Abstract
Although inbred mouse strains have been the premier model organism used in biomedical research, multiple studies and analyses have indicated that genome wide association studies (GWAS) cannot be productively performed using inbred mouse strains. However, there is one type of GWAS in mice that has successfully identified the genetic basis for many biomedical traits of interest: haplotype based computational genetic mapping (HBCGM). Here, we describe how the methodological basis for a HBCGM study significantly differs from that of a conventional murine GWAS, and how an integrative analysis of its output within the context of other ‘omic’ information can enable genetic discovery. Consideration of these factors will substantially improve the prognosis for the utility of murine genetic association studies for biomedical discovery.
Genetic association studies in mice and biomedical discovery
Genome-wide association studies (GWAS) have successfully analyzed the genetic basis for disease susceptibility or quantitative trait differences in human populations. Similar to a human association study, GWAS can also be performed in mice by correlating trait values measured in a set (usually ≤ 10) of inbred strains with alleles genotyped at single nucleotide polymorphisms (SNPs), which were selected to represent the genetic pattern within regions of the mouse genome. However, multiple modeling studies and analyses have purported to demonstrate that murine GWAS cannot identify the genetic factors affecting most biomedical traits of interest, due to low power and a high false positive rate [1–3]. The laboratory mouse has been the premier model organism used in biomedical research. It has many unique features that enable biomedical discovery, including: the availability of multiple well-characterized strains, mammalian physiology, a homozygous genome, experiments can be performed under conditions that control environmental variables, and its genome can be genetically modified, which enables the assessment of the impact that allelic variation has on phenotype. It would be very discouraging for all types of genetic research, if we truly could not perform genetic association studies using this model organism.
Despite the negative predictions, haplotype-based computational genetic mapping (HBCGM) studies have identified causative genetic factors for many biomedical traits in mice (Table 1). HBCGM results have generated potential new solutions for 21st century public health problems, including treatments for prevention of narcotic drug withdrawal symptoms [4] and for reduction of incisional pain after surgery [5, 6] that are in clinical testing. In an HBCGM experiment, a property of interest is measured in 10 or more inbred mouse strains; genetic factors are then computationally predicted by identifying genomic regions where the pattern of genetic variation (in the form of pre-assembled haplotype blocks) correlates with the distribution of trait values among the inbred strains [7, 8]. The productivity of HBCGM requires explanation, since HBCGM is a type of murine GWAS, which the published simulations and analyses indicate should fail. The reason for the discrepancy between the negative predictions from the modeling studies and the actual HBCGM results is that the modeling studies utilize standard GWAS methodology, but there are very substantial methodological differences between HBCGM and standard methods used in GWAS in mice. There are also differences between murine and human genetic association studies, which affect the design and expected outcomes from these studies. A better understanding of the methodological basis underlying HBCGM, especially how it differs from conventional murine GWAS studies and how its output can be used to enable genetic discovery, will substantially improve the prognosis for its utility for genetic discovery.
Table 1.
Biomedical models that were successfully analyzed by HBCGM.
| Biomedical model | Gene | Reference | Year |
|---|---|---|---|
| Gene expression | H2-Eα | [7] | 2004 |
| Pharmacogenetic factors | |||
| Warfarin | Cyp2c29 | [31] | 2006 |
| Irinotecan | Ugt1a | [32] | 2007 |
| P38 kinase inhibitor | Aox1 | [18] | 2010 |
| Susceptibility to infection | |||
| Invasive aspergillosis | Plg | [33] | 2008 |
| Respiratory syncitial virus | MHC (H2) | [34] | 2010 |
| Candida albicans | C1r/s | [14] | 2011 |
| Analgesic medication response | Kcnj9 | [19] | 2008 |
| Narcotic drug responses | |||
| Mechanical hyperalgesia | Adrb2 | [35] | 2006 |
| Thermal hyperalgesia | Abcb1b | [36] | 2006 |
| Analgesia | Kcnj9 | [19] | 2008 |
| Withdrawal | Ht3a | [4] | 2009 |
| Inflammatory pain responses | |||
| Early formalin response | Atp1b3 | [20] | 2009 |
| Late formalin response | Mapk8 | [37] | 2010 |
| Incisional wound biology | Nalp1b | [5], [6] | 2010 |
Distinctions that make it different
Although they are usually considered as highly similar entities, the methodological foundation, preferred phenotypes and the genotypic representations used in HBCGM and conventional murine GWAS are quite distinct (Table 2). In a murine GWAS, marker SNPs are selected to represent the pattern of genetic variation across the genome, which are utilized to identify the causative genetic loci for measured phenotypic differences in an inbred strain panel. The poor performance (low statistical power for detection) of murine GWAS in several simulations [2, 3] is partly attributable to the fact that the pattern of genetic variation within a genomic region of an arbitrarily determined size (usually 20–60 kb) is represented by a selected SNP. The selected SNP is unlikely to be the causative factor for the analyzed trait, and it may not even be in linkage disequilibrium (LD) with a potentially causative SNP in that region. The use of selected SNPs to cover an arbitrarily sized genomic region does not produce a robust genetic map [9], and some of the problems with genetic association studies result from the incorrect representation of the pattern of genetic variation [8]. We have found that some regions of the mouse genome have very low levels of linkage disequilibrium between SNPs [10], while other regions can have very high rates of polymorphism among the inbred strains [11] with frequent changes in the pattern of genetic variation, which reduces the utility of representative SNPs in these regions. Since the genetic variation within a region must be fully analyzed to know where there is a change in the genetic pattern, the poor performance of GWAS using representative SNPs is not surprising [2, 3].
