Abstract
Neurodegenerative diseases are often associated with misfolding and deposition of specific proteins in the nervous system. The prion protein, which is associated with transmissible spongiform encephalopathies (TSEs), is one of them. The normal function of the cellular form of the prion protein (PrPC) is mediated through specific signal transduction pathways and is linked to resistance to oxidative stress, neuronal outgrowth and cell survival. In TSEs, PrPC is converted into an abnormally folded isoform, called PrPSc, that may impair the normal function of the protein and/or generate toxic aggregates. To investigate these molecular events we performed a two-dimensional gel electrophoresis comparison of neuroblastoma N2a cells expressing different amounts of PrPC and eventually infected with the 22L prion strain. Mass spectrometry and peptide mass fingerprint analysis identified a series of proteins with modified expression. They included the chaperones Grp78/BiP, protein disulfide-isomerase A6, Grp75 and Hsp60 which had an opposite expression upon PrPC expression and PrPSc production. The detection of these proteins was coherent with the idea that protein misfolding plays an important role in TSEs. Other proteins, such as calreticulin, tubulin, vimentin or the laminin receptor had their expression modified in infected cells, which was reminiscent of previous results. Altogether our data provide molecular information linking PrP expression and misfolding, which could be the basis of further therapeutic and pathophysiological research in this field.
Key words: chaperones, neuroblastoma, prion, proteomics
Introduction
Transmissible spongiform encephalopathies (TSEs), also called prion diseases, are fatal neurodegenerative disorders including Kuru, Creutzfeldt-Jakob disease (CJD), Gerstmann-Sträussler-Scheinker syndrome and fatal familial insomnia in humans, scrapie in sheep and goats and bovine spongiform encephalopathy in cattle.1,2 The physiological function of the cellular prion protein, PrPC, is still elusive but seems linked in various models through specific signal transduction pathways with resistance to oxidative stress, neuronal outgrowth or cell survival.3 PrPC expression is necessary for the development of TSEs as demonstrated by the resistance of PrP knock-out animals to prions, while the overexpression of the protein accelerates the disease.4 In TSEs, this protein is converted into an abnormal isoform named PrPSc 2 that accumulates in the central nervous system. This conversion could impair PrPC function and/or generate toxic aggregated entities like those present in other neurodegenerative disorders. The exact involvement of these gain- or loss-of-function mechanisms in prion disease pathogenesis is still under debate.
Few proteomics studies have been made in cultured cells for investigating molecular pathogenesis of prion disorders.5–8 Proteomic differential protein expression analysis represents however an interesting approach that could complete previous genomic studies.9–12 In this article we performed a proteomic investigation in an integrated cellular model where both differential PrPC expression and conversion could be obtained. We relied on a PrPC tetracycline-inducible expression system that had been previously established in the murine neuroblastoma cells N2a.13 In these inducible cells, named iN2a, PrPC expression levels can be controlled by varying the concentration of the tetracycline-derivative doxycycline acting as effector. For this work, we successfully infected these cells with the 22L prion strain. We then performed a proteomic two-dimensional gel electrophoresis (2DE) comparison of iN2a cells expressing different concentrations of PrPC and either infected or non-infected. Differential proteins were identified by mass spectrometry using a peptide mass fingerprint analysis. We could identify several proteins showing a modified expression pattern in relation with a PrPC expression and/or PrPSc production. Some proteins had an opposite behavior in these two situations suggesting a relationship between the physiological function of PrPC and its conversion.
Results
Cellular model and PrPC/PrPSc expression.
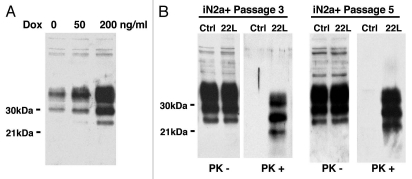
In this study, we used a murine neuroblastoma cell line (iN2a) model to investigate the proteomic consequences of both PrPC expression and prion infection. This approach was original as it relied on an integrated cellular model. We paid special attention to analyze cells with the same number of sub-passages and cultured in the same medium. The tetracycline-inducible expression system responsible here for PrPC overexpression has been used in other 2DE proteomics studies of cultured cells where it did not modified in a detectable way proteome analysis.17 As expected from previous work,13 iN2a cultured in the presence of doxycycline (iN2a+) showed an increased expression of the three major PrPC bands that corresponded to un-, mono- and diglycosylated isoforms of the protein (Fig. 1A). A concentration of 200 ng/ml of doxycycline was chosen as it gave a stable and robust PrPC expression over repeated passages without modification of the western blot profile suggesting that PrPC trafficking/biology was not affected by the overexpression. These iN2a+ cells were susceptible to the 22L prion strain (Fig. 1B). In fact, these cells named: iN2a+22L, accumulated significant amount of PrPSc by the third passage following infection and thereafter (up to 20 passages at least, data not shown). Interestingly, even though tetracycline compounds have been identified as potent anti-prion molecules,18 the amount of doxycycline (<0.5 µM) that we used did not prevent in our paradigm the accumulation of PrPSc by the cells (Fig. 1B).
Figure 1.
Western blot of the cell extracts after PrPC induction & infection. (A) iN2a cells were cultured for three days with increasing amounts of doxycycline. Cells were lysed at 80% of confluence and equal amounts of protein were analyzed by western blotting for PrPC using the antibody SAF32. (B) iN2a cultured for three passages with 200 ng/mL of doxycycline (iN2a+) were infected with 22L brain prion homogenate as detailed in the Materials and Methods section. After passage 3 or 5, control (Ctrl) and infected cells (22L) were analyzed for PrPC and PrPSc expression (PK− and PK+). Infected iN2a+ cells (iN2a+22L) accumulated significant amounts of PrPSc.
Two-dimensional electrophoresis (2DE), selection and identification of spots.
