Abstract
M. tuberculosis ArfA (Rv0899) is a membrane protein encoded by an operon that is required for supporting bacterial growth in acidic environments. Its C-terminal domain (C domain) shares significant sequence homology with the OmpA-like family of peptidoglycan-binding domains, suggesting that its physiological function in acid stress protection may be related to its interaction with the mycobacterial cell wall. Previously, we showed that ArfA forms three independently structured modules and we reported the structure of its central domain (B domain). Here we describe the high-resolution structure and dynamics of the C domain, we identify ArfA as a peptidoglycan-binding protein, and elucidate the molecular basis for its specific recognition of diaminopimelate (DAP) type peptidoglycan. The C domain of ArfA adopts the characteristic fold of the OmpA-like family. It exhibits pH-dependent conformational dynamics (with significant hereogeneity at neutral pH and a more ordered structure at acidic pH), which could be related to its acid-stress response. The C domain associates tightly with polymeric peptidoglycan isolated from M. tuberculosis and also associates with a soluble peptide intermediate of peptidoglycan biosynthesis. This enabled us to characterize the peptidoglycan binding site where five highly conserved ArfA residues, including two key arginines, establish the specificity for DAP- but not Lys-type peptidoglycan. ArfA is the first peptidoglycan-binding protein to be identified in M. tuberculosis. Its functions in acid stress protection and peptidoglycan binding suggest a link between the acid stress response and the physico-chemical properties of the mycobacterial cell wall.
Keywords: structure, NMR, virulence, membrane, periplasm
INTRODUCTION
Mycobacterium tuberculosis, the causative agent of tuberculosis, infects approximately one third of the world’s population and kills two million people each year. Its high resistance to many drugs is partly due to the nature of its cell envelope, a highly complex structure composed of lipids, polysaccharides, and proteins, that protects the organism from external stress and harmful compounds 1; 2. The mycobacterial envelope consists of an inner lipid bilayer plasma membrane and a cell wall, formed by a peptidoglycan-arabinogalactan polymeric network occupying the periplasmic space and an outer lipid membrane enriched in mycolic acids covalently linked to the arabinogalactan layer 3; 4; it is further enclosed by an exterior capsular layer of glycans, lipids and proteins 5. Most antibiotics developed for M. tuberculosis target the cell envelope, therefore identifying its components and understanding how they interact to provide a mechanically strong, highly impermeable barrier, that also maintains communication with the outside world, is important for identifying new drug targets 1; 6.
Rv0899 (also known as OmpATb) has been proposed to function as an outer membrane porin that protects M. tuberculosis against acid stress, based on its sequence homology with E. coli OmpA, which forms a membrane-spanning β-barrel in the bacterial outer membrane 7; 8; 9; 10. However, the three-dimensional structure of Rv0899 does not contain a transmembrane β-barrel and is not compatible with porin function 11; 12. Indeed, recent studies showed that Rv0899 is not a porin 13. Rather, the rv0899 gene is part of an operon, including rv0900 and rv0901, also encoding membrane proteins 13; 14, that is required for fast ammonia secretion, rapid pH neutralization and growth of M. tuberculosis in acidic environments 13. Genes with sequences and genetic context similar to rv0899, rv0900 and rv0901, are widespread in bacteria that specialize in nitrogen fixation or metabolism, adaptation to nutrient poor environments, and/or establishing symbiosis with the host organism 15, further supporting their function in M. tuberculosis ammonia secretion.
In light of these findings, we propose to name the rv0899-rv0901 operon arf (ammonia release facilitator)* and its three genes, arfA, arfB, and arfC. In mycobacteria, the arf operon is found exclusively in organisms associated with tuberculosis (M. tuberculosis, M. bovis) and other diseases (M. marinum, M. ulcerans, M. kansasii), suggesting that it may have a role in pathogenicity and be an attractive candidate for the development of chemotherapeutic agents. However, the mechanism whereby ArfA (Rv0899), ArfB, (Rv0900) and ArfC (Rv0901) contribute to physiological function is not known.
Previously, we showed that ArfA forms three independently structured domains and reported the high-resolution structure of its central domain 12. The N-terminus (M domain; residues 1–80) includes a membrane-anchoring sequence of 20 hydrophobic amino acids (residues 28–50) that is required for membrane translocation but is not cleaved 10. The central region (B domain; residues 81–195) contains two consecutive repeats of the BON (Bacterial OsmY and Nodulation) domain (pfam04972), a conserved, putative lipid binding sequence that is found in some bacterial osmotic shock protection proteins and nodulation specificity proteins 16. Finally, the C-terminal region (C domain; residues 196–326) shares significant sequence homology with the OmpA-like superfamily (pfam00691) of peptidoglycan-binding proteins, including the C-terminus of the outer membrane protein OmpA from E. coli. OmpA-like domains are ubiquitous in Gram negative bacteria and also found in some Gram positive bacteria 17, however ArfA appears to be the only OmpA-like protein of M. tuberculosis.
Owing to its homology with OmpA, most functional studies have focused on establishing the activity of ArfA as a porin, while its potential interaction with the peptidoglycan layer has not received attention. However, the physiological functions of ArfA in facilitating bacterial growth at acidic pH may well be related to its interactions with the cell wall. Indeed, the cell envelope plays an important role in bacterial adaptation and survival, and recent studies of M. tuberculosis acid stress response genes indicate that the same mechanisms involved in acid resistance are also involved in cell wall functions and provide protection against other forms of extracellular stress (reviewed by Ehrt and coworkers 18).
The peptidoglycan layer is a complex polymer of cross-linked sugar and polypeptide units, that provides both the strength and plasticity required to maintain cell shape, withstand the high osmotic pressure of the protoplast, and enable bacterial growth 1; 2; 19; 20. It also regulates molecular diffusion across the cell, participates in the regulation of cell division, and provides an anchoring scaffold for cell-envelope lipids, proteins and polysaccharides. In this study, we identify ArfA as the first known peptidoglycan-binding protein of M. tuberculosis, and describe both the high-resolution solution structure and dynamics of its C domain (ArfA-c) as well as the binding site responsible for the specific recognition of the peptide stem of M. tuberculosis peptidoglycan. The function of ArfA in acid adaption may be related to its peptidoglycan-binding ability, conferring structural strength to the bacterial cell envelope under acid or other stress conditions.
