tucson, AZ). Assays were done by using the Dual-Glo Luciferase Assay System (Promega) according to the manufacturer’s protocol.
GST pull down assays
V5-HIF-2α and truncations were generated by using TNT Quick Coupled Transcription/Translation Systems (Promega). Pull down assays were conducted as described previously (25).
CD133+ cell sorting
CD133+ cells were labeled by using CD133/2 (293C3)-APC or APC isotype control antibody according to the manufacturer’s protocol (Miltenyi Biotec Inc.). GFP+/CD133+ cells were isolated by using a BD FACSAria II Cell Sorter (Becton Dickinson Immunocytometry Systems).
Invasion assays
These were done by using either the Laminin I–coated Cultrex Cell Invasion Assay Kit (24-well; Trevigen), or the BD BioCoat Matrigel (GFR) Invasion Chambers (BD Biosciences), according to the manufacturer’s protocol. Cells were serum-starved overnight before seeding, whereas serum-containing media was added to the bottom wells as a chemoattractant.
Three-dimensional assays
Three-dimensional (3D) assays were done either by using 8-well chamber slides (Nunc) or high-binding 96-well NanoCulture plates with a microsquare pattern (SCIVAX Corp., B-Bridge International). Matrigel assays were conducted by adding cell suspensions in 2% Matrigel on chamber slides precoated with 100% Matrigel (BD). NanoCulture plate assays were done by seeding 3 × 104 cells per well. Both were left a further 3 days in hypoxia or normoxia. For siRNA experiments, cells were transfected with siRNA for 72 hours, trypsinized and reseeded into NanoCulture plates. Colonies were imaged by using the IN Cell Analyzer 1000 (GE Healthcare) and measured by using the Oxford Optronix GelCount System.
Intracranial xenografting of U87 GBM cells
Floating U87 cells were pelleted and combined with adherent U87 cells that were detached by trypsinization and injected at desired concentrations in 5 μL into the right frontal lobe of nude mice as described previously (28). Animals were sacrificed when moribund.
Results
HAF differentially regulates HIF-1α and HIF-2α
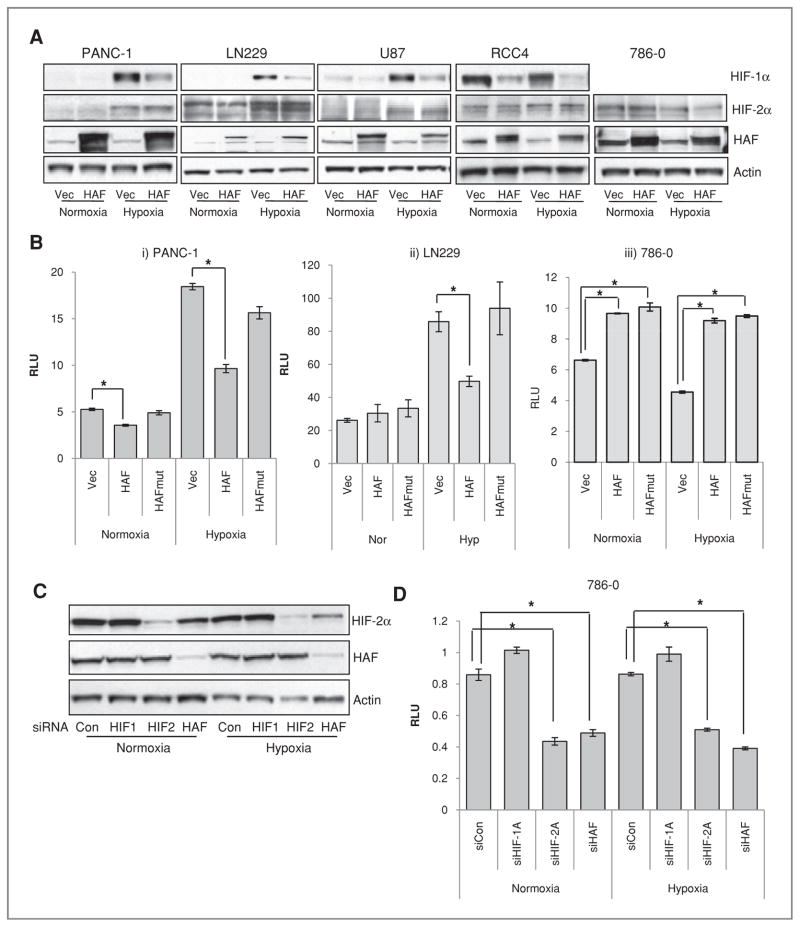
We first confirmed that HAF overexpression decreased HIF-1α levels in a panel of cell lines including PANC-1 pancreatic cancer cells, LN229 and U87 GBM cells, and pVHL-deficient RCC4 and 786-0 RCC cells, but did not affect levels of HIF-2α in any of the cells either in normoxia or hypoxia (1% O2; Fig. 1A). It should be noted that 786-0 RCC cells constitutively express HIF-2α even in normoxia but do not express HIF-1α (29). Overexpression of HAF significantly decreased HRE-luciferase reporter activity in hypoxia (and to a lesser extent, in normoxia) in PANC-1 and LN229 cells, but increased HRE-luciferase activity in 786-0 cells (Fig. 1B). Overexpression of a HAF mutant construct lacking the E3 ligase domain (HAFmut) prevented the HAF-induced decrease in HRE-lucifease activity observed in the PANC-1 and LN229 cells, but still significantly increased HRE-luciferase activity in 786-0 cells, showing that the HAF induced activation of HIF-2α in 786-0 cells is independent of its E3 ligase activity (Fig. 1B). Knockdown of HAF in 786-0 cells decreased HIF-2α protein (Fig. 1C) and HRE-luciferase activity (Fig. 1D) without affecting HIF-2α mRNA (data not shown). The decrease in HRE-luciferase was comparable to that achieved by complete knockdown of HIF-2α. The results suggest that in the absence of HIF-1α, HAF may be required for maintenance of HIF-2α levels and transactivation. Thus, HAF decreases HIF-1α protein levels and activity but is required for HIF-2α activity through a mechanism independent of the HAF E3 ligase domain.
Figure 1.