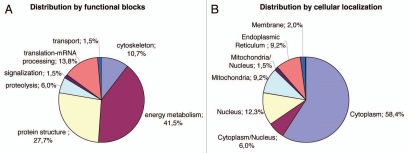
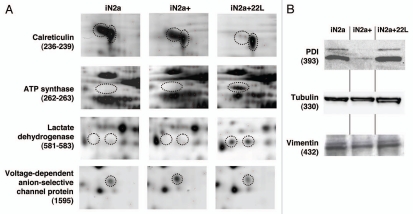
Consecutive lysates of iN2a/iN2a+/iN2a+22L cultures were analyzed by 2DE as detailed in the Materials and Methods section. An iN2a+22L gel that had the maximum number of detected spots (Fig. 2) was used as the reference gel when using the Melanie 5.0 quantitation software. 544 protein spots were detected in this gel (Suppl. Table 1). The matching of this gel to the other gels generated raw data, which were the basis for the statistical analysis. Spots that had a differential expression between iN2a, iN2a+ and iN2a+22L cells, as well as spots with the same expression localized in different sections of the gel—to have spots of references—were selected for mass spectrometry identification. A total of 65 protein spots were eventually identified (Suppl. Table 2 and Table 1). A good correlation between the observed and the theoretical pI and molecular weight (MW) standard loaded on a gel was observed. For some spots the same identification was found corresponding therefore to post-translational modified isoforms of the same protein (e.g., calreticulin, alpha enolase, lactate dehydrogenase). Classification of the identified proteins in different functional blocks6 (Fig. 3A) revealed, as often found in 2DE studies,15,19,20 that many of them belonged to the energetic metabolism (41.5%), followed by the protein structure block (27.7%), then translation-mRNA processing (13.8%), cytoskeleton (10.7%), proteolysis (6%), transport and signal transduction blocks (1.5%). The protein distribution based on the cellular localization (Fig. 3B) showed, as expected for this type of 2DE analysis with urea lysis buffer, that 58.4% of the proteins were localized in the cytoplasm, 12.3% in the nucleus, 9.2% in mitochondria and the endoplasmic reticulum, 6% in the cytoplasm and nucleus, 2% on the membrane and 1.5% in the mitochondria and nucleus. Identified proteins were sorted in eight groups (A to H) based on their differential expression between iN2a, iN2a+ and iN2a+22L cells (Table 1). Induction of PrP expression resulted in the decreased expression of nine proteins (groups A and B). Eight proteins (groups C–E) had their expression increased following PrPC overexpression. Prion infection resulted in the decreased expression of seven proteins (groups C and F) and the increased expression of 22 (groups A, D and G). Among these 22 proteins, 11 increased between both iN2a or iN2a+ and infected iN2a+22L. Some of these differential expressions were illustrated on the Figure 4A (calreticulin, ATP synthase, lactate dehydrogenase and voltage-dependent anion-selective channel protein). Importantly, 2DE allows the detection of several post-translationally modified isoforms of a same protein (after phosphorylation, glycosylation…) which expression could then vary differently. The western blot analysis of PDIA6, tubulin and vimentin (Fig. 4B) confirmed variations observed on 2DE. However, quantitation of the bands using the “ImageJ” software (not shown) confirmed the visual indication that differences were lower than those observed on 2DE, especially for tubulin and vimentin. This might be in relation with the non-linearity of the western blot, as well as, with the incapacity of 1D gels to differentiate isoforms as 2DE.
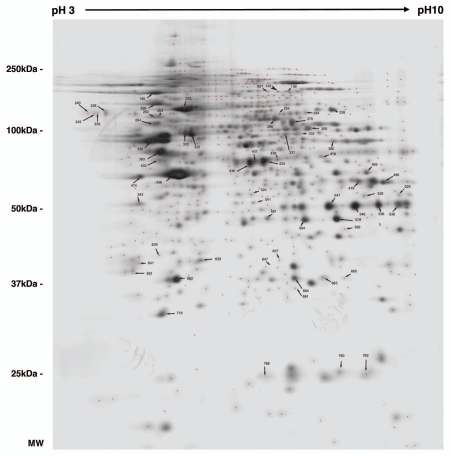
Figure 2.
2DE reference gel of iN2a cells. iN2a cell extract were separated using 2DE electrophoresis in a dry strip pH 3–10 for the first dimension, a 12% SDS-PAGE for the second dimension and silver stained. The identification of proteins, noted with their ID number (Suppl. Tables 1 and 2), was performed by peptide mass fingerprints after trypsin digestion and MALDI-TOF on Coomassie or silver stained spots.
Table 1.
Table of identified proteins (spot ID, gene, accession number from UniprotKB database, name and functional group) sorted in eight groups (A-H) based on the fold expression ratio iN2a+/iN2a and iN2a+22L/iN2a+
| Group | Spot ID | Gene | Accession number | Protein | iN2a+/iN2a fold ratio | t-test | variation | iN2a+22L/iN2a+ fold ratio | t-test | variation | Fonctional group |
| A | 200 | Hspa5 | P20029 | Grp78 / BiP (Glucose regulated protein 78kDa / heat shock 70kDa protein 5) | −1,50 | 0,029 | 1,52 | 0,018 | protein structure, modification & regulation | ||
| A | 305 | Pkm2 | P52480 | Pyruvate kinase isozyme M2 | −3,05 | 0,016 | 5,16 | 0,032 | energy metabolism | ||
| A | 325 | Prph | P15331 | Peripherin | −1,69 | 0,016 | 1,88 | 0,001 | Cytoskeleton | ||
| A | 330 | Tubb5 | P99024 | Tubulin beta-5 chain | −2,00 | 0,003 | 1,80 | 0,025 | Cytoskeleton | ||
| A | 393 | Pdia6 | Q922R8 | Pdia6 (Protein disulfide-isomerase A6) | −2,00 | 0,020 | 2,55 | 0,013 | protein structure, modification & regulation | ||
| A | 432 | Vim | P20152 | Vimentin | −2,50 | 0,095 | 6,13 | 0,015 | Cytoskeleton | ||
| B | 223 | Hspa9a | P38647 | Grp75 / Hspa9 (Stress-70 protein, heat shock protein 9A) | −1,64 | 0,047 | 1,50 | 0,075 | - | protein structure, modification & regulation | |
| B | 323 | Hspdl | P63038 | Hsp60 / chaperonin (heat shock 60kDa protein 1) | −1,88 | 0,047 | 1,14 | 0,294 | - | protein structure, modification & regulation | |
| B | 456 | Actb | P60710 | Actin beta | −1,45 | 0,013 | 1,33 | 0,162 | - | Cytoskeleton | |
| C | 595 | Vdad | Q60932 | Voltage-dependent anion-selective channel prot1 | 1,38 | 0,014 | −1,94 | 0,005 | protein structure, modification & regulation | ||
| D | 431 | Eno1 | P17182 | alpha enolase | 1,47 | 0,009 | 1,33 | 0,015 | energy metabolism | ||
| D | 524 | Stoml2 | Q99JB2 | Stomatin-like protein 2 (SLP-2) | 1,45 | 0,029 | 2,71 | 0,014 | cytoskeleton | ||
| E | 486 | Aldoa | P05064 | Fructose biphosphate Aldolase | 1,64 | 0,018 | 1,12 | 0,293 | - | energy metabolism | |
| E | 528 | Hrnpa3 | Q8BG05 | heterogeneous nuclear ribonucleoprotein A3 | 1,35 | 0,034 | −1,11 | 0,476 | - | translation-mRNA processing | |
| E | 546 | Gapdh | P16858 | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | 1,24 | 0,014 | −1,20 | 0,066 | - | energy metabolism | |
| E | 681 | Psma2 | P49722 | Proteasome subunit alpha type2 | 4,96 | 0,003 | −1,72 | 0,130 | - | Proteolysis | |
| E | 719 | Pebpl | P70296 | Phosphatidylethanolamine binding protein | 1,46 | 0,042 | −1,45 | 0,067 | - | protein structure, modification & regulation | |
| F | 239 | Calr | P14211 | Calreticulin | −1,23 | 0,163 | - | −3,05 | 0,004 | protein structure, modification & regulation | |