RESULTS AND DISCUSSION
Structure of the C-terminal domain of ArfA
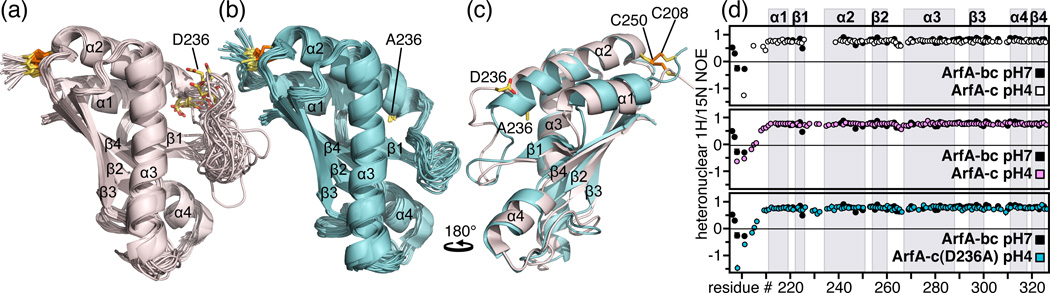
The C domain of wild-type ArfA folds into four β-strands and four α-helices, arranged in the topological order αβαβαβαβ (Fig. 1a). Three parallel (β1, β2, β3) and one antiparallel (β4) β-strands form a four-stranded β-sheet (β1–β4-β2–β3) that packs against three α-helices (α1, α2, α3), while a fourth helix (α4) extends from the N-terminus of β4. A disulfide bond between C208 and C250 connects the N-terminus of α1 to the C-terminus of α2. All ArfA orthologs from mycobacteria, as well as from Kribbella and α-proteobacteria, have Cys residues at similar positions, suggesting that the disulfide bond is conserved in all their structures 15.
Figure 1. Structures and backbone dynamics of wild-type ArfA-c and ArfA-c(D236A).
(a, b) Backbone representations of the 20 lowest energy structures of (a) wild-type ArfA-c and (b) ArfA-c(D236A). (c) Superimposed lowest energy structures of ArfA-c (pink) and ArfA-c(D236A) (cyan). The structures were determined at pH7. The average pairwise RMSDs for the structured core of the protein (residues 207–326) are reported in Table S1. (d) Heteronuclear 1H/15N NOEs reflecting backbone dynamics of: ArfA-bc at pH7 (black), ArfA-c at pH7 (white), ArfA-c at pH4 (pink), and ArfA-c(D236A) at pH7 (cyan). For wild-type ArfA-c, peaks from residues in the β1-α2 loop could not be observed at pH7. Residues in this loop gave weak, broad peaks at pH4, but gave peaks of normal intensity in the D236A mutant.
The disulfide bond stabilizes the structure (see below) but is not required to maintain the overall fold, and the protein can be reversibly reduced and oxidized by adding reducing DTT (dithiotheritol) or oxidizing GSSG (glutathione disulfide). Indeed, the 1H/15N NMR spectra obtained after treatment with DTT or GSSG show only minor changes in peaks from residues located near C208 and C250, while peaks from the rest of the protein are not affected (Fig. S1a)§. The structure is held together by a network of hydrophobic contacts among side chains in α1, α2 and β4 (L211, I215, V243, L247, Ile323 and V325) and is further stabilized by a hydrogen bond between the backbone amide of V325 and the side chain carbonyl of Q212.
The absence of NMR peaks from residues G226 to E238 in the loop linking β1 to α2 suggests that the loop undergoes conformational exchange between two or more states on an intermediate timescale relative to NMR, and precluded complete structure determination. Although several of these peaks manifest upon acidification (Fig. S1b), they remain significantly broadened, and no 1H/1H NOEs can be observed to connect residues G226 to E238 with other polypeptide sites, resulting in significant conformational heterogeneity in the β1-α2 loop. This is in contrast to a recent structural study, which also noted the pH dependence of these peaks but, nevertheless, reported a hybrid pH7.2 / pH3.5 solution structure similar to ours, albeit with a fully ordered loop 11.
Upon closer examination of wild-type ArfA we noted that a negatively charged residue (D236) at the start of helix α2 clashes with several hydrophobic residues in the β1-α2 loop (L232), in α2 (L240, A244), in α3 (V281, L285), and in β1 (I223, F225). Indeed, when we mutated D236 to Ala, all of the β1-α2 loop peaks were present at neutral pH (Fig. S2a), and several NOEs connecting the loop to α3 and to the β-sheet could be measured, enabling us to determine a high resolution structure that is very well defined over the entire length of the protein sequence (Fig. 1b). In the structure of the D236A mutant, A236 packs comfortably against L232, I233, L240 and V281, confirming that the negative charge of D236 disrupts this hydrophobic cluster in the wild-type protein at neutral pH, leading to conformational heterogeneity in the β1-α2 loop. Furthermore, the N-terminus of α2 in the mutant is longer by one turn and the β1-α2 loop forms a flap, hinged at the C-terminus of β1 and the N-terminus of α2, that folds over the β-sheet and closes at α3 (Figs. 1b, 1c).
pH-Dependent Conformational Disorder in the β1-α2 Loop
To determine whether the β1-α2 loop adopts a similar, albeit dynamically disordered, conformation in wild-type ArfA at pH7, we mutated L232, in the middle of the loop, to Gly, and examined its effects on other structured regions of the protein by mapping the peak changes in the 1H/15N NMR spectrum of ArfA(L232G). The L232G mutation in the loop causes several significant (≥ 0.03 ppm) changes in peaks from residues in α3 and in the β-sheet (Fig. S2b). In ArfA(D236A), the loop packs against the β-sheet and the presence of strong NOEs establishes close contacts between the methyl protons of L232 and the amide protons of A224 and V281. Since the peaks from A224 and V281 are significantly perturbed by the L232G mutation, we conclude that the loop of wild-type ArfA-c is not dissociated from the rest of the polypeptide at pH7 but maintains close contact with the globular structure, notwithstanding a significant degree of conformational exchange.