Table 2.
Comparison of the properties of murine HBCGM and GWAS.
| GWAS | HBCGM | |
|---|---|---|
| Unit of analysis | Marker SNPs | Haplotype blocks |
| Genetic map | Representative SNPs | All genetic variants |
| Selected for regions of same size | Actual genetic pattern represented by blocks | |
| Causative SNPs | Usually absent | Included |
| PoPVg(a) | Under-estimate | Actual |
| Analysis preferences | ||
| Phenotypic pattern | Dichotomous | 3 or more phenotypic states |
| Genetic control | Single SNP | Composite SNPs within a region |
PoPvg: Percent of population variance explained by a genetic locus.
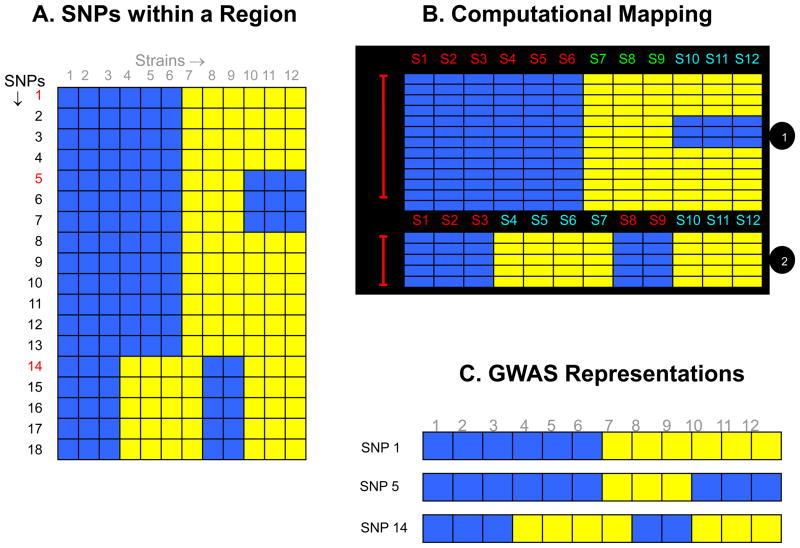
In contrast, HBCGM divides the mouse genome into discrete regions, which are based upon the extent of linkage disequilibrium among all identified SNPs in a region. A new haplotype block is produced within a region when there is a change in the pattern of genetic variation among the inbred strains. A genetic map that accurately reflects the fine structure of genetic variation has multiple advantages over representative SNPs (Figure 1): (1) the dimensions of and strain groupings within each region are based upon complete knowledge of the pattern of genetic variation; (2) since all genetic variation is analyzed, the causative SNPs are included in the haplotype map; (3) the small sizes of the correlated genomic regions enable the formulation of a precise hypothesis about how a genetic variant could impact a trait value. For example, a haplotype block affecting H2-Eα gene expression was only 1 kb in size [7], which enabled an allelic effect on gene expression to be quickly characterized. If a phenotype is entirely determined by alleles at a single SNP, then GWAS and HBCGM are methodologically equivalent and will have the same detection power.
Figure 1.
(A) A diagrammatic representation of the pattern of genetic variation within a region of the mouse genome. Each of the 18 identified SNPs within this genomic region is represented as a row, and the blue and yellow colored boxes indicate different alleles for each of the 12 strains analyzed. (B) The computational method for haplotype block formation will organize these SNP alleles into two haplotype blocks that accurately represent the pattern of genetic variation within this region. The first haplotype block has three different strain groupings (haplotypes), while the 2nd block has two different haplotypes. Strains 1–6, strains 7–9 and strains 10–12 have distinct genetic patterns, which gives rise to the three different haplotypes in the first haplotype block. While in the 2nd block, strains 1–3 and 7–8 have one allelic pattern, and strains 4–7 and 10–12 have the other allelic pattern, which produce the two haplotypes in this block. (C) In contrast, if the genetic variation is represented by the alleles at a single SNP-without knowing the true pattern of genetic variation within this region the allelic pattern and strain groupings will vary, depending upon the marker SNP that is selected to represent this region.