| F | 240 | Calr | P14211 | Calreticulin | 1,20 | 0,283 | - | −5,90 | 0,004 | protein structure, modification & regulation | |
| F | 242 | Calr | P14211 | Calreticulin | 1,38 | 0,182 | - | −6,66 | 0,006 | protein structure, modification & regulation | |
| F | 284 | Hnrnpk | P61979 | Heterogeneous nuclear ribonucleoprotein K | −1,10 | 0,278 | - | −3,50 | 0,349 | translation-mRNA processing | |
| F | 478 | Aldoa | P05064 | Fructose biphosphate Aldolase | 1,23 | 0,060 | - | −1,60 | 0,009 | energy metabolism | |
| F | 766 | Ppia | P17742 | Peptidyl-prolyl-cis-trans isomerase A | −1,11 | 0,310 | - | −1,97 | 0,038 | protein structure, modification & regulation | |
| G | 145 | Hsp90ab1 | P11499 | Heat shock protein(HSP90B) | −1,37 | 0,154 | - | 1,67 | 0,047 | protein structure, modification & regulation | |
| G | 244 | Hnrnpl | Q8R081 | heterogeneous nuclear ribonucleoprotein L | −1,26 | 0,234 | - | 2,44 | 0,053 | translation-mRNA processing | |
| G | 262 | Tcl1b1 | P56480 | ATP synthase beta chain, mitochondrial precursor | - | >2 | <0.001 | energy metabolism | |||
| G | 263 | Tcl1b1 | P56480 | ATP synthase beta chain, mitochondrial precursor | - | >2 | <0.01 | energy metabolism | |||
| G | 331 | cct5 | P80316 | T-complex protein 1 epsilon (TCP1) | −1,13 | 0,305 | - | 1,94 | 0,017 | protein structure, modification & regulation | |
| G | 416 | Eno1 | P17182 | alpha enolase | −1,31 | 0,241 | - | 3,68 | 0,007 | energy metabolism | |
| G | 425 | Eno1 | P17182 | alpha enolase | −1,07 | 0,300 | - | 1,99 | 0,001 | energy metabolism | |
| G | 426 | Eno1 | P17182 | alpha enolase | −1,01 | 0,489 | - | 2,11 | 0,009 | energy metabolism | |
| G | 474 | Rpsa | P38983 | Laminin receptor 40Sribosomal protein SA | −1,04 | 0,404 | - | 1,63 | 0,034 | translation-mRNA processing | |
| G | 525 | Hrnpa3 | Q8BG05 | Heterogenousnuclear ribonucleoprotein A3 | - | >2 | <0.01 | translation-mRNA processing | |||
| G | 579 | Ldha | P06151 | Lactate dehydrogenase | 1,14 | 0,277 | - | 2,23 | 0,012 | energy metabolism | |
| G | 581 | Ldha | P06151 | Lactate dehydrogenase | −1,54 | 0,082 | - | 3,68 | 0,001 | energy metabolism | |
| G | 629 | Actb | P60710 | Actin beta | 1,16 | 0,186 | - | 3,95 | 0,120 | Cytoskeleton | |
| G | 647 | Pgaml | Q9DBJ1 | Phosphoglycerate mutase 1 | - | >2 | <0.05 | energy metabolism | |||
| H | 139 | Khsrp | Q3UOV1 | KH-type splicing regulatory protein | 1,07 | 0,434 | - | 1,71 | 0,207 | - | translation-mRNA processing |
| H | 143 | Khsrp | Q3UOV1 | KH-type splicing regulatory protein | 1,06 | 0,391 | - | 1,19 | 0,319 | - | translation-mRNA processing |
| H | 226 | Tkt | P40142 | Transketolase | −1,05 | 0,407 | - | 1,50 | 0,075 | - | energy metabolism |
| H | 236 | Calr | P14211 | Calreticulin | −1,06 | 0,399 | - | 1,15 | 0,238 | - | protein structure, modification & regulation |
| H | 254 | Hnmpl | Q8R081 | Heterogenousnuclear ribonucleoprotein L | 1,13 | 0,391 | - | −1,15 | 0,053 | - | translation-mRNA processing |
| H | 268 | STI1 | Q60864 | Stress-induced-protein 1 | −1,03 | 0,405 | - | 1,12 | 0,202 | - | protein structure, modification & regulation |
| H | 275 | cct6a | P80317 | T-complex prot1-zeta (TCP1) | −1,05 | 0,405 | - | 1,37 | 0,096 | - | protein structure, modification & regulation |
| H | 332 | Lap3 | Q9CPY7 | Cytosol leu aminopeptidase | −1,51 | 0,176 | - | 1,90 | 0,066 | - | protein structure, modification & regulation |
| H | 360 | Atp5a1 | Q03265 | ATP synthase alpha mitoc prec. | 1,05 | 0,457 | - | 1,37 | 0,234 | - | energy metabolism |
| H | 430 | Eno1 | P17182 | alpha enolase | −1,33 | 0,069 | - | 1,28 | 0,157 | - | energy metabolism |
| H | 462 | Pgk1 | P09411 | Phosphoglycerate Kinase | 1,40 | 0,100 | - | 1,71 | 0,060 | - | energy metabolism |
| H | 536 | Gapdh | P16858 | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | 1,22 | 0,166 | - | 1,45 | 0,117 | - | energy metabolism |
| H | 538 | Gapdh | P16858 | Glyceraldehyde-3-phosphate dehydrogenase(GAPDH) | 1,20 | 0,179 | - | 1,01 | 0,489 | - | energy metabolism |
| H | 541 | Laspl | Q61792 | LIM & SH3 domain prot1 | 1,13 | 0,286 | - | −1,03 | 0,445 | - | cytoskeleton |
| H | 543 | Npm1 | Q61937 | Nucleophosmin | 1,25 | 0,062 | - | −1,17 | 0,145 | - | transport |
| H | 547 | Gapdh | P16858 | Glyceraldehyde-3-phosphate dehydrogenase(GAPDH) | 1,09 | 0,258 | - | −1,40 | 0,086 | - | energy metabolism |
| H | 584 | Ldha | P06151 | Lactate dehydrogenase | −1,22 | 0,226 | - | 1,18 | 0,287 | - | energy metabolism |
| H | 637 | Cacybp | Q9CXW3 | Calcyclin-binding prot | 1,40 | 0,120 | - | −1,19 | 0,244 | - | proteolysis |
| H | 639 | Pgls | Q9CQ60 | 6-Phosphogluconolactonase(6PG L) | 1,08 | 0,389 | - | 1,40 | 0,091 | - | energy metabolism |
| H | 641 | Rab35 | Q6PHN9 | Ras-related protein Rab-35 | 1,16 | 0,199 | - | −1,06 | 0,431 | - | signalisation |
| H | 651 | Psma5 | Q9Z2U1 | Proteasome subunit alpha type 5 | −1,09 | 0,159 | - | 1,06 | 0,373 | - | proteolysis |
| H | 662 | Uch.11 | Q9R0P9 | 5-ubiquitin carboxyterminal hydrolase | −1,06 | 0,271 | - | −1,14 | 0,129 | - | proteolysis |
| H | 663 | Tpi1 | P17751 | Those Phosphate Isomerase | 1,23 | 0,239 | - | 1,24 | 0,269 | - | energy metabolism |
| H | 664 | Tpi1 | P17751 | Those Phosphate Isomerase | 1,20 | 0,200 | - | 1,06 | 0,368 | - | energy metabolism |
| H | 665 | Gstm2 | P15626 | GlutathionS-transferase Mu 2 | 1,22 | 0,123 | - | −1,12 | 0,267 | - | energy metabolism |
| H | 760 | Ppia | P17742 | Peptidyl-prolyl-cis-trans isomerase A | 1,20 | 0,326 | - | 1,28 | 0,256 | - | protein structure, modification & regulation |
| H | 762 | Ppia | P17742 | Peptidyl-prolyl-cis-trans isomerase A | 1,35 | 0,138 | - | 1,08 | 0,402 | - | protein structure, modification & regulation |
| H | 821 | Khsrp | Q3UOV1 | KH-type splicing regulatory protein | −1,12 | 0,365 | - | 1,61 | 0,250 | - | translation-mRNA processing |
Fold ratio above 1.5 or below −1.5, as well as t-test below 0.05 were used as thresholds for the main proteins studied.
Figure 3.
Distribution of the identified proteins per functional blocks (A) and cellular origin (B). (References in Suppl. Table 2.)
Figure 4.
Illustration of protein differential expression. (A) Regions of 2DE silver stained gel of the different cell types illustrating the variation of calreticulin, ATP synthase, lactate dehydrogenase and voltage-dependent anion-selective channel protein. (B) Western blot detection after 1D gel electrophoresis of PDIA6, tubulin beta and vimentin in iN2a, iN2a+ and iN2a+22L. Numbers in parenthesis refer to protein ID numbers (Suppl. Tables 1 and 3).