The effect of HAF overexpression on levels and activity of HIF-1α and HIF-2α. A, Western blots showing the effect of FLAG-HAF overexpression on HIF-1α and HIF-2α levels in normoxia or hypoxia. B, the effect of HAF overexpression on HRE-Luciferase (Luc) activity normalized to constitutive Renilla Luc. C, Western blot showing the effect of HIF-1α, HIF-2α, or HAF siRNA transfection on HIF-2α and HAF levels with accompanying HRE-luc activity (D) in 786-0 cells stably expressing HRE-Luc and Renilla Luc. Results shown as relative light units (RLU) of a representative experiment from at least 3 independent experiments and are the mean ± SE (*, P < 0.05).
Characterization of the HAF–HIF-2α interaction
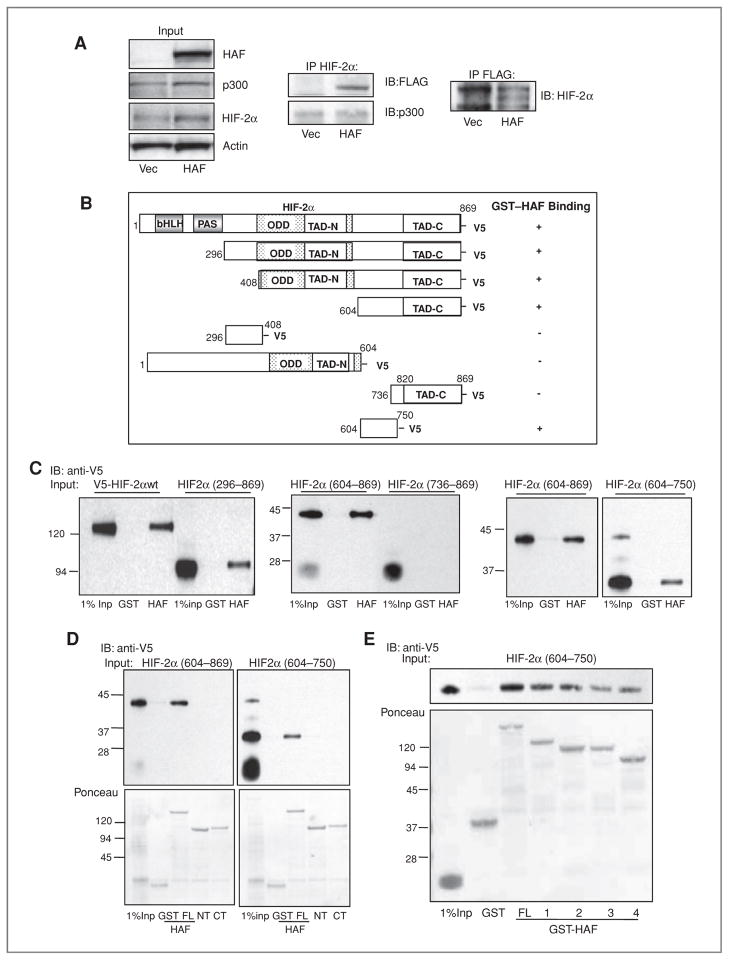
We previously showed that HAF and HIF-1α bind within the HIF-1α (298–400) and HAF (654–800) domains (25). To determine whether HAF can also bind to HIF-2α, we conducted IP studies in U87 GBM cells transfected with FLAG-HAF. We found that FLAG-HAF could be detected associated with immunoprecipitated HIF-2α and vice versa, showing that HAF and HIF-2α interact in cells (Fig. 2A). HAF overexpression did not affect the recruitment of the transcriptional coactivator p300/CBP to HIF-2α (Fig. 2A).
Figure 2.
Mapping of the HAF–HIF-2α interaction domains. A, Western blot showing the co-IP of FLAG-HAF with HIF-2α and vice versa in U87 cells stably overexpressing FLAG-HAF. B, schematic diagram showing HIF-2α deletion mutants used in pull down assays and their ability to bind GST-HAF as detected by Western blot. C, Western blots showing results of pull down assays depicted in B. The binding of GST-HAF to V5-HIF-2α was detected by using anti-V5 antibodies. D, Western blot showing the results of pull down assays by using GST-HAF (full length, FL), GST-HAF 1–422 (N-terminus, NT), or GST-HAF 396–800 (C-terminus, CT) and in vitro transcribed/translated V5-HIF-2α deletion mutants. E, mapping of the HAF-binding region of HIF-2α (604–750). HAF truncations: FL – full length; 1–HAF (200–600); 2–HAF (300–600); 3–HAF (200—500); 4–HAF (300–500). IB, immunoblotting.
To identify the interaction domains between HAF and HIF-2α, we used recombinant GST-HAF to pull down in vitro transcribed/translated V5-HIF-2α and V5-HIF-2α deletion mutants (schematic depiction in Fig. 2B). We first found that HAF does not bind to HIF-2α (298–400), although this region has 56% similarity to residues in HIF-1α (298–400), the HAF-binding domain within HIF-1α (Supplementary Data S1A). Instead, we found that the minimal binding domain of HAF to HIF-2α lies within residues HIF-2α (604–750; Fig. 2C–E, with representative Ponceau staining shown in Supplementary Data S1B). Further pull down studies by using full-length HAF (HAF FL), HAF (1–422; HAF N-terminus, NT), or HAF (396–800 HAF C-terminus, CT) showed that HIF-2α (604–750) binds to HAF FL but not to HAF NT or HAF CT (Fig. 2D). This suggested that HAF may bind to HIF-2α within the boundary between HAF NT and HAF CT, which was eliminated in both HAF NT and HAF CT. Indeed, further pull downs showed that the minimal HAF interaction domain of HIF-2α (604–750) is HAF (300–500; Fig. 2E). Hence, HAF–HIF-2α binding occurs between HAF (300–500) and HIF-2α (604–750).