To examine backbone dynamics, we measured heteronuclear 1H/15N NOEs for both wild-type and D236A mutant ArfA-c, at neutral and acidic pH (Fig. 1d). The dynamics of isolated ArfA-c are similar to those observed in the connected B and C domains 12. For both wild-type protein and D236A mutant, negative NOEs, reflecting rapid (ps-ns) backbone motions, are observed at the N-terminus corresponding to the flexible inter-domain linker. Beyond the N-terminus, both wild-type ArfA-c at acidic pH, and D236A mutant at neutral pH, exhibit very similar, positive values (~0.8) of the 1H/15N NOE, reflecting a rigid backbone for the entire protein sequence. Slightly, but visibly lower 1H/15N NOEs for residues in the loops (β1-α2; β2-α3; β3-α4) indicate the presence of some restricted motions and may reflect the importance of the loops in protein/ligand interactions.
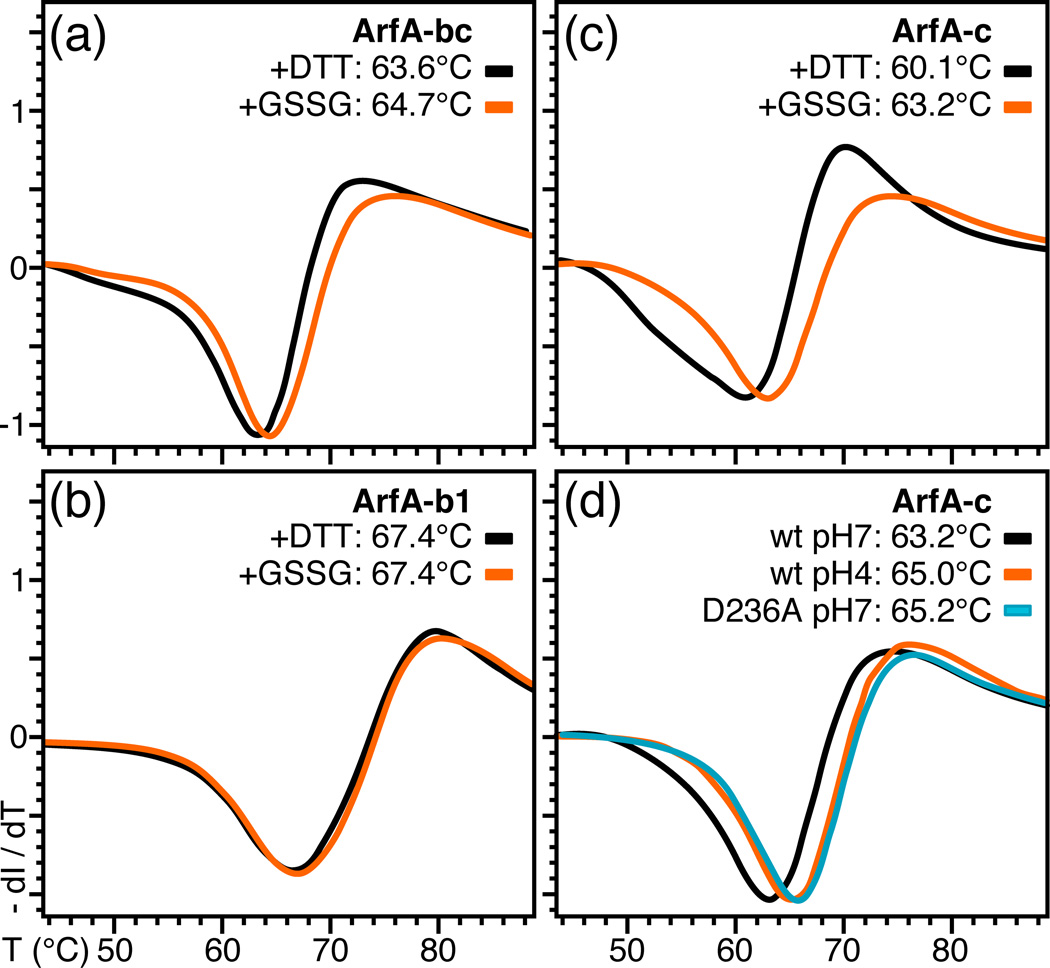
To further dissect the stabilizing effects of acidic pH and of the C208–C250 disulfide bond, we performed DSF (differential scanning fluorimetry) studies comparing the thermal stability of wild-type and mutant ArfA, under various conditions of pH and oxidation state. After treatment with DTT, ArfA-c undergoes thermal unfolding at 60.1°C, while disulfide bond formation by oxidation with GSSG has a significant stabilizing effect and increases the melting temperature to 63.2°C (Fig. 2c). A similar effect is observed for the joint B and C domains (ArfA-bc) where the disulfide bond provides about 1°C of stabilization (Fig. 2a), while the melting temperature of ArfA-b, which does not contain Cys, is unaffected by oxidation state (Fig. 2b). Notably, ArfA-b has a significantly higher melting temperature (67.4°C) than either the C or combined BC domains, reflecting its overall greater content of secondary structure relative to loops 12.
Figure 2. Differential scanning fluorimetry traces showing the effect of the disulfide bond, pH, and D236A mutation on the thermal stability of ArfA.
(a–c) Protein was treated with DTT or GSSG to break or form the C208–C250 disulfide bond. ArfA-b1 encompasses residues 73 to 220. (d) Traces were obtained for wild-type ArfA-c at pH7 (black) or pH4 (orange) and for ArfA-c(D236A) at pH7 (cyan).
Lowering the pH from 7 to 4 further increases the temperature of thermal unfolding of ArfA-c by 2°C (from 63.2°C to 65.0°C; Fig. 2d). The effect of pH is mirrored by the D236A mutation, which increases the unfolding temperature of ArfA-c at pH7 by a similar amount (from 63.2°C to 65.2°C; Fig. 2d). We conclude that the stabilizing effect of acidic pH on the conformational exchange of the β1-α2 loop of ArfA-c is primarily through neutralization of the negative charge of D236, which enables α2 and the loop to pack against α3 and the β-sheet. Whether this pH-dependent conformational heterogeneity correlates with the pH-dependent physiological function of ArfA is not known.
ArfA binds M. tuberculosis peptidoglycan
While the structure of M. tuberculosis ArfA-b was unprecedented in the database, ArfA-c shares the same βαβαβαβ core structure and significant amino acid conservation with other domains of the OmpA-like family (Fig. S3), some of which have been shown to bind peptidoglycan 21; 22; 23. To determine whether ArfA also shares this function we tested its ability to associate with intact peptidoglycan isolated from M. tuberculosis.