Discussion
Proteomic profiling and differential protein expression analyses in the prion field were performed in a small number of studies on experimental mice and cellular models,5–8,21,22 or on biological fluids in order to find new diagnosis biomarkers.23,24 Work focused on identifying PrP partners has been also conducted on purified and recombinant molecules.25–27 In a study similar to the one presented here, Chich et al. compared GT1 cells chronically infected with prions to control cells6 while Biaisini and Massignan5,8 reported interesting results in cultured cells using a 2D proteomic approach, but they focused on the impact of mutated PrP rather than on its conversion into PrPSc. The number of proteins identified in common in the different studies is limited which could be easily explained by the different cellular models used and by distinct paradigms. Importantly in our model, PrPC induction and infection was obtained in the same cell clone with a parallel analysis of the infected cells, while in Chich et al.6 a comparison of control and chronically infected subclones was undertaken. Two recent publications analyzed using 2DE gels the influence of PrP expression on the proteome of mice21 and cultured cells.7 The latter work focused mainly on HEK 293 cells and revealed the modification of proteins involved in cell death and in response to oxidative stress which was reminiscent of our results. The other interesting study did not identify significant differential proteins between brain homogenates of Prnp−/− and Prnp+/+ mice. It is however difficult to compare these results with ours as the starting material (tissue) and the paradigm (transgenic animals) is very different. Finally, the recent study of Moore et al.22 on 22L-infected brain homogenates did not report protein specifically associated with PrPSc, suggesting that the selection of our candidates related to a metabolic (rather than to a physical) effect of this abnormal isoform.
Genomic investigations of cells expressing or not PrPC or after prion infection have also been realized.9–12 Comparison of the list of differential genes generated in these studies with our differential proteins was however not informative as it is known that such global comparison between genomics and proteomics are often disappointing. In addition, a recent work concluded in a transcriptional stability of cultured cells upon prion infection,28 which was in discrepancy with previous works as a possible result of differences in experimental stringency of the study design to minimize genetic drifts. Genomic investigations have been also conducted on brain tissues, for example on cattle during the courses of TSEs.29 Comparison with our data was also difficult in these cases since these studies evaluated a global pathophysio-logical tissue response, and not only that of prion-infected cells.
To evaluate the interest and the relevance of our data, we therefore focused on proteins that have already been involved in prion biology. A first group of interesting proteins was represented by chaperones molecules which have been already identified as differential proteins in prion infected cells.30 These molecules assist protein folding and handle misfolded polypeptide chains by breaking up aggregates and assisting in the refolding process.31 These proteins were often identified in whole proteome studies due to their high representation in 2D gels.19,20 The differential expression of these chaperones was interesting since prion conversion is believed to be associated with protein misfolding (this is particularly well documented in yeast).19,20 In our study five chaperones (Grp78/BiP, Pdia6, Grp75/ Hspa9, Hsp60/chaperonin, Hsp90B) had their expression decreased along with PrPC induction and/or increased following prion infection. This could be in relation with PrPC acting as a protector of cellular stress while its misfolding conversion could affect this physiological function. Several of these chaperones (or their close isoforms) have been already identified as important players in prion replication, both in vitro and in cell culture. For example, Grp78/BiP bound to mutated PrPs and was induced by prion infection.32 Grp58, which corresponds to a protein disulfide-isomerase closely related to Pdia6 (84% of homology), also followed closely the formation of PrPSc at all stages of the disease in different brain areas in mice and in scrapie infected N2a.32 In addition Grp58 and PrP were shown to interact by co-immunoprecipitation and inhibition of the chaperone with small interfering RNA led to a significant enhancement of PrPSc toxicity.32 As a matter of fact, the role of heat shock proteins (Hsp) in prion response (for our study Hspa5, Hspa9, Hsp60, Hsp90B) has been put forward in many studies.32–37 Some of these molecules could be associated with PrP26,27 as shown in particular for Hsp60,37 or be part of the stress response in relation with the oxidative stress resulting from prion propagation.38 We also observed that isoforms of two additional chaperones were under-expressed in infected cells: calreticulin and peptidyl-prolylcis-trans isomerase A (Ppia). Calreticulin promotes correct glycoprotein synthesis, supports quality control and it was shown that it binds a specific hydrophobic polypeptide stretch in PrP.39 Modification of PrP folding during prion infection could explain the low expression of calreticulin, which might be a new therapeutic target of TSEs, by restoring a normal PrP folding along with its glycosylation. Ppia, also called cyclophilin A, has been already implicated in the metabolism of PrP which contains many proline residues. Inhibition of Ppia activity by cyclosporine A resulted in the formation of aggresomes containing PrP40 that were associated to caspase activation and apoptosis. These structures were also detected in prion-infected cells34 and in fact they could be linked to the decreased expression of Pdia that we observed. Aggresomes with PrP can also result from the inhibition of the proteasome which was involved in PrP degradation.41,42 Indeed, we detected an increased expression of the alpha 2 major proteasome sub-unit, which could be in relation with PrPSc production.