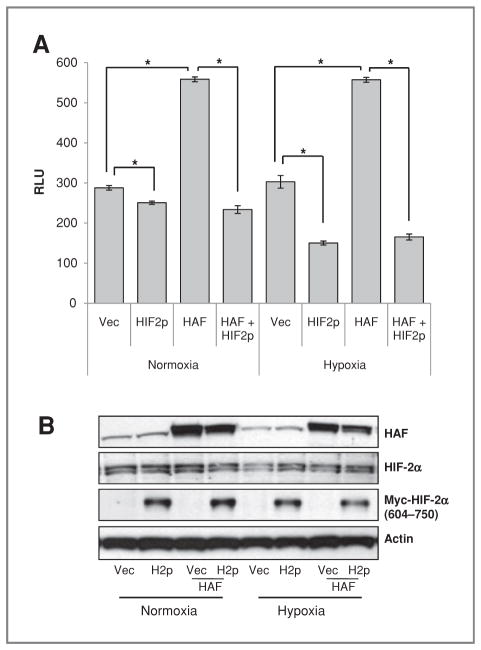
To investigate the effect of the HAF–HIF-2α interaction on HIF-2α activity, HIF-2α (604–750; H2p) was overexpressed in 786-0 cells to act as a dominant negative inhibitor of the interaction between HAF and HIF-2α. H2p overexpression decreased both endogenous and HAF-induced HRE-luciferase activity under both normoxic and hypoxic conditions without changing levels of HIF-2α protein, suggesting that the interaction of HAF with HIF-2α is necessary for HIF-2α transactivation (Fig. 3A and B).
Figure 3.
The effect of inhibition of HAF–HIF-2α binding on HIF-2α transactivation. A, HRE-Luciferase assay showing the effect of inhibition of the HAF–HIF-2α interaction by overexpression of HIF-2α (604–750; HIF2p) on endogenous and HAF-induced HRE activity in 786-0 cells. Cells were transiently transfected (equal total DNA transfected in each condition) for 48 hours and then incubated for a further 16 hours in normoxia or hypoxia. Results are the mean of a representative experiment from at least 2 independent experiments ± SE. *, P < 0.05. B, Western blot for representative experiment. RLU, relative light units.
The implications of these data are 2-fold; first that HAF induces the activation of HIF-2α through an interaction between HAF (300–500) and HIF-2α (604–750), and second that endogenous HAF is a necessary coactivator of HIF-2α. The distinct HAF-binding regions on HIF-1α (298–400) and on HIF-2α (604–750) may provide a mechanism for the differential effects of HAF, resulting in HIF-1α degradation and increased HIF-2α transactivation.
HAF overexpression shifts hypoxia-dependent transcription from HIF-1α to HIF-2α
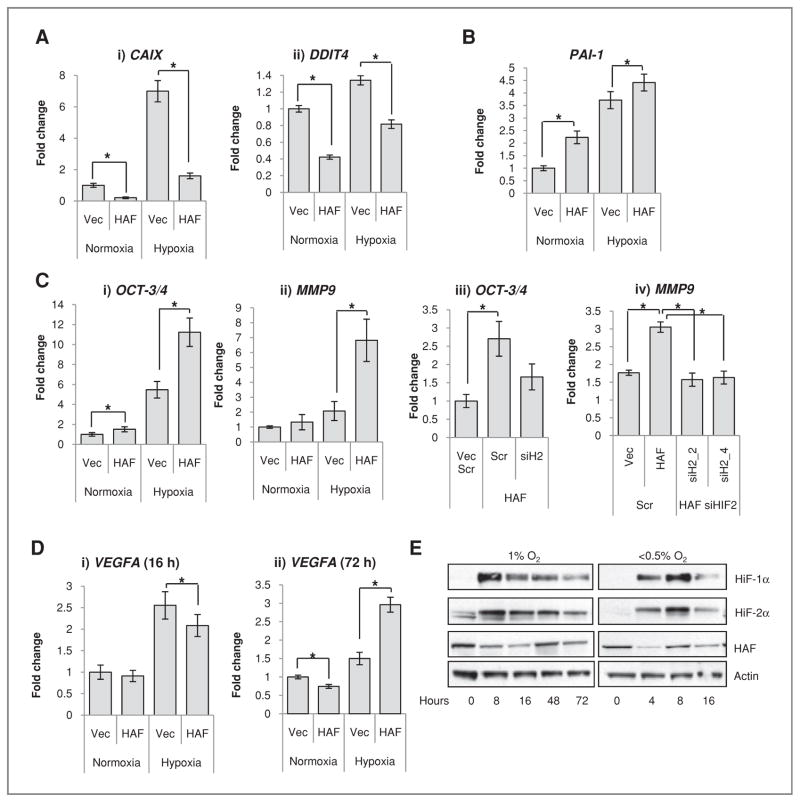
To identify specific HIF-1α and HIF-2α target genes in GBM cells, we carried out microarray analyses of LN229 cells transfected with siRNA to HIF-1α, HIF-2α, or nontargeting control. siRNA transfection resulted in a more than 90% decrease in respective HIF-1α and HIF-2α protein (Supplementary Data S2A). HAF mRNA and protein levels were unaffected by knockdown of either HIF-1α or HIF-2α (not shown). We found 82 hypoxia-inducible genes that were regulated by both HIF-1α and HIF-2α, 190 genes that were specifically induced by HIF-1α, whereas only 1 gene, plasminogen activator inhibitor 1 (PAI-1), was specifically induced by HIF-2α. Intriguingly, only 9 hypoxia-inducible genes were unchanged by either HIF-1α or HIF-2α knockdown. An abbreviated list of specific HIF-1α and HIF-2α target genes is shown in Supplementary Data S2B. This bias toward HIF-1α signaling is similar to that observed previously in other non-RCC cell lines (30, 31). By using these lists as identifiers for HIF-1α-or HIF-2α–dependent genes, we found that HAF overexpression decreased levels of the HIF-1α–specific target genes carbonic anhydrase 9 (CAIX) and DNA damage–inducible transcript 4 (DDIT4) in both normoxia and hypoxia in U87 cells (Fig. 4Ai, ii). In contrast, HAF overexpression significantly increased levels of the sole HIF-2α–dependent gene PAI-1 in both normoxia and hypoxia (Fig. 4B). Similar results were obtained in the LN229 cells (not shown).
Figure 4.
HAF overexpression shifts cells from HIF-1α– to HIF-2α–dependent transcription. TaqMan qRT-PCR showing the effects of HAF overexpression on transcription of A, HIF-1α–dependent genes (i) CAIX and (ii) DDIT4; B, the HIF-2α–specific target PAI-1 after 16 hours 1% O2; and C, (i) OCT-3/4 and (ii) MMP9 after 72 hours 1% O2. C, iii and iv, the effects of HAF overexpression on OCT-3/4 and MMP9 after transfection with Scr nontargeting control or HIF-2α siRNA (siH2) after 72 hours 1% O2. siH2_2 and siH2_4 denote different siRNA duplexes. D, TaqMan qRT-PCR showing the effects of hypoxic duration on the HAF-induced modulation of VEGFA transcription. Data were obtained in U87 cells with similar results in LN229 cells and are the mean of at least 3 independent experiments ± SE. *, P < 0.05. E, Western blot showing the effects of hypoxic durations and intensities on levels of HIF-1α, HIF-2α, and HAF in U87 cells. Western data are representative of at least 3 separate experiments.