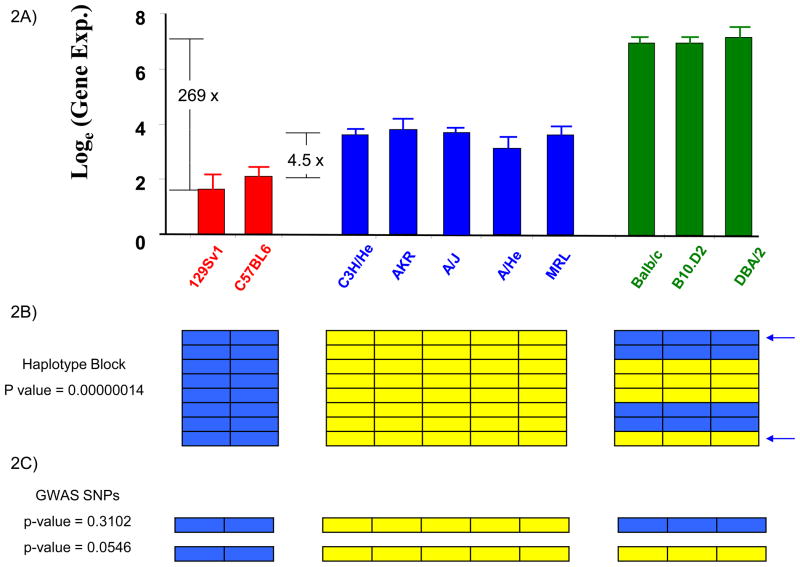
However, the genetic control of most biological traits is usually not this simple. HBCGM cannot evaluate complex genetic traits that are affected by multiple alleles located in discrete regions of the genome, but can evaluate phenotypic traits that are predominantly regulated by allelic differences within a contiguous genomic region. These allelic differences can produce a spectrum of phenotypes that can match the haplotype structure within a region. In these situations, HBCGM is better able than a GWAS to analyze phenotypes with multiple different states (Figure 2). For example, the composite effect of alleles at 2 SNPs contributed to three discrete levels of H2-Eα gene expression in an inbred strain panel. HBCGM produced a block with three distinct haplotypes, which maximized detection power for analyzing this gene expression difference [7]. Since GWAS methodology can analyze only one SNP at a time, it cannot distinguish the three groups with distinct gene expression levels.
Figure 2.
The difference in perspective when pulmonary H2-Eα mRNA expression is analyzed by linkage analysis in 2 strains, or in 10 inbred strains using HBCGM or GWAS methodology. (A) Graph showing pulmonary H2-Eα gene expression as the natural logarithm of the average of 3 independent measurements for each of 10 analyzed strains. The strains are divided into 3 distinct groups with high, intermediate or low levels of H2-Eα expression, which are indicated by different colored bars. The expression level of the DBA/2 strain (highest) is 269-fold greater than that of 129Sv (lowest), and is 37-fold greater than that of C3H/He (intermediate); while the two strains (C3H/He and C57BL6) analyzed by linkage analysis in [3] differ by only 4.5-fold (reproduced [7] with permission from Science). (B) HBCGM identified a haplotype block with 8 SNPs within the H2-Eα gene. Each SNP is represented as a row, and the colored boxes indicate the allele for each of the 10 analyzed strains. There were 3 different haplotypes within this region, and the one-way ANOVA model [7] showed a very strong correlation (p=1×10−7) between the haplotypic strain groupings and H2-Eα mRNA expression. (C) In contrast, a GWAS uses a two-sample t-test to assess the correlation between alleles at one SNP and the phenotypic data. The poor (p=0.054) or insignificant (p=0.31) correlation between individual selected SNP alleles (indicated by arrows) and the H2-Eα expression makes it impossible to detect the causal genetic locus if other spurious loci with smaller p-values were produced by the analysis, or after correction for multiple comparisons. This demonstrates the advantage that HBCGM has over GWAS methodology when multiple SNPs exert a composite effect on the phenotype.
HBCGM is the most appropriate method for analyzing phenotypic data obtained from larger numbers of inbred strains, which can have 3 or more distinct phenotypic states. These advantages were illustrated when 3 phenotypic datasets (the response to aromatic hydrocarbons, H2-Eα gene expression, and survival after Candida albicans infection) were analyzed using another computational method, which is used to analyze genetic association studies in mice. The efficient mixed-model association (EMMA) method [12] analyzes the correlation between phenotypic data measured across a set of inbred strains and the alleles at a single SNP, and its ability to correct for population structure and genetic relatedness among the inbred strains has been shown to reduce the false positive rate [13]. Each of these datasets was previously analyzed by HBCGM, and the allelic effect for the gene with the highest correlation was experimentally verified. However, the causative gene for only the aromatic hydrocarbon response could be identified using EMMA [14], because it is a binary response (either present or absent) phenotype. If a binary phenotype is entirely determined by alleles at a single SNP, then GWAS and HBCGM will have the same detection power. For example, conventional GWAS methods have been used to identify causative genetic factors for some traits in outbred strains of mice [15]. However, the poor performance of EMMA when analyzing the H2-Eα gene expression and survival after fungal infection data was striking: EMMA identified >516,000 SNPs (corresponding to 783 Mb of the genome) with a higher correlation than the causative gene effecting survival after fungal infection, and ~10,000 SNPs had a higher correlation with the H2-Eα gene expression data in Figure 2 than the experimentally verified cis-acting SNPs within H2-Eα. Since EMMA can analyze only one SNP at a time, it is not the optimal method for analyzing phenotypic traits with 3 discrete phenotypic states. Although there are many situations were HBCGM will be unable to identify the genetic basis for trait differences, it can be productively utilized to evaluate traits that are controlled by multiple polymorphisms within a contiguous region, especially when there are 3 or more discrete phenotypic responses.