We also noticed that five cytoskeleton proteins: vimentin, tubulin beta, actin beta, stomatin-like protein 2 and peripherin had their expression decreased following PrPC induction and/or increased after infection. Vimentin is a member of the intermediate filaments family and a component of aggresome linked to prion replication as mentioned above.34 PrP has been also identified as an inhibitor of microtubule assembly by inducing formation of stable tubulin oligomers43 while actin was involved in yeast prion aggregation.44 Modification of cytoskeleton upon PrP expression may also reveal a role of the protein in cell morphology.45
As often in 2DE differential analysis, proteins involved in energy metabolism were detected. In our case, the most remarkable observation was a clear increase upon prion infection of several proteins such as: pyruvate kinase, alpha enolase, aldolase, glyceraldehyde-3-phosphate dehydrogenase, ATP synthase or lactate dehydrogenase. Overexpression of these enzymes may well relate to the increased requirement of both energy and protein synthesis pathways upon PrP expression or conversion. It is also possible that PrP interacts with some of these proteins and in particular with aldolase as detected by far-western immunoblotting.25 Interestingly, in the latter study, heterogeneous nuclear ribonucleoproteins have also been selected as PrP partners and some of these proteins had their expression modified in our study which was therefore reminiscent of the possible role of the protein in RNA trafficking and metabolism.46
Finally, we could observe an increase of the 37 kDa laminin receptor (Rpsa) upon infection. This result was consistent with previous work47,48 and confirmed the importance of this protein that could serve as a prion receptor. The Stress-induced-protein 1, which was identified as an important partner of PrP49, had its expression not modified by either PrP expression or conversion. The recent article of Massignan et al.,8 which investigated in 2DE the molecular alterations induced by expression of mutant PrP in N2a cells, identified 11 proteins (out of 42) that were also detected in our study. Importantly, many varied in the same manner in both studies (in iN2a: hspa, peripherin, fructose aldolase; in iN2a+22L: heterogenius nuclear ribonucleoproteins L, pyruvate kinase, enolase, lactate deshydrogenase, voltage-dependant anion-selective channel 1). This suggested that the impact of mutated PrPs is analogous to that of genuine PrPSc.
Materials and Methods
Culture and prion infection of N2a cells.
Generation of the murine inducible N2a neuroblastoma cell line (N2a-rtTA-74) stably transfected with the tetracycline-controlled transactivator and the murine PrP gene under the control of the transactivator-responsive promoter is described elsewhere.13 To simplify the nomenclature, we referred to these cells as “iN2a” in the manuscript. They were cultured in DMEM with glutamax, 1 mM sodium pyruvate and 1% L-glutamine supplemented with 10% heat-inactivated fetal calf serum in an atmosphere of 5% CO2, 95% air and collected at 80% confluence for protein analysis. They were cultured in the presence of doxycycline to induce PrPC expression. In this case, we referred them as “iN2a+.” Prion infection of iN2a+ cells was realized using the 22L brain homogenates as described before.14 Briefly, subconfluent cultures of iN2a+ (induced for 3 passages) were incubated for 4 hr with 0.1% of 22L brain homogenate diluted in Opti-MEM. Fresh complete medium was then added overnight and the cells were split at 1/3 thereafter. The infected status of the cells was assessed by proteinase K digestion analysis followed by western blot detection as described previously.14 22L prion infected iN2a+ cells were referred as “iN2a+22L.”
Whole cell extract for 2DE analysis.
2 × 106 cells at 80% of confluence were washed twice with PBS containing a cocktail of 0.1% protease and 0.1 mmol/L phosphatase inhibitors (P8340 and P6508, Sigma respectively) and lysed in 400 µL of a 2DE lysis buffer (urea 8 M, thiourea 1 M, Chaps 4.8%, dithiothreitol 50 mM). 1% Spermin 24 mM (Fluka 85588) was then added for 1 h at room temperature and the sample centrifuged 1 h at 20,000 g to precipitate DNA.15 Supernatant were then collected and stored at −80°C until use. Protein concentration of these lysates was determined using the PlusOne 2-D Quant Kit (GE Healthcare) following the supplier recommendation. Extracts were prepared at the same time for control and infected cells.
2D-Electrophoresis.
The following conditions for 2DE resulted from a previous study15 and adapted to murine N2a cell extract (not shown). For the first dimension, 20 µg of proteins per sample were added to 250 µL of rehydration buffer (9.8 M urea 4% CHAPS 50 mM DTT and 0.5% IPG buffer 3–10). IPG strips (13 cm), covering a pH range from 3–10 were rehydrated with this solution during 9 h at 20°C covered by low viscosity paraffin oil. For focalization, the following voltage/time profile was used: 30 V for 3 h, 200 V for 2 h, 1,000 V for 1 h, 3,000 V for 1 h, a gradient between 3,000 V and 8,000 V during 2 h and 8,000 V for 7 h. A total of 75,000 v.h was used. Focused strips were frozen at −20°C. For the second dimension, strips were equilibrated for 30 min in 6 M urea, 30% glycerol, 2% SDS, 50 mM Tris pH 8.8, 1% DTT and then for an additional 30 min in the same solution except that DTT was replaced by 5% iodoacetamide. After equilibration, proteins were separated in the second dimension by an SDS-PAGE method using 12% acrylamide gel with a ratio of acrylamide/bisacrylamide of 37.5:1.
Identification of protein expression and statistical test.