Previous studies have suggested that HIF-2α, rather than HIF-1α, is the major transcriptional regulator of responses to prolonged hypoxia (15, 16). As our microarray analyses were conducted after a relatively short duration (16 hours) of hypoxia, we sought to investigate the effect of HAF overexpression on the previously reported HIF-2α target genes OCT-3/4 (POU5F1) and MMP9 after longer durations of hypoxia. OCT3/4 and MMP9 levels peaked after 48 to 72 hours of hypoxia, suggesting that 16 hours may have been insufficient for monitoring gene inductions (Supplementary Fig. S3A and B). This pattern of hypoxic induction differed to that observed for VEGFA (regulated by both HIF-1α and HIF-2α; ref. 15), which reached maximal induction after 8 hours of hypoxia and decreased thereafter (Supplementary Fig. S3C). By monitoring gene transcription during 48 to 72 hours of continuous hypoxia, we found that HAF overexpression increased the transcription of OCT-3/4 and MMP9 in a HIF-2α–dependent manner (Fig. 4C). In contrast, HAF overexpression decreased VEGFA transcription after 16 hours of hypoxia but increased VEGFA transcription after 72 hours of hypoxia (Fig. 4D), during which time VEGFA transcription has been reported to be HIF-2α dependent (15). Investigating the effect of continuous hypoxia on the HIFs, we found that 1% O2 for 8 to 16 hours caused an initial increase in HIF-1α and HIF-2α protein levels, which then decreased despite continued hypoxia (Fig. 4E). In contrast, HAF protein levels showed an initial decrease in 1% O2, which slowly recovered with continued hypoxia, peaking after 48 hours (Fig. 4E). The increase in HAF levels after prolonged hypoxia is similar to the kinetics of MMP9 and OCT-3/4 activation (Supplementary Fig. S3), suggesting that HAF is necessary, together with HIF-2α, for induction of HIF-2α target genes. However, HAF mRNA levels were not induced by prolonged hypoxia (not shown), so the mechanism for the recovery in HAF protein levels is unclear but may involve translocation from the soluble to insoluble nuclear fractions (currently under investigation). When U87 cells were subjected to less than 0.5% O2, the recovery in HAF protein levels was complete by 8 hours compared with 48 hours at 1% O2; Fig. 4E). Similar fluctuations in HAF levels were also observed in PANC-1 cells (Supplementary Fig. S4), suggesting that the modulation in HAF protein levels in response to hypoxia may be a general phenomenon.
Taken together, the data suggest that HAF switches cells from HIF-1α- to HIF-2α–dependent gene transcription. HAF is elevated after prolonged and/or intense hypoxia in multiple cells types and is necessary for the induction of the HIF-2α target genes OCT-3/4 and MMP9.
Physiologic outcomes of HAF-induced HIF-2α activation
Colony growth and invasion
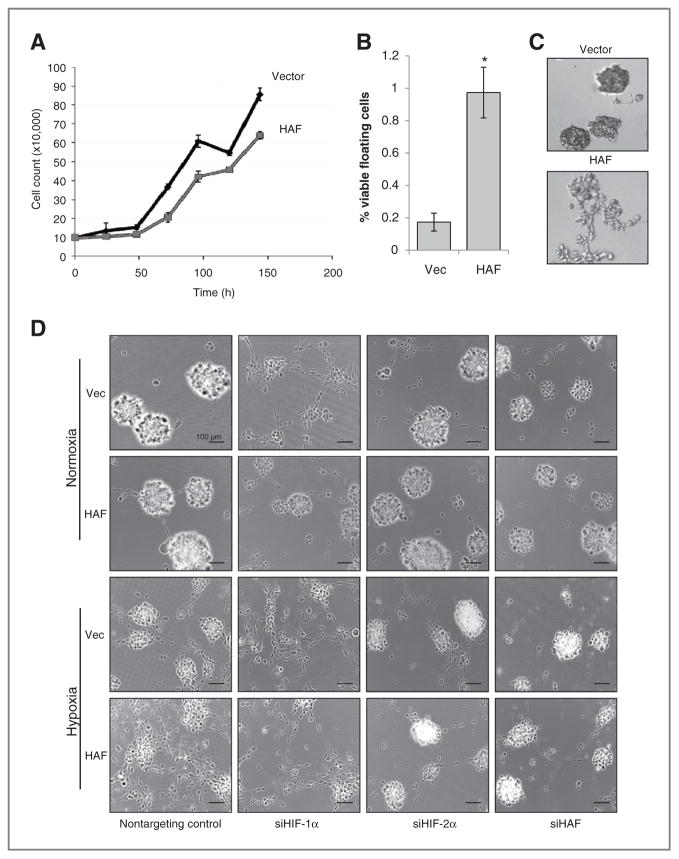
To investigate the physiologic consequences of HAF overexpression, we focused on GBM in which HIF-2α has been linked to increased angiogenesis and stem cell maintenance (3, 32). We found that U87 cells overexpressing HAF grew slower and had an increased number of viable floating cells than the vector control cells (Fig. 5A and B). When grown in 3D culture (by using Matrigel or NanoCulture matrix-free plates), the HAF cells showed more diffuse colony growth compared with the vector cells in hypoxia, suggesting increased motility and/or invasiveness, whereas no obvious differences were observed in normoxia (Fig. 5C and D). When transfected with HIF-1α siRNA, the vector cells recapitulated the diffuse HAF morphology, whereas transfection with HIF-2α siRNA promoted spheroid formation and inhibited the diffuse morphology in both the vector and the HAF cells (Fig. 5D), suggesting that changes induced by HAF overexpression are a result of impaired HIF-1α–dependent spheroid formation and increased HIF-2α–dependent cell spreading. In contrast, knockdown of HAF resulted in growth arrest and cell death, and ultimately fewer and smaller spheroids (Fig. 5D and Supplementary Data S5).
Figure 5.