There is also a fundamental difference in the way that GWAS and HBCGM results are interpreted. The results from a conventional murine GWAS are usually evaluated without considering other types of data, and a very small genome-wide significance cutoff is applied in order to strictly control the false positive rate. Therefore, a large sample size is required to reduce the number of SNPs that will randomly correlate with a trait value to enable a true causative locus to be identified. However, the sample size used in prior murine GWAS studies was always less than 20 strains (and often 6–10 strains are used), which makes it difficult for even a true causative locus to have a p-value that achieves genome-wide significance level. Thus, the need to control the false positive rate leads to a high probability that the study will produce a false negative result, which explains why investigators have concluded that murine association studies can’t work. In contrast, HBCGM results are viewed within the context of integrated analysis of a biomedical trait. As a consequence, a less stringent filtering criterion is used to evaluate HBCGM results, which increases the number of false positives but ensures that the true positives are retained. Then, the causative genetic candidates are selected from among the many correlated genes by applying orthogonal criteria [16], such as gene expression and metabolomic [17] or curated biologic data [18], or using the genomic regions delimited by prior QTL analyses [19, 20]. This integrated approach evaluates genetic candidates using multiple criteria, even though less stringent cutoffs are used for identification of genetic candidates. This has proven to be a better method for murine genetic analysis, than that of a typical GWAS that is performed using a single highly stringent criterion to identify candidates.
More may not be the same
When considering the relative utility of HBCGM, it is reasonable to ask whether 3 distinct phenotypic states will be a rare or common occurrence when a trait is evaluated across a large panel of inbred strains. Of direct relevance, mouse laboratory strains are reproductively isolated populations, which were derived from at least 3 subspecies (Mus musculus domesticus, M. m. musculus, and M. m. casteneus) that diverged ~1 million years ago (reviewed in [21]). Analysis of murine SNPs indicated that 4 distinct subspecies contributed to the genetic variation in the inbred strains, the vast majority of genetic variation is derived from the founding subspecies, and 40% of murine genes contain 3 or more different haplotypic patterns [22]. Since both mouse genealogy and murine SNP analysis indicates that many genomic regions could have 3 or more haplotypic patterns, the presence of 3 different phenotypic states may be a common occurrence when phenotypes are analyzed across a larger number of strains. Since over 450 inbred mouse strains have been described [23], a substantial number of inbred strains are available for phenotypic analyses.
Beyond the differences between the two association-based (HBCGM and GWAS) methods, the more extreme differences between association- and linkage-based methods were not factored into comparative modeling studies, which can produce misleading conclusions about the capabilities of murine GWAS. For example, one modeling study [3] used 723 loci affecting gene expression that were identified in linkage studies involving 2 inbred strains as the ‘gold standard’ for assessing the ability of GWAS involving 15 strains to identify causative loci. The poor correlation between the loci identified by the two different methods led them to incorrectly conclude that murine GWAS would not be productive. However, there is no reason to believe that genetic factors identified by analysis of gene expression differences across >10 inbred strains would be identical to those identified by linkage analyses involving two strains. For example, analysis of only two strains would present a myopic picture that would not reveal that there are three distinct levels of H2-Eα mRNA expression among the 10 strains that we analyzed [7] (Figure 2). H2-Eα expression levels varied by 268-fold across the 10 strains, but only differed by 4.5 fold between the two strains used in the linkage study [3]. The novel enhancer element identified by analysis of 10 strains [7] would not have been found in the linkage study, and would be regarded as a false positive in the published modeling study [3].
Representative genetics
It has been pointed out that each of the genetic loci that were identified by analysis of inbred strains [1, 24] accounted for only a small fraction (usually ~5%) of the overall trait variation. This analysis has fueled the concern that murine GWAS will be futile endeavors for most traits of interest, since these studies cannot identify genetic loci with a small phenotypic effect. For example, our simulations indicate that genetic loci must be responsible for at least 15% of the overall trait difference to be reliably identified by HBCGM (80% power) [8]. However, murine genetic studies have systematically under-estimated the percent of phenotypic variance that can be explained by a causative genetic locus (PoPVg) for a number of reasons. First, the PoPVg has previously been calculated from analyses that evaluated a very small number of strains. Only two strains are evaluated in conventional linkage studies, while genomic regions from only 6–8 founder strains are evaluated in linkage analyses using heterogeneous stock [24] or collaborative cross [25] mice. Analysis of a small number of strains does not represent the actual extent of phenotypic variation present in the mouse population, and significantly under-estimates the variation that would be observed if a larger strain panel was evaluated.