Gels were stained with a silver nitrate procedure16 and scanned at 300 dots per inch using the Labscan 3 software (GE Healthcare) after a procedure of calibration using the kaleidoscope LaserSoft Imaging (Kodak Ref: R020123). Spot detection and quantitation were performed with Melanie 5.0 software. The volume of each spot (integrated optical density) was calculated as the product of spot area and spot intensity. To take into account experimental variations, 2DE gels were normalized to the volume of all spots detected. Each gel was matched with others using at least ten landmarks per pairs and a minimal percentage of matches of 70% was obtained between each pairs. The reference gel used was that of the iN2a+22L cells. The experimental molecular weight (MW) and iso-electric point (pHi) were obtained based on the migration of a 2D protein standard (Bio-Rad, Richmond, CA, USA) spiked into a whole iN2a extract (not shown). Four to seven replicates with independent samples were performed to ensure reproducibility of the results (Suppl. Table 1). For comparison, the iN2a+22L gel was used as reference. Monovariate statistical analysis (Mann-Whiteney test) was performed using Tanagra software version 1.4.36.
Protein isolation and identification by mass spectrometry.
Protein spots were excised from either silver or PageBlue stained 2D gels. In the latter case the gel was ran with 300 µg of proteins. Spots were washed in 15 µL of 100 mM NH4HCO3 for 10 min. After addition of 15 µL of acetonitril and incubation for 10 min, the supernatant was removed and the procedure repeated. After 5 min in a centrivap concentrator, spots were re-hydrated in 10 µL of trypsin solution (15 ng/µL, Promega) and digested overnight at 25°C in 10 µL of 100 mM NH4HCO3 5 mM CaCl2 buffer. The tryptic peptides were extracted in a two steps procedure: the first step was composed of addition of 10 µL of 100 mM NH4HCO3 followed by 10 min of 10 µL acetonitril. This step was repeated twice and the supernatants were pooled. The second step was an incubation of 10 min with 10 µl of 5% formic acid and an addition of 10 µl of acetonitril for 10 min. This step was repeated twice and the supernatants were pooled with those from the previous step then dried in the centrivap concentrator. The resulting pellets were put in 20% formic acid solution and desalted using millipore C18 columns.
Analyses were performed using an UltraFlex I MALDI TOF mass spectrometer (Bruker Daltonics, Bremen, Germany) in the reflectron mode with a 26 kV accelerating voltage and a 50 ns delayed extraction. Mass spectra were acquired manually or automatically using the AutoXecute™ module of Flexcontrol™ (Bruker Daltonics) (laser power ranged from 22–85%, 600 shots). Spectra were analyzed using FlexAnalysis™ software (Bruker Daltonics) and calibrated internally with the autoproteolysis peptides of trypsin (m/z 842,51; 1045,56; 2211,10). Peptides were selected in the mass range of 900–3,000 Da.
Peptide Mass Fingerprint identification of proteins was performed by searching against the Mus_musculus entries of either SwissProt and/or TrEMBL databases (www.expasy.ch) and by using the Mascot v2.2 algorithm (www.matrixscience.com) with trypsin enzyme specificity and one trypsin missed cleavage. Carbamidomethyl was set as fixed cystein modification and oxidation was set as variable methionine modification for searches. A mass tolerance of 50 ppm was allowed for identification. Matching peptides with one missed cleavage were considered as pertinent when there were two consecutives basic residues or when arginine and lysine residues were in an acidic context. Mascot scores higher than 60 were considered as significant (p < 0.05) for SwissProt and TrEMBL databases interrogation. These MALDI-TOF analyses were performed on the “Plate-forme de Protéomique Fonctionnelle” (CNRS/INSERM, Montpellier, France).
Expasy proteomics server (www.expasy.org), online Mendelian Inheritance in Man, OMIM™, McKusick-Nathans Institute for Genetic Medicine, Johns Hopkins (Baltimore, MD) and National Center for Biotechnology Information, National Library of Medicine (Bethesda, MD), 2000 (www.ncbi.nlm.nih.gov/omim/) and the Gene Ontology database (www.geneontology.org) using the fatigo tool (www.fatigo.org) were also used to complete information.
Western blot analysis.
Sample migration and transfer on a PVDF membrane were performed as recommended by the supplier using the NuPAGE system (including buffers and Bis/Tris 12% gels) from Invitrogen. PVDF membranes were blocked for 1 h in 5% (w/v) nonfat dry milk in PBS-T (1% Tween-20 in PBS) and incubated for 90 min with 1/500 dilution of the following primary antibodies: PDI (SPA 891D, Stressgen), Tubulin beta (T8660, Eurogentec) and Vimentin (clone V9, Dako). After several washes in PBS-T, the secondary antibody anti-mouse IgG coupled to HRP from Jackson was incubated for 90 min (1/25,000) and after washes, the signal was revealed using enhanced chemiluminescent (ECL) (SuperSignal West Pico, Ref: 34078) from Pierce using a camera system (ChemiDoc XRS from Bio-Rad).
Conclusion
Both increased expression of PrPC and conversion into PrPSc in N2a cells induced significant and partially opposite changes in the proteome. Among the 65 identified proteins, many chaperones, enzymes of the energetic pathway, as well as cytoskeleton proteins had their expression modified. These results were mostly coherent with previous findings and knowledge of prion biology. Finally, we observed a very interesting opposite expression variation trend of proteins upon PrPC expression and conversion suggesting that the infection rendered PrPC less available for molecular partners and/or to fulfill its normal cellular function.
Acknowledgements
This work is supported by grants from the “GIS Infections à Prion”, the EU Commission program NEUROPRION (FOOD-CT-2004-506579), the DEFRA project SE2002 and the CNRS. We thank Jacques Grassi and colleagues (CEA Saclay, France) for providing anti-prion antibodies and Patrick Jouin and colleagues from the “Plate-forme de Protéomique Fonctionnelle, IFR3, CNRS-UMR 5203, INSERM-U661” for MALDI-TOF proteomic analysis.