Downstream effects of the HAF-mediated switch from HIF-1α to HIF-2α. A, growth curve of U87 cells in regular culture in normoxia. B, numbers of floating versus adherent cells in HAF and vector U87 cells. Viability was determined by trypan blue exclusion. *, P < 0.05. C, 3D colony appearance of vector or HAF U87 cells in Matrigel-coated chamber slides after 72 hours hypoxia (1% O2). D, 3D colony appearance of vector or HAF U87 cells grown on NanoCulture 96-well plates after 72 hours normoxia or hypoxia ± siRNA transfection. Images were taken with a 40× objective and are representative of at least 4 replicate wells with 6 fields taken per well. Scale bars indicate 100 μm.
By using Matrigel or LamininI–coated transwells, we found that hypoxic incubation alone increased the invasiveness of U87 cells by 2-fold, whereas HAF overexpression increased invasiveness by 2- to 5-fold over the vector control cells in both normoxia and hypoxia, which was accompanied by elevated MMP9 and MMP2 activity in the growth media (Fig. 6A). Similar results were obtained by using 786-0 and LN229 cells (not shown).
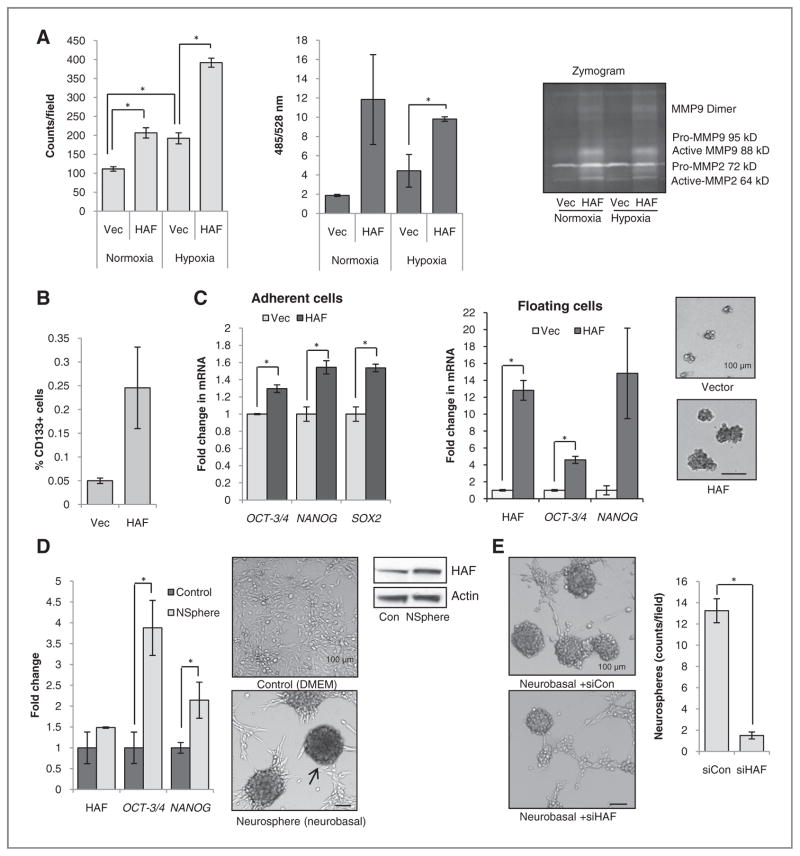
Figure 6.
Effect of HAF overexpression on invasion and self-renewal. A, effect of HAF overexpression on invasion through Matrigel- (L) or Laminin I–coated (R) transwell membranes, respectively. Inset, Zymogram assay by using supernatants of cells grown in normoxia or hypoxia. B, % CD133+ cells in vector or HAF overexpressing U87 cells as determined by fluorescence-activated cell sorting analysis. C, levels of indicated stem cell markers in HAF relative to vector U87 adherent or floating cells, respectively, as determined by TaqMan qRT-PCR. Photos of neurospheres generated by continuous culture of HAF or vector U87 floating cells in neurobasal media are shown inset. D, levels of HAF, OCT3/4, and NANOG in U87 neurospheres generated by continuous culture in neurobasal media normalized to cells grown in regular media with representative photomicrographs (arrow indicates neurosphere) and Western blots. E, the effect of control or HAF siRNA transfection on neurosphere formation. Data are the mean of at least 2 independent experiments ± SE. Scale bars indicate 100 μm; *, P < 0.05.
Tumor stem cell phenotype
Because HIF-2α has been associated with stem cell maintenance, particularly in GBM (3, 14), we investigated the effects of HAF overexpression on the tumor stem cell phenotype. The U87 HAF cells contained a 5-fold higher number of cells staining positive for CD133+, a neural stem cell marker (2, 33) and increased levels of the stem cell factors OCT-3/4, NANOG, and SOX2 compared with the vector cells (Fig. 6B and C and Supplementary Data S7). In addition, the floating HAF cell population expressed higher levels of OCT-3/4 and NANOG compared with the floating vector cell population and were able to form neurospheres when maintained in serum-free neurobasal media (Fig. 6C, inset), suggesting that the floating HAF subpopulation was enriched in stem cells. The proliferation of the tumor stem cell population can also be induced by the continuous maintenance of wild-type U87 cells in serum-free neurobasal media (34), which results in the formation of floating neurospheres with elevated levels OCT-3/4 and NANOG compared with adherent cells in regular growth media (Fig. 6D). These neurospheres expressed higher levels of HAF protein than control adherent cells (Fig. 6D), whereas transfection with HAF siRNA significantly decreased the number of neurospheres obtained, suggesting that HAF is required for stem cell proliferation and neurosphere formation (Fig. 6E).
Taken together, the data suggest that HAF promotes tumor progression by driving the HIF-2α–dependent transcription of genes that promote invasion and the proliferation of tumor stem cells.