Allelic variants that could have a large effect on a phenotype may not be present in a small and unrepresentative strain set. The inbred laboratory strains are reproductively isolated populations, which are derived from several different ancestral founders that diverged ~1 million years ago [21]; and thus contain a substantial amount of genetic variation that could affect many phenotypic traits. The observed phenotypic variance would be increased if a larger percentage of the 450 different inbred strains [23] were evaluated. Thousands of unrelated individuals are randomly sampled in a human association study, which ensures that phenotypic and genetic variation present in the human population is well represented. In contrast, the inbred mouse strains used in linkage or GWAS studies are not a random sample of the mouse population. These studies usually examine only a limited number of strains, and have a strong selection bias toward strains analyzed in previous studies, which may not be relevant for the current phenotype. Analysis of a small number of strains with a restricted phenotypic range will produce a reduced estimate of the genetic effect on a trait value. In addition, the use of marker SNPs to represent the genetic pattern in murine GWAS also reduces the PoPVg estimate. The underlying haplotype structure among inbred mouse strains is ignored in a conventional GWAS that uses selected marker SNPs. When a small number of strains are analyzed, there are many cases where a marker SNP can have very high LD with a causative SNP, but the marker and causative SNP alleles will produce different GWAS results; and the marker SNP will provide a reduced estimate of the PoPVg (Box 1). Even when the marker and causative SNPs have significant LD (r2=0.8), the use of the marker SNP will reduce the calculated PoPVg by 20% relative to the causative SNP (see supplement).
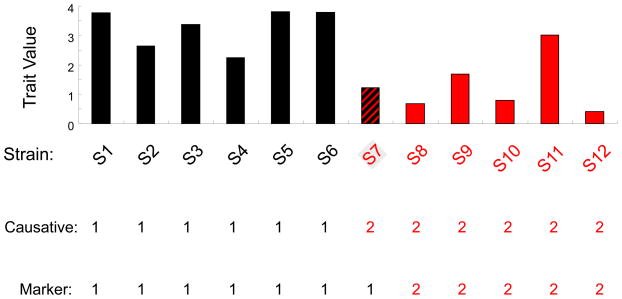
Box 1. The effect of marker SNPs on GWAS results.
A simulation study was used to investigate the impact of using a marker SNP as a surrogate in a genetic association study. We assume that a bi-allelic causative SNP exists that divides the 12 analyzed strains into two groups of equal size (S1-S6 and S7-S12); the trait values in both groups are normally distributed and have the same standard deviation, and the difference between the two group means is 3 times the standard deviation (i.e. the causative SNP has an effect size of 3). Assume that a nearby representative SNP is selected as a surrogate marker in an association study, and that the representative and the (un-genotyped) causative SNP have only one discordant allele among the 12 strains, which is found in strain 7 (S7). Although both SNPs are in very high LD (D′ = 1 and r = 0.85), the calculated p-values (averaged from among 1000 simulations) for the association of the causative and marker SNPs with the trait values are 0.002 and 0.02, respectively. The PoPVg explained by the causative SNP was 0.73, while that of the marker SNP was only 0.52. Table I examines these differences when the number of strains (12 or 16, which are still divided into 2 groups of equal size) or the genetic effect size (2–3) is varied, when there is one single allelic difference between the causative SNP and the marker (i.e. the allele for one strain in one group is altered to that in the other group). Although the SNPs are in very high LD (D′ =1, and r=0.8–0.9) in all cases, the marker SNP significantly underestimates the actual PoPVg. If a causative SNP has a large genetic effect, the significant difference in the calculated p-values indicates that it is more difficult to detect a causative genetic factor in murine GWAS using representative SNPs.
There is another subtle, yet fundamental difference in the way that the PoPVg is calculated in GWAS and HBCGM studies, which also impacts the PoPVg estimate. A quantitative trait can be modeled by the equation “trait value ~ G + E + G*E + residual”; where G, E and G*E represents the genetic effect, the environmental effect and their interaction, and the “residual” represents the variation that can’t be explain by G or E. Many murine or human traits can be highly variable, even when repeatedly assayed in the same human subject or mouse strain. (The environmental variation in a human study can be exceedingly large.) As a result, the unexplained variation can be large, which leads to a small genetic effect (G). Multiple phenotypic measurements are made for each strain under controlled conditions, and HBCGM uses only the average value of the strain replicates. As a result, the environmental effects are eliminated (E and G*E) in a murine study, and the un-explained variation is also minimized. These factors increase the PoPVg in a HBCGM study, which increase the range of traits that can successfully evaluated by HBCGM.
‘Next-generation’ HBCGM: future directions and limitations
Improvements in HBCGM methodology should enable a wider range of biomedical traits to be evaluated. The previous HBCGM algorithm [7] had significant limitations that inhibited our ability to analyze many phenotypes. (1) The genetic map only covered ~15% of the genes in the murine genome. (2) The haplotype block construction algorithm did not allow for overlapping blocks within a region, which enabled a causative block to easily be missed (producing false negative results) if the algorithm selected another block with fewer haplotypes and fewer SNPs that was preferred by the algorithm. (3) All analyses used a single haplotype map that incorporated all available allelic data for all strains, yet phenotypic data was usually available for a subset of the strains in a typical mapping experiment. Inclusion of irrelevant alleles can disrupt haplotypic patterns that are uniform among the strains of interest. To overcome these problems, a ‘next-generation’ computational genetic mapping program with three advanced features was developed [14]. (1) By merging two large SNP datasets, which included SNPs generated from analysis of the complete genomic sequence of multiple inbred strains [26], a high quality haplotype map with ~3 million SNPs was produced that covers virtually (>95%) the entire genome for 16 inbred strains. (2) A new haplotype block construction method was developed that allows haplotype blocks within a region to overlap, which enables all patterns of genetic variation within a region to be identified. (3) A 30,000-fold improvement in the computational efficiency enables customized haplotype blocks to be dynamically produced for the strains with available phenotypic data. The next generation method identified a causative genetic factor that would have been missed if the previous genetic mapping method was used [14].