Abbreviation
- PrPC
cellular prion protein
- PrPSc
scrapie prion protein
- N2a
neuroblastoma 2A cell line
- TSE
transmissible spongiform encephalopathy
- iN2a
inducible N2a
- 2DE
two-dimensional gel electrophoresis
Footnotes
Previously published online: www.landesbioscience.com/journals/prion/article/13435
Supplementary Material
References
- 1.Aguzzi A, Glatzel M, Montrasio F, Prinz M, Heppner FL. Interventional strategies against prion diseases. Nat Rev Neurosci. 2001;2:745–749. doi: 10.1038/35094590. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner SB, Scott MR, DeArmond SJ, Cohen FE. Prion protein biology. Cell. 1998;93:337–348. doi: 10.1016/s0092-8674(00)81163-0. [DOI] [PubMed] [Google Scholar]
- 3.Linden R, Martins VR, Prado MA, Cammarota M, Izquierdo I, Brentani RR. Physiology of the prion protein. Physiol Rev. 2008;88:673–728. doi: 10.1152/physrev.00007.2007. [DOI] [PubMed] [Google Scholar]
- 4.Weissmann C, Flechsig E. PrP knock-out and PrP transgenic mice in prion research. Br Med Bull. 2003;66:43–60. doi: 10.1093/bmb/66.1.43. [DOI] [PubMed] [Google Scholar]
- 5.Biasini E, Massignan T, Fioriti L, Rossi V, Dossena S, Salmona M, et al. Analysis of the cerebellar proteome in a transgenic mouse model of inherited prion disease reveals preclinical alteration of calcineurin activity. Proteomics. 2006;6:2823–2834. doi: 10.1002/pmic.200500620. [DOI] [PubMed] [Google Scholar]
- 6.Chich JF, Schaeffer B, Bouin AP, Mouthon F, Labas V, Larramendy C, et al. Prion infection-impaired functional blocks identified by proteomics enlighten the targets and the curing pathways of an anti-prion drug. Biochim Biophys Acta. 2007;1774:154–167. doi: 10.1016/j.bbapap.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 7.Ramljak S, Asif AR, Armstrong VW, Wrede A, Groschup MH, Buschmann A, et al. Physiological role of the cellular prion protein (PrPC): protein profiling study in two cell culture systems. J Proteome Res. 2008;7:2681–2695. doi: 10.1021/pr7007187. [DOI] [PubMed] [Google Scholar]
- 8.Massignan T, Biasini E, Lauranzano E, Veglianese P, Pignataro M, Fioriti L, et al. Mutant prion protein expression is associated with an alteration of the Rab GDP dissociation inhibitor alpha (GDI)/Rab11 pathway. Mol Cell Proteomics. 2010;9:611–622. doi: 10.1074/mcp.M900271-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fasano C, Campana V, Griffiths B, Kelly G, Schiavo G, Zurzolo C. Gene expression profile of quinacrine-cured prion-infected mouse neuronal cells. J Neurochem. 2008;105:239–250. doi: 10.1111/j.1471-4159.2007.05140.x. [DOI] [PubMed] [Google Scholar]
- 10.Greenwood AD, Horsch M, Stengel A, Vorberg I, Lutzny G, Maas E, et al. Cell line dependent RNA expression profiles of prion-infected mouse neuronal cells. J Mol Biol. 2005;349:487–500. doi: 10.1016/j.jmb.2005.03.076. [DOI] [PubMed] [Google Scholar]
- 11.Satoh J, Kuroda Y, Katamine S. Gene expression profile in prion protein-deficient fibroblasts in culture. Am J Pathol. 2000;157:59–68. doi: 10.1016/s0002-9440(10)64517-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Satoh J, Yamamura T. Gene expression profile following stable expression of the cellular prion protein. Cell Mol Neurobiol. 2004;24:793–814. doi: 10.1007/s10571-004-6920-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Windl O, Lorenz H, Behrens C, Romer A, Kretzschmar HA. Construction and characterization of murine neuroblastoma cell clones allowing inducible and high expression of the prion protein. J Gen Virol. 1999;80:15–21. doi: 10.1099/0022-1317-80-1-15. [DOI] [PubMed] [Google Scholar]
- 14.Nishida N, Harris DA, Vilette D, Laude H, Frobert Y, Grassi J, et al. Successful transmission of three mouse-adapted scrapie strains to murine neuroblastoma cell lines overexpressing wild-type mouse prion protein. J Virol. 2000;74:320–325. doi: 10.1128/jvi.74.1.320-325.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roche S, Delorme B, Oostendorp RA, Barbet R, Caton D, Noel D, et al. Comparative proteomic analysis of human mesenchymal and embryonic stem cells: towards the definition of a mesenchymal stem cell proteomic signature. Proteomics. 2009;9:223–232. doi: 10.1002/pmic.200800035. [DOI] [PubMed] [Google Scholar]
- 16.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 17.Kim S, Lee YZ, Kim YS, Bahk YY. A Proteomic approach for protein-profiling the oncogenic ras induced transformation (H-, K- and N-Ras) in NIH/3T3 mouse embryonic fibroblasts. Proteomics. 2008;8:3082–3093. doi: 10.1002/pmic.200800106. [DOI] [PubMed] [Google Scholar]
- 18.Forloni G, Iussich S, Awan T, Colombo L, Angeretti N, Girola L, et al. Tetracyclines affect prion infectivity. Proc Natl Acad Sci USA. 2002;99:10849–10854. doi: 10.1073/pnas.162195499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petrak J, Ivanek R, Toman O, Cmejla R, Cmejlova J, Vyoral D, et al. Deja vu in proteomics. A hit parade of repeatedly identified differentially expressed proteins. Proteomics. 2008;8:1744–1749. doi: 10.1002/pmic.200700919. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Hanash S. Electrospray mass spectrometry for quantitative plasma proteome analysis. Methods Mol Biol. 2009;564:227–242. doi: 10.1007/978-1-60761-157-8_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crecelius AC, Helmstetter D, Strangmann J, Mitteregger G, Frohlich T, Arnold GJ, et al. The brain proteome profile is highly conserved between Prnp−/− and Prnp+/+ mice. Neuroreport. 2008;19:1027–1031. doi: 10.1097/WNR.0b013e3283046157. [DOI] [PubMed] [Google Scholar]
- 22.Moore RA, Timmes A, Wilmarth PA, Priola SA. Comparative profiling of highly enriched 22L and Chandler mouse scrapie prion protein preparations. Proteomics. 2010;10:2858–2869. doi: 10.1002/pmic.201000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brechlin P, Jahn O, Steinacker P, Cepek L, Kratzin H, Lehnert S, et al. Cerebrospinal fluid-optimized two-dimensional difference gel electrophoresis (2-D DIGE) facilitates the differential diagnosis of Creutzfeldt-Jakob disease. Proteomics. 2008;8:4357–4366. doi: 10.1002/pmic.200800375. [DOI] [PubMed] [Google Scholar]