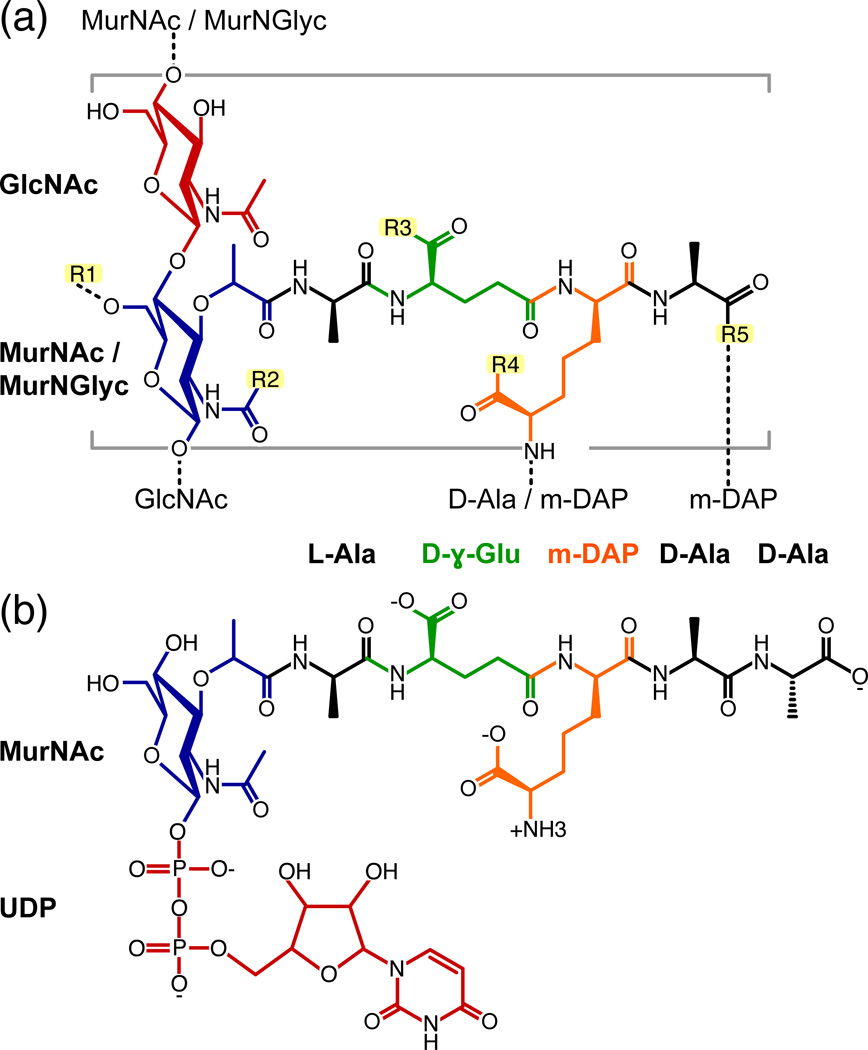
The peptidoglycan of M. tuberculosis (Fig. 3a) is composed of linear chains of GlcNAc (N-acetyl-D-glucosamine) and Mur (muramic acid) that can be both N-acetylated (MurNAc) and N-acylated with glycolic acid (MurNGlyc) 1; 2; 19; 20. The sugars are substituted with a heavily cross-linked peptide, containing L-Ala, D-γ-Glu, m-DAP (meso-diaminopimelate), and D-Ala, similar to the composition found Gram-negative bacteria (e.g. E. coli) and Gram-positive Bacillus, but distinct from the peptidoglycan that is more typically found in Gram-positive bacteria, where m-DAP is usually replaced by L-Lys. OmpA-like proteins have been identified exclusively in organisms with DAP-type peptidoglycan 17, suggesting specificity for DAP in their structures.
Figure 3. Chemical structures of (a) M. tuberculosis peptidoglycan and (b) the peptidoglycan biosynthesis intermediate, UMDP.
Dashed lines designate cross links to arabinogalactan or to the sugar and peptide stems of neighboring monomeric units. GlcNAc: N-acetyl-glucosamine; MurNGlyc: N-glycolyl-muramic acid; MurNAc: N-acetyl-muramic acid; m-DAP: meso-diaminopimelic acid. UDP: uridine-5'-diphosphate; R1: H or the linker unit of arabinogalactan; R2: H, CH3 (N-acetyl) or CH2OH (N-glycolyl); R3, R4: OH, NH2 or OCH3; R5: OH or amidated cross link to m-DAP.