HAF promotes tumor progression in vivo
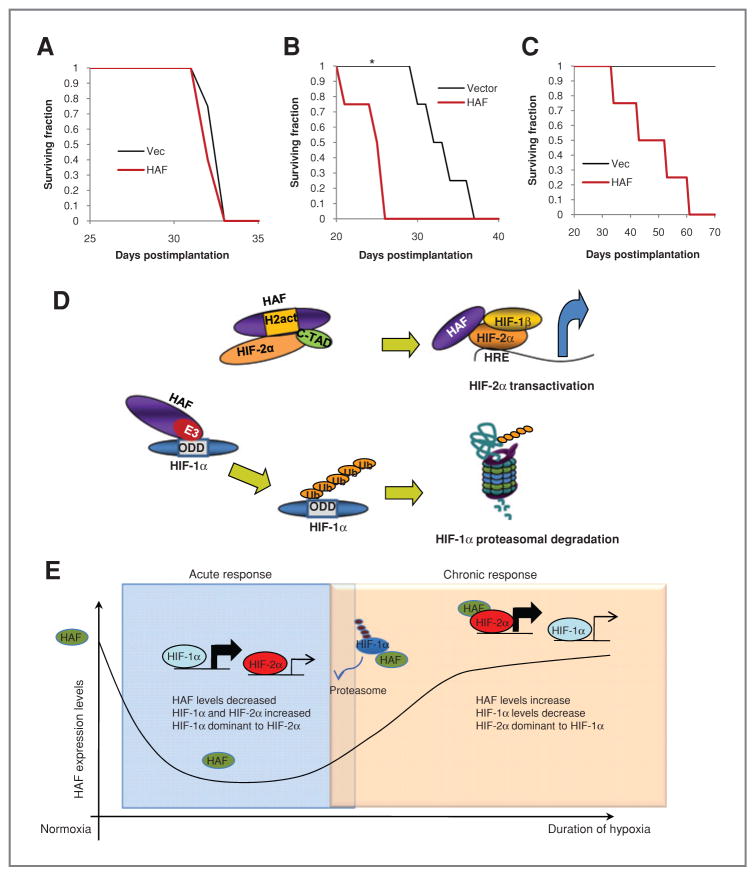
To investigate the effects of HAF on GBM progression in vivo, we used the intracranial GBM model. When U87 cells (adherent cells only) were injected, we found no significant differences in survival between the HAF and vector control mice (mean = 32.5 ± 0.3 days; Fig. 7A). However, when 1 × 105 of the pooled floating and adherent U87 populations of the vector and HAF cells were injected, the survival time of mice injected with HAF cells was significantly reduced compared with vector mice (mean survival = 25.5 ± 0.3 vs. 33.3 ± 1.5 days; P = 0.00425; Fig. 7B). When only 5,000 cells of the pooled population was injected, there was 100% morbidity within the mice injected with HAF cells (mean survival = 47.5 ± 6 days), whereas none of the vector mice exhibited signs of neurologic deficits up to 70 days after injection (Fig. 7C). This suggests that HAF overexpression promotes self-renewal particularly within the floating cell population, hence increasing GBM initiation and progression in vivo.
Figure 7.
Effect of HAF on tumor growth in vivo. A, survival curve when 500,000 vector or HAF U87 cells (adherent population only) were injected intracranially. B and C, survival curve of mice injected intracranially with 100,000 or 5,000 pooled floating and adherent cell populations (*, P = 0.00425). D, diagram showing mechanism of the HAF-induced switch from HIF-1α to HIF-2α signaling. The C-terminal E3 ubiquitin ligase domain of HAF binds to HIF-1α close to its oxygen-dependent degradation domain (ODD) and ubiquitinates it, targeting it for proteasomal degradation, whereas the central domain of HAF binds close to the C-TAD of HIF-2α, hence promoting HIF-2α transactivation. E, diagram showing the proposed role of HAF in regulating the kinetics of the HIF-1/2α activation in response to continuous hypoxia. During the early/acute hypoxic response, HAF protein levels decrease which facilitates the activation of HIF-1α but limits HIF-2α–dependent transcription. During prolonged/chronic hypoxia, HAF levels gradually increase in which it cooperates with HIF-2α to promote transcription.
Discussion
Tumor hypoxia is well recognized as a major driving factor for tumor growth and resistance to therapy (1). In addition to promoting tumor cell survival during hypoxic stress by shifting cells toward anerobic metabolism, neovascularization, and resistance to apoptosis, the hypoxia response may drive other responses that contribute to tumor aggressiveness such as increased genetic instability, invasion, metastasis, and an undifferentiated phenotype (35–37). Hence hypoxia, rather than acting as a simple on-off switch for the hypoxia response as once thought, initiates a complex cellular response that involves multiple players, including the HIFs, depending on the duration and intensity of the hypoxia. Indeed, it has been suggested that HIF-1α, because of its rapid induction and negative feedback regulation by prolonged hypoxia (16), provides a swift response to acute or transient hypoxia. On the other hand, prolonged or chronic hypoxia seems to favor activation of HIF-2α in most cell types, possibly because of differential affinities for specific regulators of stability and activity, such as the prolyl hydroxlases, factor inhibiting HIF, and also Hsp70, which promotes the ubiquitination and proteasomal degradation of HIF-1α but not HIF-2α (7, 38, 39). Intriguingly, chronic hypoxia has been implicated as a causal factor for the increased aggressiveness of tumors that develop resistance to antiangiogenic therapy such as to VEGF inhibition (40, 41), underscoring the need for a clearer understanding of the mechanisms that regulate the responses of the chronic versus acute hypoxia.
We have previously reported that the HAF C-terminus (residues 654–800) binds and ubiquitinates HIF-1α within residues 298–400, targeting HIF-1α for proteasomal degradation (25). We now report the ability of HAF to bind differentially to the 2 HIF-α isoforms resulting in HIF-1α degradation and promoting HIF-2α transactivation. In this regard, HAF (residues 300–500) binds to HIF-2α within residues 604–750 to increase HIF-2α transactivation. Thus, HAF plays a critical role in hypoxia signaling by turning off the HIF-1α response and turning on the HIF-2α response (Fig. 7D). Indeed, HAF knockdown inhibits HIF-2α–dependent HRE activity in 786-0 cells, suggesting that HAF is necessary and sufficient for HIF-2α activity. Furthermore, HAF overexpression inhibits the transcription of HIF-1α–specific target genes during acute hypoxia (16 hours) but promotes the transcription of the HIF-2α–dependent genes after prolonged hypoxia (72 hours).
HAF overexpression in cells grown in 3D culture resulted in the formation of colonies with a more diffuse, invasive phenotype compared with vector control cells. This phenotype is consistent with HIF-1α knockdown, which abrogated colony formation, and with increased HIF-2α, which stimulated invasiveness in hypoxia. Intriguingly, long-term knockdown of HAF decreased the size of colonies, both in normoxia and hypoxia, which may be due to previously reported roles of HAF in pre-mRNA splicing and mitosis (42, 43). Significantly, HAF overexpression was associated with the increased incidence and morbidity of intracranial U87 tumor xenografts, possibly because of the involvement of HAF in promoting self-renewal, which enabled tumor formation from cell numbers that in the vector control cells were insufficient to form tumors. The possibility exists that the availability of HAF may be the limiting factor for the full activation of HIF-2α and further work will show whether HAF levels may be a potential biomarker of highly undifferentiated, HIF-2α–driven tumors. Indeed, the unique hypoxia-dependent modulation of HAF levels may be a determinant of HIF-2α activation in response to varying intensities and durations of hypoxia (Fig. 7E).