Although it reduces the probability of producing false negative results, the new method produces a very large number of haplotype blocks, and some blocks will correlate with trait values purely by chance. Raising the significance threshold could reduce the number of correlated genes, but, as discussed above, this increases the chance of producing a false negative result. It has been previously concluded that these “spurious associations” render HBCGM unable to identify a true causative genetic factor [2, 3]. However, HBCGM results are only one component of a comprehensive data analysis package that is used for biomedical trait analysis. Causative genetic candidates have been selected from among the many correlated genes by applying orthogonal criteria [16], such as gene expression and metabolomic [17] or curated biologic data [18], or using the genomic regions delimited by prior QTL analyses [19, 20]. An integrated approach, where HBCGM output is analyzed within the context of multiple ‘omic’ (metabolomic, proteomic, or gene expression) datasets, will become an increasingly important part of 21st century biomedical discovery. This requires a paradigm shift, since it is current practice to use genetic analysis to identify a single major candidate gene, which will then undergo subsequent testing. In contrast, the integrated approach uses genetic analysis to identify groups of potential candidate genes – and the causative factor may not even have the highest correlation – which are then filtered using other criteria. This approach is certainly not without precedent. It has been an accepted standard for human GWAS that a replicate analysis (validation study) must be performed in a different population “to separate true associations from the blizzard of false positives” [27]. HBCGM output has the same filtering requirement, but other types of data (rather than a replicate study) are used as the filtering mechanism. Given the large number of available inbred strains, it is also possible to perform a ‘replicate’ association study using a different set of strains.
For all of the reasons discussed above, phenotypes must be characterized across a larger number of inbred strains to facilitate genetic discovery. Our acetaminophen toxicity study [17], where the only resistant inbred strain was not in our genetic database, indicates that many 21st century biomedical problems may not be solved using the inbred strains that were commonly studied in the 20th century. Each inbred strain has unique genetic variants, and possibly phenotypic responses, which could enable genetic discovery. Our simulated datasets indicate that genetic loci with an effect size as low as 0.15 could be identified if 40 inbred strains were analyzed [8], which would certainly overcome prior criticism that computational genetic mapping cannot analyze genes of small effect size [28]. Given the multiple factors that contribute to an under-estimation of the genetic effect size (discussed above), it is likely that many different phenotypes can be analyzed by HBCGM when a larger number of strains are characterized. Since additional strains must be genetically analyzed, the new computational tool was used to examine the impact that incorporating allelic data from an additional strain had on the genetic map. Allelic data derived from whole genome sequencing data obtained from an additional strain resulted in the formation of ~30,000 additional haplotype blocks, which represented 5–6% of the total number of blocks formed. Surprisingly, new genetic variation present in the added strain was responsible for ~50% of the newly formed blocks [14]. Genotyping arrays that characterize known SNP alleles can provide useful information for QTL analysis [13, 29], but ~15,000 additional blocks produced by new sites of genetic variation would not be identified with array-based genotyping data, which only characterizes previously known SNPs. The unique genotypic variants that could be responsible for outlier phenotypic responses would be missed if the allelic data generated by these genotyping arrays was used [14]. We have already demonstrated that whole genome sequencing data can be used to produce comprehensive genetic maps, which can be used for HBCGM studies [14]. Since the pattern of genetic variation across a large number of inbred strains can be characterized at a reasonable cost by whole genome sequencing, it is feasible to produce genetic maps that enable a large number of strains to be used in HBCGM studies.
HBCGM is one of several methods that are being utilized to advance murine genetic analysis. For example, large arrays of ~1000 recombinant inbred mouse strains are being produced as a genetic-mapping resource [30]. HBCGM methodology could be used to analyze phenotypic datasets obtained from these strains. However, despite the large number of recombinants, this panel only contains the genetic variation present within eight founder strains. Although some phenotypes that are not found in the parental strains could appear within recombinant strains, it will have a limited ability to analyze many disease traits whose causative genes are not variable within the limited set of founder strains. However, the acetaminophen study [17] also illustrates a very significant limitation of using these recombinant panels. Because the strain that was uniquely resistant to acetaminophen-induced liver toxicity was not among the founder strains, this panel could offer little insight into this important problem. If a SNP of interest happens to be in the recombinant strain panel, HBGCM of data obtained from analysis of a panel of inbred strains could be used to quickly identify the candidate genes located within an identified QTL interval, as we have already demonstrated [19, 20].