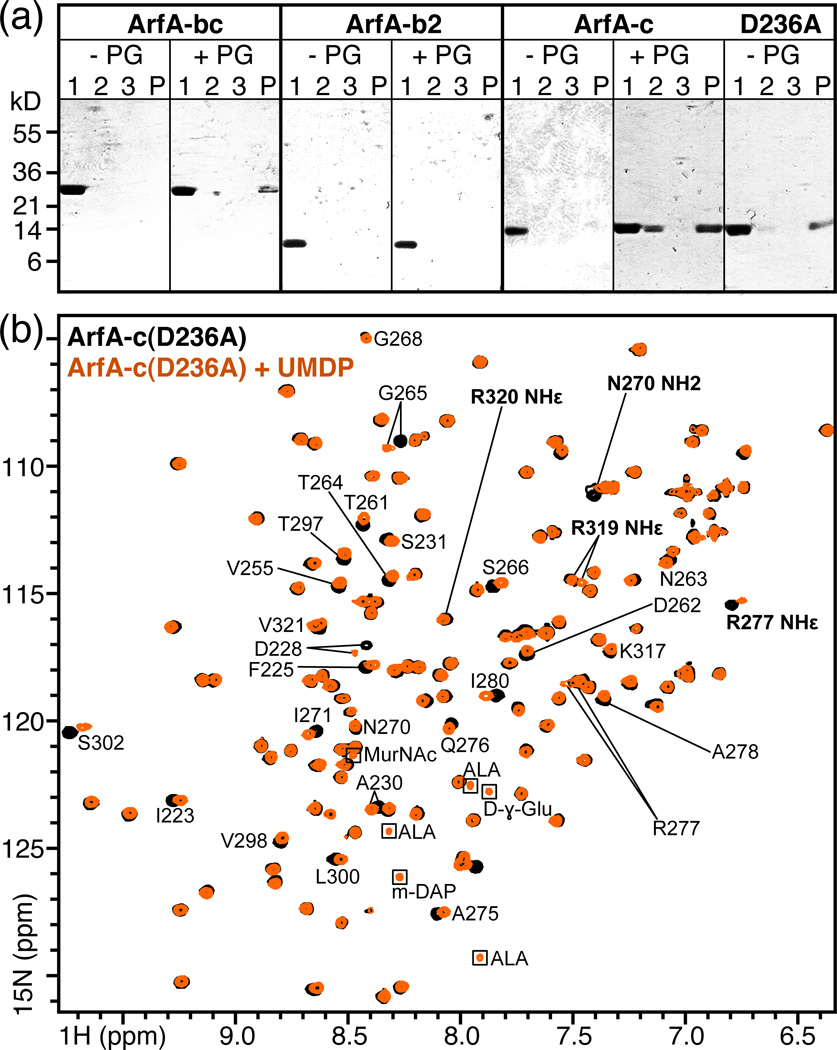
Incubation of soluble ArfA polypeptides with insoluble polymeric M. tuberculosis peptidoglycan caused a significant amount of ArfA-bc, ArfA-c and ArfA-c(D236A) to separate with the insoluble fraction after centrifugation, while all three proteins remained in the supernatant in the absence of peptidoglycan (Fig. 4a). In contrast, ArfA-b2 (residues 73–197) remained primarily in the supernatant both in the presence and absence of peptidoglycan. We conclude that ArfA binds polymeric M. tuberculosis peptidoglycan via its C domain. No appreciable difference (within the limits of this pull-down assay) was detected between the peptidoglycan-binding abilities of wild-type ArfA-c and ArfA-c(D236A), indicating that the mutant is active in this respect and that conformational heterogeneity in the β1-α2 loop does not affect peptidoglycan-binding.
Figure 4. Association of ArfA-c with peptidoglycan.
(a) Soluble, purified ArfA polypeptides: ArfA-bc, ArfA-b2 (residues 73–197), ArfA-c and ArfA-c(D236A) were mixed with purified peptidoglycan from M. tuberculosis (+PG). As negative controls, proteins were incubated without peptidoglycan (−PG). After 2 hr incubation, the supernatant (Lane 1) and insoluble fraction were separated by centrifugation, the pellet was washed with buffer and again separated from the supernatant (Lane 2) by centrifugation. After a second wash and centrifugation step to generate a third supernatant (Lane 3) and final pellet (Lane P), all fractions were analyzed by SDS-PAGE and visualized with Coomassie blue. ArfA-bc, ArfA-c and ArfA-c(D236A) bind peptidoglycan and separate with it in the insoluble fraction, while ArfA-b2 does not bind peptidoglycan and remains in solution. For each ArfA polypeptide, the result of one out of two or more independent experiments is shown. (b) NMR 1H/15N HSQC spectra of ArfA-c(D236A) obtained at pH7, 25°C, with (orange) or without (black) ~20 molar equivalents of UMDP. Examples of peaks sensitive (e.g. D228) or insensitive (e.g. G268) to addition of UMDP are labeled. Peaks from side chains are labeled in bold. Peaks from the natural abundance peptide are enclosed in boxes.
Peptidoglycan Recognition by ArfA
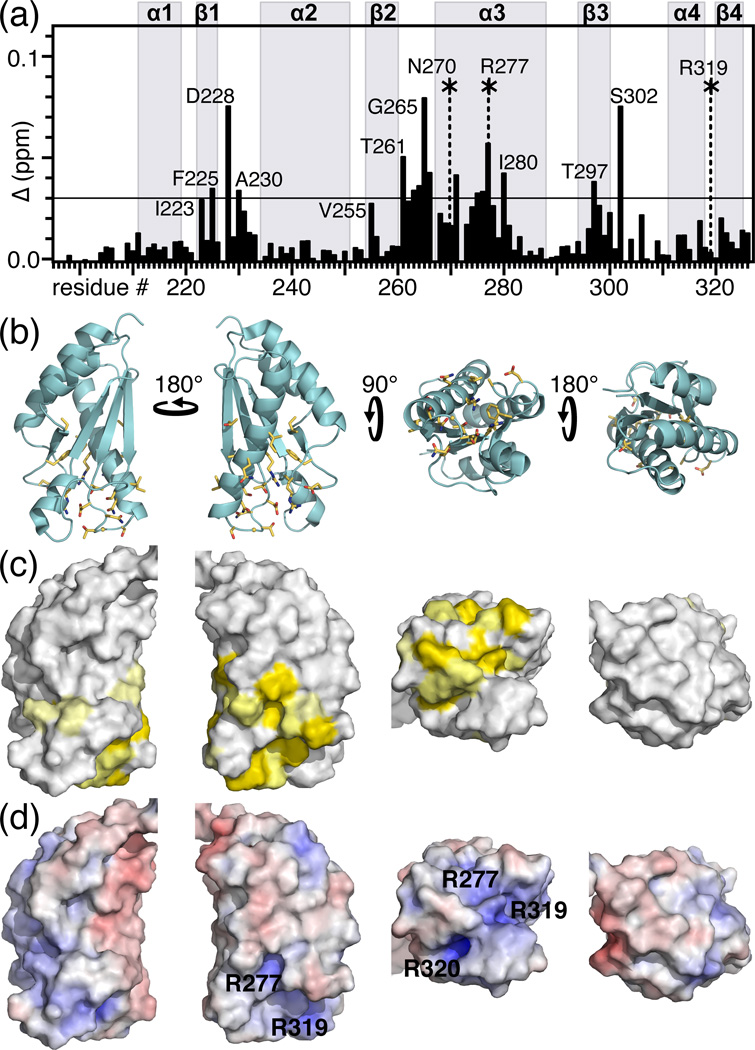
To further characterize the peptidoglycan binding site of ArfA we examined its interaction with a soluble intermediate of peptidoglycan biosynthesis, UMDP (also known as Park's nucleotide, Fig. 3b), similar to the peptide found in association with H. influenzae Pal 21. Addition of UMDP to 15N-labeled ArfA-c(D236A) produced several peak frequency and intensity changes in the 1H/15N NMR spectrum of the protein (Figs. 4b). The changes are specific as they map to a surface cavity formed by residues in the β1-α2 and β2-α3 loops, the N-terminus of helix α3 and the C-terminus of helix α4 (Fig. 5), similar to the binding site identified for Pal 21. Similar effects of UMDP were observed for wild-type ArfA-c at both pH7 and pH4. This is consistent with the observation that both wild-type and D236A mutant ArfA-c bind intact peptidoglycan (Fig. 4a), indicating that the acid-dependent structural heterogeneity of the β1-α2 loop does not affect peptidoglycan binding by ArfA. The most striking peak perturbations are observed for residues with high sequence conservation in the OmpA-like family (D228, A230, T261, D262, N263, T264, G265, N270, R277, I280, T297, S302, R319). Notably, mutations of residues corresponding to T261, D262, R277, and R319 all render the OmpA-like protein MotB nonfunctional 24, underscoring their importance.