- 24.Huzarewich RL, Siemens CG, Booth SA. Application of “omics” to prion biomarker discovery. J Biomed Biotechnol. 2010;2010:613504. doi: 10.1155/2010/613504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strom A, Diecke S, Hunsmann G, Stuke AW. Identification of prion protein binding proteins by combined use of far-western immunoblotting, two dimensional gel electrophoresis and mass spectrometry. Proteomics. 2006;6:26–34. doi: 10.1002/pmic.200500066. [DOI] [PubMed] [Google Scholar]
- 26.Petrakis S, Sklaviadis T. Identification of proteins with high affinity for refolded and native PrPC. Proteomics. 2006;6:6476–6484. doi: 10.1002/pmic.200600103. [DOI] [PubMed] [Google Scholar]
- 27.Giorgi A, Di Francesco L, Principe S, Mignogna G, Sennels L, Mancone C, et al. Proteomic profiling of PrP27–30-enriched preparations extracted from the brain of hamsters with experimental scrapie. Proteomics. 2009;9:3802–3814. doi: 10.1002/pmic.200900085. [DOI] [PubMed] [Google Scholar]
- 28.Julius C, Hutter G, Wagner U, Seeger H, Kana V, Kranich J, et al. Transcriptional stability of cultured cells upon prion infection. J Mol Biol. 2008;375:1222–1233. doi: 10.1016/j.jmb.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 29.Tang Y, Xiang W, Hawkins SA, Kretzschmar HA, Windl O. Transcriptional changes in the brain of cattle orally infected with BSE precede detection of infectivity. J Virol. 2009;83:9464–9473. doi: 10.1128/JVI.00352-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tatzelt J, Zuo J, Voellmy R, Scott M, Hartl U, Prusiner SB, et al. Scrapie prions selectively modify the stress response in neuroblastoma cells. Proc Natl Acad Sci USA. 1995;92:2944–2948. doi: 10.1073/pnas.92.7.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.True HL. The battle of the fold: chaperones take on prions. Trends Genet. 2006;22:110–117. doi: 10.1016/j.tig.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 32.Hetz C, Russelakis-Carneiro M, Walchli S, Carboni S, Vial-Knecht E, Maundrell K, et al. The disulfide isomerase Grp58 is a protective factor against prion neurotoxicity. J Neurosci. 2005;25:2793–2802. doi: 10.1523/JNEUROSCI.4090-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wacker JL, Huang SY, Steele AD, Aron R, Lotz GP, Nguyen Q, et al. Loss of Hsp70 exacerbates pathogenesis but not levels of fibrillar aggregates in a mouse model of Huntington's disease. J Neurosci. 2009;29:9104–9114. doi: 10.1523/JNEUROSCI.2250-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kristiansen M, Messenger MJ, Klohn PC, Brandner S, Wadsworth JD, Collinge J, et al. Disease-related prion protein forms aggresomes in neuronal cells leading to caspase activation and apoptosis. J Biol Chem. 2005;280:38851–38861. doi: 10.1074/jbc.M506600200. [DOI] [PubMed] [Google Scholar]
- 35.Gauczynski S, Hundt C, Leucht C, Weiss S. Interaction of prion proteins with cell surface receptors, molecular chaperones and other molecules. Adv Protein Chem. 2001;57:229–272. doi: 10.1016/s0065-3233(01)57024-2. [DOI] [PubMed] [Google Scholar]
- 36.DebBurman SK, Raymond GJ, Caughey B, Lindquist S. Chaperone-supervised conversion of prion protein to its protease-resistant form. Proc Natl Acad Sci USA. 1997;94:13938–13943. doi: 10.1073/pnas.94.25.13938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edenhofer F, Rieger R, Famulok M, Wendler W, Weiss S, Winnacker EL. Prion protein PrPc interacts with molecular chaperones of the Hsp60 family. J Virol. 1996;70:4724–4728. doi: 10.1128/jvi.70.7.4724-4728.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milhavet O, Lehmann S. Oxidative stress and the prion protein in transmissible spongiform encephalopathies. Brain Res Brain Res Rev. 2002;38:328–339. doi: 10.1016/s0165-0173(01)00150-3. [DOI] [PubMed] [Google Scholar]
- 39.Sandhu N, Duus K, Jorgensen CS, Hansen PR, Bruun SW, Pedersen LO, et al. Peptide binding specificity of the chaperone calreticulin. Biochim Biophys Acta. 2007;1774:701–713. doi: 10.1016/j.bbapap.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 40.Cohen E, Taraboulos A. Scrapie-like prion protein accumulates in aggresomes of cyclosporin A- treated cells. EMBO J. 2003;22:404–417. doi: 10.1093/emboj/cdg045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma J, Lindquist S. Wild-type PrP and a mutant associated with prion disease are subject to retrograde transport and proteasome degradation. Proc Natl Acad Sci USA. 2001;98:14955–14960. doi: 10.1073/pnas.011578098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deriziotis P, Tabrizi SJ. Prions and the proteasome. Biochim Biophys Acta. 2008;1782:713–722. doi: 10.1016/j.bbadis.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 43.Nieznanski K, Nieznanska H, Skowronek KJ, Osiecka KM, Stepkowski D. Direct interaction between prion protein and tubulin. Biochem Biophys Res Commun. 2005;334:403–411. doi: 10.1016/j.bbrc.2005.06.092. [DOI] [PubMed] [Google Scholar]
- 44.Ganusova EE, Ozolins LN, Bhagat S, Newnam GP, Wegrzyn RD, Sherman MY, et al. Modulation of prion formation, aggregation and toxicity by the actin cytoskeleton in yeast. Mol Cell Biol. 2006;26:617–629. doi: 10.1128/MCB.26.2.617-629.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin SF, Herva ME, Espinosa JC, Parra B, Castilla J, Brun A, et al. Cell expression of a four extra octarepeat mutated PrPC modifies cell structure and cell cycle regulation. FEBS Lett. 2006;580:4097–4104. doi: 10.1016/j.febslet.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 46.Silva JL, Lima LM, Foguel D, Cordeiro Y. Intriguing nucleic-acid-binding features of mammalian prion protein. Trends Biochem Sci. 2008;33:132–140. doi: 10.1016/j.tibs.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 47.Rieger R, Edenhofer F, Lasmezas CI, Weiss S. The human 37 kDa laminin receptor precursor interacts with the prion protein in eukaryotic cells. Nat Med. 1997;3:1383–1388. doi: 10.1038/nm1297-1383. [DOI] [PubMed] [Google Scholar]
- 48.Gauczynski S, Peyrin JM, Haik S, Leucht C, Hundt C, Rieger R, et al. The 37 kDa/67 kDa laminin receptor acts as the cell-surface receptor for the cellular prion protein. EMBO J. 2001;20:5863–5875. doi: 10.1093/emboj/20.21.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lopes MH, Hajj GN, Muras AG, Mancini GL, Castro RM, Ribeiro KC, et al. Interaction of cellular prion and stress-inducible protein 1 promotes neuritogenesis and neuroprotection by distinct signaling pathways. J Neurosci. 2005;25:11330–11339. doi: 10.1523/JNEUROSCI.2313-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.