The differences in specific HIF-1α and HIF-2α downstream targets, when compounded with the genetic aberrations present within the diverse tumor population, may determine which HIF-α isoform may provide the greatest growth advantage to a tumor as a whole. Many tumor cells develop resistance to apoptosis, which could counteract the proapoptotic factors induced by HIF-1α, effectively harnessing protumorigenic outcomes of HIF-1α, while negating its anti-tumorigenic effects (44). Hence, HAF overexpression inhibits the growth of HT29 tumor xenografts that express high HIF-1α and low HIF-2α (25). In contrast, the unique ability of HIF-2α to collaborate with oncogenes such as c-Myc, EGFR, and K-Ras may provide a growth advantage to tumor cells with deregulation of these pathways (14, 18, 21, 45, 46). Our present finding that HAF overexpression in the intracranial GBM model promotes poor survival provides a context in which HIF-1α inhibition and HIF-2α activation may promote tumor progression.
In conclusion, this study characterizes a new mechanism for the differential hypoxic regulation of HIF-1α and HIF-2α by HAF. HAF is overexpressed in many cancers (22–24), and its availability may be critical for HIF-2α–driven tumor progression. The HIF-binding domains of HAF which are able to differentially bind to HIF-1/2α may provide novel avenues for modulation of the HIF-1/2α balance for therapeutic benefit.
Supplementary Material
Acknowledgments
The authors thank Taly Spivak-Kroizman and Geoffrey Bartholomeuz for helpful discussions, and Geoffrey Grandjean and Mayur Gupta for technical support.
Grant Support
The work was financially supported by NIH Grants CA095060, CA17094, CA098920, and CA127001.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Vaupel P, Mayer A. Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev. 2007;26:225–39. doi: 10.1007/s10555-007-9055-1. [DOI] [PubMed] [Google Scholar]
- 2.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 3.Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501–13. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Koh MY, Spivak-Kroizman TR, Powis G. HIF-1alpha and cancer therapy. Recent Results Cancer Res. 2010;180:15–34. doi: 10.1007/978-3-540-78281-0_3. [DOI] [PubMed] [Google Scholar]
- 6.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995:925510–4. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo W, Zhong J, Chang R, Hu H, Pandey A, Semenza GL. Hsp70 and CHIP selectively mediate ubiquitination and degradation of hypoxia-inducible factor (HIF)-1alpha but not HIF-2alpha. J Biol Chem. 2010;285:3651–63. doi: 10.1074/jbc.M109.068577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohh M, Park CW, Ivan M, Hoffman MA, Kim TY, Huang LE, et al. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol. 2000;2:423–7. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 9.Maxwell PH, Pugh CW, Ratcliffe PJ. Activation of the HIF pathway in cancer. Curr Opin Genet Dev. 2001;11:293–9. doi: 10.1016/s0959-437x(00)00193-3. [DOI] [PubMed] [Google Scholar]
- 10.Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol. 2003;23:9361–74. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang V, Davis DA, Haque M, Huang LE, Yarchoan R. Differential gene up-regulation by hypoxia-inducible factor-1alpha and hypoxia-inducible factor-2alpha in HEK293T cells. Cancer Res. 2005;65:3299–306. doi: 10.1158/0008-5472.CAN-04-4130. [DOI] [PubMed] [Google Scholar]
- 12.Petrella BL, Lohi J, Brinckerhoff CE. Identification of membrane type-1 matrix metalloproteinase as a target of hypoxia-inducible factor-2 alpha in von Hippel-Lindau renal cell carcinoma. Oncogene. 2005;24:1043–52. doi: 10.1038/sj.onc.1208305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oda N, Abe M, Sato Y. ETS-1 converts endothelial cells to the angiogenic phenotype by inducing the expression of matrix metalloproteinases and integrin beta3. J Cell Physiol. 1999;178:121–32. doi: 10.1002/(SICI)1097-4652(199902)178:2<121::AID-JCP1>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 14.Covello KL, Kehler J, Yu H, Gordan JD, Arsham AM, Hu CJ, et al. HIF-2alpha regulates Oct-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev. 2006;20:557–70. doi: 10.1101/gad.1399906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmquist-Mengelbier L, Fredlund E, Lofstedt T, Noguera R, Navarro S, Nilsson H, et al. Recruitment of HIF-1alpha and HIF-2alpha to common target genes is differentially regulated in neuroblastoma: HIF-2alpha promotes an aggressive phenotype. Cancer Cell. 2006;10:413–23. doi: 10.1016/j.ccr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 16.Uchida T, Rossignol F, Matthay MA, Mounier R, Couette S, Clottes E, et al. Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha expression in lung epithelial cells: implication of natural antisense HIF-1alpha. J Biol Chem. 2004;279:14871–8. doi: 10.1074/jbc.M400461200. [DOI] [PubMed] [Google Scholar]
- 17.Franovic A, Holterman CE, Payette J, Lee S. Human cancers converge at the HIF-2alpha oncogenic axis. Proc Natl Acad Sci U S A. 2009;106:21306–11. doi: 10.1073/pnas.0906432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scrideli CA, Carlotti CG, Jr, Mata JF, Neder L, Machado HR, Oba-Sinjo SM, et al. Prognostic significance of co-overexpression of the EGFR/IGFBP-2/HIF-2A genes in astrocytomas. J Neurooncol. 2007;83:233–9. doi: 10.1007/s11060-007-9328-0. [DOI] [PubMed] [Google Scholar]