Figure 5. Peptidoglycan binding site of M. tuberculosis ArfA-c(D236A).
(a) Chemical shift changes in the spectrum of ArfA-c(D236A) induced by UMDP. The total chemical shift change (Δ) for each peak was calculated by adding the changes in 1H (ΔH) and 15N (ΔN) using the equation Δ = [(ΔH)2 + (ΔN/5)2]1/2. Asterisks indicate side chain NH peaks that undergo significant chemical shift or intensity changes. Values of Δ≥0.03 ppm (horizontal line) were mapped on the structure. (b, c) Molecular backbone and surface representations of ArfA-c(D236A) showing residues with Δ≥0.03 ppm (yellow) in the peptidoglycan binding site of ArfA. (d) Molecular surface representations colored by electrostatic potential, with isocontours shown at +8kBT (blue) and −8kBT (red), where kB is the Boltzmann constant, and T is the temperature.
The interaction is specific for m-DAP at position 3 of the peptide stem, since addition of the L-Lys analog UMKP, more typical of Gram-positive bacteria but absent from M. tuberculosis, had no effect on the spectrum of ArfA-c (Fig. S4b), and neither did addition of the dipeptide GMAG, lacking m-DAP (S4c). Thus, the presence of MurNAc alone is insufficient for binding while m-DAP is required, although we cannot exclude the possibility that MurNGlyc, which is abundant in M. tuberculosis, may be important. Finally, addition of UMDP had no effect on the spectrum of ArfA-b (Fig. S4d), confirming that ArfA binds peptidoglycan solely through its C domain.
Unlike the Pal study, where NOEs connecting the peptide to the protein enabled the structure of the complex to be determined 21, the interaction of ArfA-c with UMDP did not yield any intermolecular NOEs. However, a number of key experimental observations, described below, enabled us to construct a structural model that sheds light on peptidoglycan recognition by ArfA (Fig. 6).
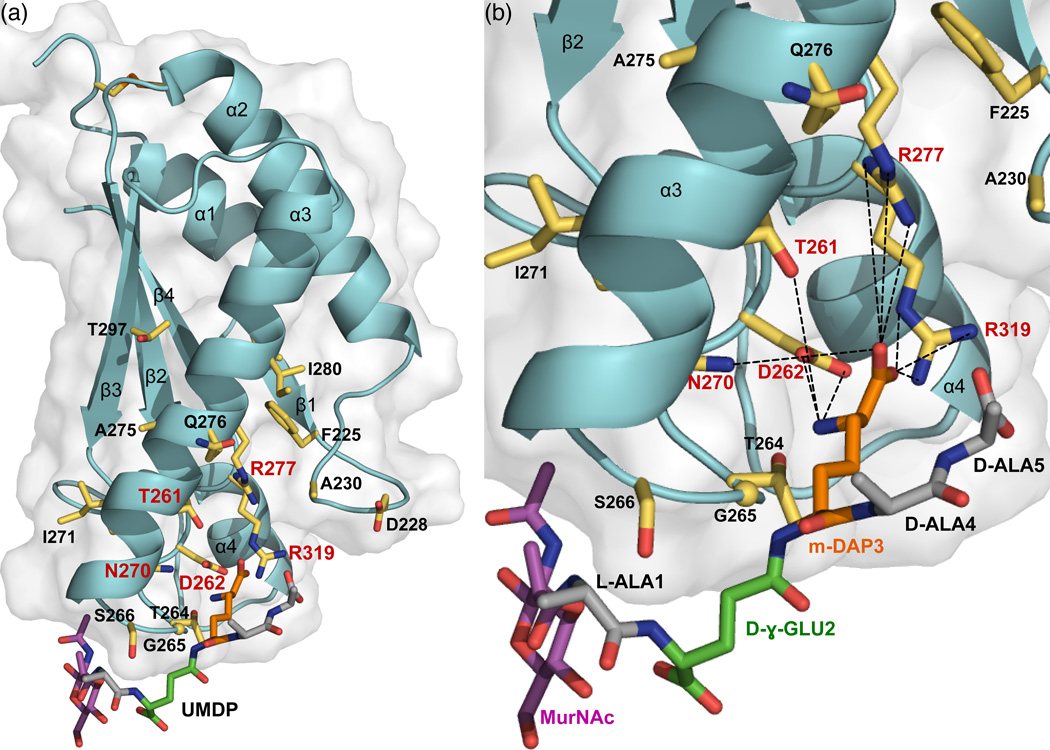
Figure 6. Peptidoglycan recognition by ArfA-c.
(a) Full and (b) close-up views of the structure of ArfA-c(D236A) associated with a structural model of the peptidoglycan peptide MDP (MurNAc–L-Ala–D-γ-Glu–m-DAP–D-Ala–D-Ala). Residues with NMR peaks most affected by UMDP (Δ≥0.03 ppm) are shown as yellow sticks. Residues that form the m-DAP recognition site are labeled in red. MDP is color coded with MurNAc in magenta, Ala in gray, D-γ-Glu in green, and m-DAP in orange.