A major limitation of HBCGM is that it cannot analyze traits with a complex genetic architecture, which is a major strength for linkage analysis. However, it currently costs only ~$6,000 to sequence the genome of an additional inbred strain with a recently discovered phenotype of interest, which enables it to be used in a HBCGM experiment. In contrast, multiples of $10 million are required to create and maintain new collaborative cross panels that incorporate new strains. Moreover, identification of genetic modifiers affecting a strain-specific phenotype produced by transgene expression or by a gene knockout will be of increased importance in 21st century studies. While it would be prohibitively difficult to introduce a knockout or transgene onto a large panel of recombinant inbred strains, it can be bred onto a set of inbred strains to enable haplotype-based computational genetic studies.
Concluding remarks
Murine genetic association studies can identify genetic factors affecting a wide range of 21st century biomedical phenotypes. However, some alterations to the methodology, which was developed in the 20th century, is required to ensure that murine genetic association studies produce useful results. To facilitate genetic discovery, investigators should: 1) assess a phenotype across a large number (≥20) of inbred strains; 2) perform the genetic analysis using recently developed methods (such as ‘next generation’ HBCGM) that can optimally analyze the phenotypic data obtained from a large inbred strain panel; 3) use comprehensive genetic maps produced by analysis of whole genome sequencing data; and 4) integrate the analysis of the genetic association results with gene expression, metabolomic or other types of ‘omic’ datasets that are relevant to the phenotype. Next generation sequencing enables a genetic map covering a very large number of inbred strains to be assembled. Although not all biomedical traits can be analyzed by this approach, these four modifications should greatly increase the number of genetic discoveries that emerge from murine genetic association studies.
Supplementary Material
Table I.
Comparison of GWAS results obtained using marker or causative SNPs.
| GWAS P-values | GWAS PoPVg | |||||
|---|---|---|---|---|---|---|
| # strains | D′ | r | Causative | Marker | Causative | Marker |
| Genetic effect size = 2 | ||||||
| 12 | 1 | 0.85 | 0.03 | 0.08 | 0.54 | 0.40 |
| 16 | 1 | 0.88 | 0.01 | 0.03 | 0.53 | 0.42 |
| Genetic effect size = 3 | ||||||
| 12 | 1 | 0.85 | 0.002 | 0.02 | 0.73 | 0.53 |
| 16 | 1 | 0.88 | 0.0003 | 0.004 | 0.72 | 0.57 |
Acknowledgments
Funding Source
G.P. was partially supported by funding from a transformative RO1 award (1R01DK090992-01) from the NIDDK.
Abbreviations
- HBCGM
Haplotype-Based computational Genetic Mapping
- GWAS
Genome-wide Association Study
- PoPVg
Percent of Population Variance Explained by a Genetic locus
Footnotes
Disclosure Statement
The authors do not have any conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Flint J, et al. Strategies for mapping and cloning quantitative trait genes in rodents. Nat Rev Genet. 2005;6:271–286. doi: 10.1038/nrg1576. [DOI] [PubMed] [Google Scholar]
- 2.Payseur BA, Place M. Prospects for association mapping in classical inbred mouse strains. Genetics. 2007;175:1999–2008. doi: 10.1534/genetics.106.067868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su WL, et al. Assessing the prospects of genome-wide association studies performed in inbred mice. Mamm Genome. 2010;21:143–152. doi: 10.1007/s00335-010-9249-7. [DOI] [PubMed] [Google Scholar]
- 4.Chu LF, et al. From Mouse to Man: The 5-HT3 Receptor Modulates Physical Dependence on Opioid Narcotics. Pharmacogenetics and Genomics. 2009;19:193–205. doi: 10.1097/FPC.0b013e328322e73d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Y, et al. The Role of IL-1 in Wound Biology Part I: Murine in Silico and In vitro Experimental Analysis. Anesthesia & Analgesia. 2010;111:1525–1533. doi: 10.1213/ANE.0b013e3181f5ef5a. [DOI] [PubMed] [Google Scholar]
- 6.Hu Y, et al. The Role of IL-1 in Wound Biology Part II: In vivo and Human Translational Studies. Anesthesia & Analgesia. 2010;111:1534–1542. doi: 10.1213/ANE.0b013e3181f691eb. [DOI] [PubMed] [Google Scholar]
- 7.Liao G, et al. In Silico Genetics: Identification of a Functional Element Regulating H2-Ea Gene Expression. Science. 2004;306:690–695. doi: 10.1126/science.1100636. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, et al. Computational Genetics: From Mouse to Man? Trends in Genetics. 2005;21:526–532. doi: 10.1016/j.tig.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 9.Yalcin B, et al. Unexpected complexity in the haplotypes of commonly used inbred strains of laboratory mice. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9734–9739. doi: 10.1073/pnas.0401189101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, et al. Computational Genetics and Genomics: New Tools for Understanding Disease. Humana Press Inc; 2005. Haplotypic Structure of the Mouse Genome; pp. 71–83. [Google Scholar]
- 11.Wade CM, et al. The mosaic structure of variation in the laboratory mouse genome. Nature. 2002;420:574–578. doi: 10.1038/nature01252. [DOI] [PubMed] [Google Scholar]