- 19.Giatromanolaki A, Koukourakis MI, Sivridis E, Turley H, Talks K, Pezzella F, et al. Relation of hypoxia inducible factor 1 alpha and 2 alpha in operable non-small cell lung cancer to angiogenic/molecular profile of tumours and survival. Br J Cancer. 2001;85:881–90. doi: 10.1054/bjoc.2001.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raval RR, Lau KW, Tran MG, Sowter HM, Mandriota SJ, Li JL, et al. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol Cell Biol. 2005;25:5675–86. doi: 10.1128/MCB.25.13.5675-5686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim WY, Perera S, Zhou B, Carretero J, Yeh JJ, Heathcote SA, et al. HIF2alpha cooperates with RAS to promote lung tumorigenesis in mice. J Clin Invest. 2009;119:2160–70. doi: 10.1172/JCI38443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imaizumi T, Kuramoto T, Matsunaga K, Shichijo S, Yutani S, Shigemori M, et al. Expression of the tumor-rejection antigen SART1 in brain tumors. Int J Cancer. 1999;83:760–4. doi: 10.1002/(sici)1097-0215(19991210)83:6<760::aid-ijc11>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 23.Kawamoto M, Shichijo S, Imai Y, Imaizumi T, Koga T, Yanaga H, et al. Expression of the SART-1 tumor rejection antigen in breast cancer. Int J Cancer. 1999;80:64–7. doi: 10.1002/(sici)1097-0215(19990105)80:1<64::aid-ijc13>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 24.Sasatomi T, Yamana H, Shichijo S, Tanaka S, Okamura T, Ogata Y, et al. Expression of the SART1 tumor-rejection antigens in colorectal cancers. Dis Colon Rectum. 2000;43:1754–8. doi: 10.1007/BF02236863. [DOI] [PubMed] [Google Scholar]
- 25.Koh MY, Darnay BG, Powis G. Hypoxia-associated factor, a novel E3-ubiquitin ligase, binds and ubiquitinates hypoxia-inducible factor 1alpha, leading to its oxygen-independent degradation. Mol Cell Biol. 2008;28:7081–95. doi: 10.1128/MCB.00773-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamothe B, Besse A, Campos AD, Webster WK, Wu H, Darnay BG. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of IkappaB kinase activation. J Biol Chem. 2007;282:4102–12. doi: 10.1074/jbc.M609503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polek TC, Talpaz M, Darnay BG, Spivak-Kroizman T. TWEAK mediates signal transduction and differentiation of RAW264.7 cells in the absence of Fn14/TweakR. Evidence for a second TWEAK receptor. J Biol Chem. 2003;278:32317–23. doi: 10.1074/jbc.M302518200. [DOI] [PubMed] [Google Scholar]
- 28.Lal S, Lacroix M, Tofilon P, Fuller GN, Sawaya R, Lang FF. An implantable guide-screw system for brain tumor studies in small animals. J Neurosurg. 2000;92:326–33. doi: 10.3171/jns.2000.92.2.0326. [DOI] [PubMed] [Google Scholar]
- 29.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cock-man ME, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–5. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 30.Carroll VA, Ashcroft M. Role of hypoxia-inducible factor (HIF)-1alpha versus HIF-2alpha in the regulation of HIF target genes in response to hypoxia, insulin-like growth factor-I, or loss of von Hippel-Lindau function: implications for targeting the HIF pathway. Cancer Res. 2006;66:6264–70. doi: 10.1158/0008-5472.CAN-05-2519. [DOI] [PubMed] [Google Scholar]
- 31.Sowter HM, Raval RR, Moore JW, Ratcliffe PJ, Harris AL. Predominant role of hypoxia-inducible transcription factor (Hif)-1alpha versus Hif-2alpha in regulation of the transcriptional response to hypoxia. Cancer Res. 2003;63:6130–4. [PubMed] [Google Scholar]
- 32.Khatua S, Peterson KM, Brown KM, Lawlor C, Santi MR, LaFleur B, et al. Overexpression of the EGFR/FKBP12/HIF-2alpha pathway identified in childhood astrocytomas by angiogenesis gene profiling. Cancer Res. 2003;63:1865–70. [PubMed] [Google Scholar]
- 33.Beier D, Hau P, Proescholdt M, Lohmeier A, Wischhusen J, Oefner PJ, et al. CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–5. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- 34.Heddleston JM, Li Z, McLendon RE, Hjelmeland AB, Rich JN. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle. 2009;8:3274–84. doi: 10.4161/cc.8.20.9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ, et al. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008;10:295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 36.Reynolds TY, Rockwell S, Glazer PM. Genetic instability induced by the tumor microenvironment. Cancer Res. 1996;56:5754–7. [PubMed] [Google Scholar]
- 37.Huang LE, Bindra RS, Glazer PM, Harris AL. Hypoxia-induced genetic instability–a calculated mechanism underlying tumor progression. J Mol Med. 2007;85:139–48. doi: 10.1007/s00109-006-0133-6. [DOI] [PubMed] [Google Scholar]
- 38.Appelhoff RJ, Tian YM, Raval RR, Turley H, Harris AL, Pugh CW, et al. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J Biol Chem. 2004;279:38458–65. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- 39.Koivunen P, Hirsila M, Gunzler V, Kivirikko KI, Myllyharju J. Catalytic properties of the asparaginyl hydroxylase (FIH) in the oxygen sensing pathway are distinct from those of its prolyl 4-hydroxylases. J Biol Chem. 2004;279:9899–904. doi: 10.1074/jbc.M312254200. [DOI] [PubMed] [Google Scholar]
- 40.Ebos JM, Lee CR, Cruz-Munoz W, Bjarnason GA, Christensen JG, Kerbel RS. Accelerated metastasis after short-term treatment with a potent inhibitor of tumor angiogenesis. Cancer Cell. 2009;15:232–9. doi: 10.1016/j.ccr.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paez-Ribes M, Allen E, Hudock J, Takeda T, Okuyama H, Vinals F, et al. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell. 2009;15:220–31. doi: 10.1016/j.ccr.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makarova OV, Makarov EM, Luhrmann R. The 65 and 110 kDa SR-related proteins of the U4/U6. U5 tri-snRNP are essential for the assembly of mature spliceosomes. EMBO J. 2001;20:2553–63. doi: 10.1093/emboj/20.10.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kittler R, Putz G, Pelletier L, Poser I, Heninger AK, Drechsel D, et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036–40. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- 44.Bruick RK. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc Natl Acad Sci U S A. 2000;97:9082–7. doi: 10.1073/pnas.97.16.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, et al. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394:485–90. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 46.Bertout JA, Majmundar AJ, Gordan JD, Lam JC, Ditsworth D, Keith B, et al. HIF2{alpha} inhibition promotes p53 pathway activity, tumor cell death, and radiation responses. Proc Natl Acad Sci U S A. 2009;106:14391–6. doi: 10.1073/pnas.0907357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.