Although the affinity of ArfA-c for polymeric M. tuberculosis peptidoglycan is strong (enough to precipitate the protein), its binding affinity for monomeric UMDP is weak, and a dose-response experiment, performed by mapping NMR peak changes upon titration of UMDP into the protein, yielded a Kd in the range of 1 mM. This high-specificity, low-affinity interaction suggests that electrostatics (hydrogen bonds, charge-charge contacts) play an important role in mediating UMDP recognition. A prominent surface electrostatics feature of ArfA-c is the presence of two juxtaposed clusters of positive charge at or near the peptidoglycan binding site, formed by R277, R319 and R320 (Fig. 5d). In the free protein, these arginines participate in electrostatic interactions within a hydrophobic environment, reflected in the presence of side chain guanidinium NHε peaks, with characteristic downfield chemical shift (e.g. 8.06 ppm for R320), in the NMR spectrum. Indeed, the R320 guanidinium group has polar interactions with E322 and N304, and its hydrocarbon side chain is stabilized by hydrophobic contacts with the side chains of I306 and Y260, all established by NOE connections. The R277 side chain is stabilized by hydrophobic contacts with the aromatic ring of F225, with L271 and I280, all residues with very high OmpA-like conservation. The heteronuclear 1H/15N NOEs measured for the guanidinium NHε peaks of these arginines (0.58 for R277; 0.62 for R319; 0.86 for R320) reflect a rather rigid side chain for R320, but significantly more flexibility for R277 and R319.
The R277 and R319 side chains are positioned to facilitate the formation of a hydrogen bond network that includes their guanidinium groups as well as the hydroxyl of T261, the carboxylate of D262, and the carboxamide of N270. These residues all have NMR peaks that exhibit significant changes in the presence of UMDP: the side chain NHε peaks from R277 and R319, but not R320, undergo substantial shifts, and the side chain NH2 peak of N270 is broadened beyond detection, suggesting a direct interaction with the peptide (Fig. 5a). Furthermore, the backbone amide peaks of N270, T261 and D262 are all affected by UMDP. The five T261, D262, N270, R277 and R319 side chains all converge at the peptidoglycan binding site where they provide an effective recognition site for m-DAP (Fig. 6), possibly facilitated by the side chain flexibility of R277 and R319. Peptidoglycan recognition proteins of the immune system rely on a similar, buried electrostatic interaction between conserved Arg and the m-DAP carboxyl group to select for Dap-type peptidoglycan 25, suggesting that this may be a conserved interaction in a variety of protein folds.
To confirm the identity of the peptidoglycan recognition site, we generated a mutant of ArfA-c(D236A) where R277 was replaced by Glu, and tested its ability to bind UMDP by NMR spectroscopy. Although the R277E mutation alone caused several peak changes compared to the spectrum of ArfA-c(D236A), the perturbations were largely localized to sites near residue 277, the peak movements could be easily tracked, and the overall appearance of the spectrum was maintained, indicating that the overall fold of the protein is preserved. Most importantly, no changes were observed in the spectrum of the R277E mutant upon addition of UMDP (Fig. S5), indicating that the protein has lost its ability to bind peptidoglycan. Thus, we conclude that R277 is a key residue for the recognition of peptidoglycan by ArfA.
Our model indicates that the side chain of m-DAP is stabilized by charge-charge interactions between its electronegative carbonyl group with the R277 and R319 guanidinium groups and with the N270 carboxamide, and between its electropositive amino group with the D262 carboxyl and the T261 hydroxyl. These residues are strictly conserved in the OmpA-like family and our data show that they constitute the specific recognition site for DAP-type peptidoglycan (Fig. 6b). For example, in the structure of H. influenzae Pal, D71 (corresponding to ArfA D262) forms an electrostatic interaction with the side chain NH3 group of m-DAP. Our model also indicates that UMDP could further interact with ArfA-c through contacts of its γ-Glu3 and Ala2 backbone amides with the side chain hydroxyl of S266, of its MurNac OH4 with the backbone carbonyl of G265, of its MurNac O3 with the amide proton of E267, and of its MurNac NH2 with the E267 carboxyl.
Saturation transfer difference NMR experiments show that the amide proton of S302, located in the β3-α4 loop, is in some proximity to the methyl protons of the UDMP MurNAc group (Fig. S6). The structural position of S302 is identical to that of S208 in H. pylori MotB, where it appears to stabilize the interaction with a single MurNAc sugar molecule, by forming hydrogen bonds between its side chain hydroxyl group and the O1 atom of MurNAc and between its backbone amide NH and the acetamido oxygen atom (O7) of MurNAc 22. S302 may assist the interaction of ArfA with peptidoglycan in a similarly way. A Ser or other hydroxyl-bearing residue at this position is conserved in many of the OmpA-like sequences with the exception of YiaD, where it is replaced by Pro. In all sequences, this residue is preceded by a perfectly conserved Gly, which could confer structural flexibility in the loop to further assist interaction with the peptidoglycan.
The highly specific albeit weak interaction of ArfA-c with UMDP is reminiscent of the immune response peptidoglycan recognition proteins, which achieve selective recognition of peptidoglycan type by specifically discriminating for either DAP or Lys in the third position of the peptide stem, and for the degree and type of peptide stem cross-linking 26. We note that UMDP is chemically and structurally different from polymeric M. tuberculosis peptidoglycan, whose affinity for ArfA is sufficiently strong to precipitate the protein (Fig. 4a). The interaction of ArfA with polymeric peptidoglycan may be strengthened by the highly cross-linked peptidoglycan structure, which could provide additional binding sites for ArfA (e.g. at peptide stem cross-links), cause an effective increase in protein concentration by sterically constraining its diffusion, and reduce the flexibility and conformational space of the peptidoglycan peptide stem. These effects could be particularly important for M. tuberculosis where 75% of peptidoglycan is cross-linked, compared to the 20 to 50% that is cross-linked in E. coli (reviewed by Rubin, Jackson, Brennan and coworkers 1; 2). The interaction may also be strengthened by the formation of ArfA dimers in vivo, as proposed for Pal 27. For example, MotB, and to a weaker extent RmpM, crystallize as dimers 22; 23, and the B domain of ArfA has been proposed to oligomerize 11. Pal and ArfA are monomeric in solution, both in their free and UMDP-bound states. However, the molecular surface of ArfA C domain displays a distinct electrostatic polarity (electropositive on the side where α3 packs against β3 and negative on the opposite side where α1 packs against α2), and it is tempting to speculate that this charge juxtaposition may assist the assembly of stacked, neighboring C domains on the peptidoglycan polymer as a means of strengthening the cell wall.
