Abstract
Micrococcal nuclease is shown to cleave DNA under conditions of partial digestion in a specific manner. Sequences of the type 5'CATA and 5'CTA are attacked preferentially, followed by exonucleolytic degradation at the newly generated DNA termini. GC-rich flanking sequences further increase the probability of initial attack. Unexpectedly, long stretches containing only A and T are spared by the nuclease. These results, which were obtained with spared by the nuclease. These results, which were obtained with mouse satellite DNA and two fragments from the plasmid pBR22, do not support the previous contention that it is the regions of high At-content which are initially cleaved by micrococcal nuclease. This specificity of micrococcal nuclease complicates its use in experiments intended to monitor the nucleoprotein structure of a DNA sequence in chromatin.
Full text
PDF



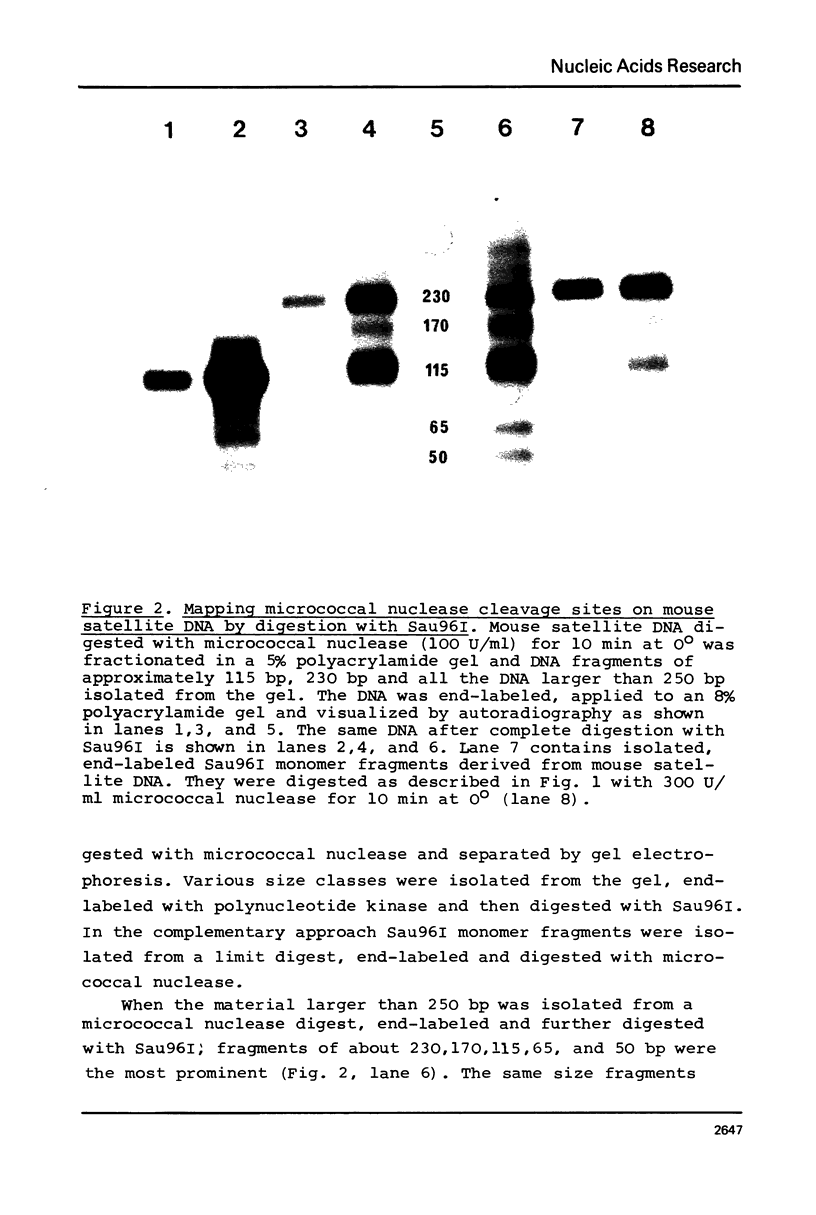








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Fittler F., Zachau H. G. Subunit structure of alpha-satellite DNA containing chromatin from African green monkey cells. Nucleic Acids Res. 1979 Sep 11;7(1):1–13. doi: 10.1093/nar/7.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray C. P., Sommer R., Polke C., Beck E., Schaller H. Structure of the orgin of DNA replication of bacteriophage fd. Proc Natl Acad Sci U S A. 1978 Jan;75(1):50–53. doi: 10.1073/pnas.75.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene P. J., Heyneker H. L., Bolivar F., Rodriguez R. L., Betlach M. C., Covarrubias A. A., Backman K., Russel D. J., Tait R., Boyer H. W. A general method for the purification of restriction enzymes. Nucleic Acids Res. 1978 Jul;5(7):2373–2380. doi: 10.1093/nar/5.7.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S. Partial purification of the Escherichia coli K-12 mec+ deoxyribonucleic acid-cytosine methylase: in vitro methylation completely protects bacteriophage lambda deoxyribonucleic acid against cleavage by R-EcoRII. J Bacteriol. 1977 Mar;129(3):1330–1334. doi: 10.1128/jb.129.3.1330-1334.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Hörz W., Altenburger W. Nucleotide sequence of mouse satellite DNA. Nucleic Acids Res. 1981 Feb 11;9(3):683–696. doi: 10.1093/nar/9.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörz W., Zachau H. G. Characterization of distinct segments in mouse satellite DNA by restriction nucleases. Eur J Biochem. 1977 Mar 1;73(2):383–392. doi: 10.1111/j.1432-1033.1977.tb11329.x. [DOI] [PubMed] [Google Scholar]
- Hörz W., Zachau H. G. Deoxyribonuclease II as a probe for chromatin structure. I. Location of cleavage sites. J Mol Biol. 1980 Dec 15;144(3):305–327. doi: 10.1016/0022-2836(80)90093-5. [DOI] [PubMed] [Google Scholar]
- Kacian D. L., Spiegelman S. Use of micrococcal nuclease to monitor hybridization reactions with DNA. Anal Biochem. 1974 Apr;58(2):534–540. doi: 10.1016/0003-2697(74)90221-8. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Mathis D., Oudet P., Chambon P. Structure of transcribing chromatin. Prog Nucleic Acid Res Mol Biol. 1980;24:1–55. doi: 10.1016/s0079-6603(08)60670-4. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- Nedospasov S. A., Georgiev G. P. Non-random cleavage of SV40 DNA in the compact minichromosome and free in solution by micrococcal nuclease. Biochem Biophys Res Commun. 1980 Jan 29;92(2):532–539. doi: 10.1016/0006-291x(80)90366-6. [DOI] [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Rosenberg H., Singer M., Rosenberg M. Highly reiterated sequences of SIMIANSIMIANSIMIANSIMIANSIMIAN. Science. 1978 Apr 28;200(4340):394–402. doi: 10.1126/science.205944. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- VONHIPPEL P. H., FELSENFELD G. MICROCOCCAL NUCLEASE AS A PROBE OF DNA CONFORMATION. Biochemistry. 1964 Jan;3:27–39. doi: 10.1021/bi00889a006. [DOI] [PubMed] [Google Scholar]
- Wingert L., Von Hippel P. H. The conformation dependent hydrolysis of DNA by micrococcal nuclease. Biochim Biophys Acta. 1968 Mar 18;157(1):114–126. doi: 10.1016/0005-2787(68)90270-0. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]