- 12.Kang HM, et al. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirby A, et al. Fine mapping in 94 inbred mouse strains using a high-density haplotype resource. Genetics. 2010;185:1081–1095. doi: 10.1534/genetics.110.115014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peltz G, et al. Next-Generation Computational Genetic Analysis: Multiple Complement Alleles Control Survival After Candida Albicans Infection. Infection and Immunity. 2011;79:4472–4479. doi: 10.1128/IAI.05666-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yalcin B, et al. Commercially available outbred mice for genome-wide association studies. PLoS Genet. 2010:6. doi: 10.1371/journal.pgen.1001085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng M, et al. Computational Genetic Mapping in Mice: ‘The Ship has Sailed’. Science Translational Medicine. 2009;1:3ps4. doi: 10.1126/scitranslmed.3000377. [DOI] [PubMed] [Google Scholar]
- 17.Liu H-H, et al. An Integrative Genomic Analysis Identifies Bhmt2 As A Diet-Dependent Genetic Factor Protecting Against Acetaminophen-Induced Liver Toxicity. Genome Research. 2010;20:28–35. doi: 10.1101/gr.097212.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, et al. In Silico and In Vitro Pharmacogenetics: Aldehyde Oxidase Rapidly Metabolizes a p38 Kinase Inhibitor. The Pharmacogenomics Journal. 2011;11:15–24. doi: 10.1038/tpj.2010.8. [DOI] [PubMed] [Google Scholar]
- 19.Smith SB, et al. Quantitative trait locus and computational mapping identifies Kcnj9 (GIRK3) as a candidate gene affecting analgesia from multiple drug classes. Pharmacogenetics and Genomics. 2008;18:231–241. doi: 10.1097/FPC.0b013e3282f55ab2. [DOI] [PubMed] [Google Scholar]
- 20.LaCroix-Fralish ML, et al. The β3 Subunit of the Na+, K+-ATPase Affects Pain Sensitivity. Pain. 2009;144:294–302. doi: 10.1016/j.pain.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guenet JL, Bonhomme F. Wild mice: an ever-increasing contribution to a popular mammalian model. Trends Genet. 2003;19:24–31. doi: 10.1016/s0168-9525(02)00007-0. [DOI] [PubMed] [Google Scholar]
- 22.Reuveni E, et al. The consequence of natural selection on genetic variation in the mouse. Genomics. 2010;95:196–202. doi: 10.1016/j.ygeno.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 23.Beck JA, et al. Genealogies of mouse inbred strains. Nature Genetics. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- 24.Valdar W, et al. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet. 2006;38:879–887. doi: 10.1038/ng1840. [DOI] [PubMed] [Google Scholar]
- 25.Valdar W, et al. Simulating the collaborative cross: power of quantitative trait loci detection and mapping resolution in large sets of recombinant inbred strains of mice. Genetics. 2006;172:1783–1797. doi: 10.1534/genetics.104.039313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keane TM, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chanock SJ, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 28.Flint J, Mott R. Applying mouse complex-trait resources to behavioural genetics. Nature. 2008;456:724–727. doi: 10.1038/nature07630. [DOI] [PubMed] [Google Scholar]
- 29.Hutchins LN, et al. CGDSNPdb: a database resource for error-checked and imputed mouse SNPs. Database (Oxford) 2010;2010:baq008. doi: 10.1093/database/baq008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iraqi FA, et al. The Collaborative Cross, developing a resource for mammalian systems genetics: a status report of the Wellcome Trust cohort. Mamm Genome. 2008;19:379–381. doi: 10.1007/s00335-008-9113-1. [DOI] [PubMed] [Google Scholar]
- 31.Guo YY, et al. In Silico Pharmacogenetics: Warfarin Metabolism. Nature Biotechnology. 2006;24:531–536. doi: 10.1038/nbt1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo YY, et al. In vitro and In silico Pharmacogenetic Analysis in Mice. Proceedings of the National Academy of Sciences. 2007;104:17735–17740. doi: 10.1073/pnas.0700724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaas AK, et al. Plasminogen Alleles Influence Susceptibility to Invasive Aspergillosis. PLoS genetics. 2008;4:e1000101. doi: 10.1371/journal.pgen.1000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tregoning JS, et al. Genetic Susceptibility to the Delayed Sequelae of RSV Infection is MHC-Dependent, but Modified by Other Genetic Loci. J Immunology. 2010;185:5384–5391. doi: 10.4049/jimmunol.1001594. [DOI] [PubMed] [Google Scholar]
- 35.Liang D, et al. A Genetic Analysis of Opioid-Induced Hyperalgesia in Mice. Anesthesiology. 2006;104:1054–1062. doi: 10.1097/00000542-200605000-00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang DY, et al. Genetic Variants of the P-Glycoprotein Gene Abcb1b Modulate Opioid-Induced Hyperalgesia, Tolerance and Dependence. Pharmacogenetics and Genomics. 2006;16:825–835. doi: 10.1097/01.fpc.0000236321.94271.f8. [DOI] [PubMed] [Google Scholar]
- 37.Li X, et al. Expression genetics identifies spinal mechanisms supporting formalin late phase behaviors. Molecular Pain. 2010;6:11. doi: 10.1186/1744-8069-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.