CONCLUDING REMARKS
ArfA is the first peptidoglycan-binding protein to be identified in M. tuberculosis. Its other established roles in fast ammonia secretion and rapid pH neutralization 13 are consistent with the presence of ArfA-like proteins in bacteria active in nitrogen fixation and/or nitrogen metabolism 15. Interestingly, M. tuberculosis is known to generate substantial quantities of ammonia, which inhibits phagosome fusion in infected macrophages 28. Taken together, the peptidoglycan binding activity of ArfA and its function in supporting growth of M. tuberculosis at low pH indicate a possible link between the physico-chemical properties of the mycobacterial cell envelope and the acid stress response, as previously noted for other stress response proteins 18.
ArfA could contribute structural strength to the bacterial cell wall during acid or other stress conditions. It could achieve this directly, by providing a physical link between the peptidoglycan layer and the bacterial outer or inner membrane, or it could help guide nascent peptidoglycan peptide stems in their structurally correct position for polymerization, to ensure solid construction of the cell wall. This could be important for maintenance and determination of the mycobacterial cell shape, as suggested for other cytoskeletal bacterial proteins 1. Additional studies examining the relationship between the peptidogycan binding and pH neutralization activities of ArfA will shed light on the acid stress response mechanism of M. tuberculosis.
ArfA is reported to reside in the outer membrane of M. tuberculosis and to be exposed to the cell surface 5; 7; 10; 13; 29; 30. However, the finding that ArfA binds M. tuberculosis peptidoglycan through its C-terminus appears to contradict its surface accessibility, since the only possible architectures allowing for an interaction with peptidoglycan would involve insertion of the ArfA N-terminal transmembrane helix across either the outer or inner membranes, placing the B and C domains into the periplasmic space. It is well known that both periplasmic and inner membrane proteins can fractionate with the bacterial cell wall due to their significant interactions with the outer membrane or with the peptidoglycan layer 31; 32. The interaction of ArfA with peptidoglycan would certainly strengthen its association with the cell wall and potentially cause it to separate with it in fractionation experiments, or to be shed with it under conditions of bacterial stress. On the other hand, ArfA could occupy distinct areas of the cell, as recently reported for the E. coli major lipoprotein Lpp, which appears to localize to different subcellular compartments depending on whether its C-terminus is covalently linked to peptidoglycan or not 33. The structure and peptidoglycan recognition of Lpp are completely different from those of ArfA. However, in the case of ArfA, the existence of two protein populations, one located in the periplasm and the other on the mycobacterial cell surface, could reconcile the surface accessibility of ArfA with its peptidoglycan binding activity. Understanding the cellular localization of ArfA, its relationships with the other arf-encoded proteins, ArfB and ArfC, and the function of its pH-dependent dynamics, will shed light on the physiological function of the arf operon and could advance the development of new drugs for the treatment of tuberculosis.
MATERIALS AND METHODS
Protein preparation
Protein preparation, and characterization were as described previously 12. All proteins were monomeric by size exclusion chromatography or analytical ultracentrifugation. The ArfA polypeptides used in this study were: ArfA-bc (residues 73–326); ArfA-b1 (residues 73–220); ArfA-b2 (residues 73–197); and ArfA-c (residues 196–326). Mutations (D236A; L232G; R277E) were introduced by site-directed mutagenesis using the QuikChange Lightning kit (Stratagene). Purified protein was stored at 4°C as a 40 µM solution in buffer A (25 mM Na/PO4, pH 6.8, 1 mM EDTA). Formation of the C208–C250 disulfide bond was induced by adding 50 mM oxidized glutathione (GSSG) to the protein in 50 mM Tris/HCl, 150 mM NaCl, pH 8.0, and incubating it for 1 h at room temperature. Reduction of the disulfide bond was obtained by adding 100 mM DTT to the protein in degassed buffer and incubating overnight at 4°C. For NMR experiments the protein solution was concentrated by ultrafiltration to 0.2–1.0 mM.
Differential Scanning Fluorimetry (DSF)
Solutions containing 15 µM protein and Sypro orange dye (500 fold dilution of the 5000X stock solution supplied by Invitrogen) were mixed to a final volume of 15 µL, loaded into a 96-well plate (Roche), and sealed with sealing foil to prevent evaporation and contamination. Protein melting experiments were performed using a LightCycler 480 instrument (Roche). The temperature was increased from 20°C to 95°C, with a linear gradient of 0.03°C per second. Fluorescence intensity was measured at every 0.3°C increment using excitation and emission wavelengths of 465 nm and 580 nm, respectively. The data were processed using the Roche Protein Melt Tool, using the first derivative method to smooth the raw data by converting each data point into a moving average of itself and 19 additional adjacent points, for an average of 20 points, as described by the manufacturer. Fluorescence intensity, as well as the change in intensity divided by the change in temperature, were plotted as a function of temperature.
Peptidoglycan binding assays
Purified peptidoglycan from Mycobacterium tuberculosis, strain H37Rv, was obtained through the NIH Biodefense and Emerging Infections Research Resources Repository supported by the NIH, NIAID (BEI #: NR-14853). Purified protein (1 µL of 40 µM stock) was added to peptidoglycan (40 µg), resuspended in 10 µM of buffer A, and incubated for 2 hours at room temperature, as described 34. The mixture was centrifuged in a microfuge (14,000 rpm, 15 min) to separate the soluble and insoluble fractions. After removing the supernatant, the pellet was washed with fresh buffer and the mixture was centrifuged again. This procedure was repeated twice. The resulting supernatant and pellet fractions were analyzed with SDS-PAGE with coomassie blue staining.
NMR chemical shift mapping experiments were performed using water-soluble peptidoglycan peptides. The peptides UMDP (uridine-5'-diphosphate–MurNAc–L-Ala–D-γ-Glu–m-DAP–D-Ala– D-Ala) and UMKP (uridine-5'-diphosphate–MurNAc–L-Ala–D-γ-Glu–L-Lys–D-Ala–D-Ala) were obtained from the UK Bacterial Cell Wall Biosynthesis Network, at Warwick University. The GMAG (GlcNAc–MurNAc–L-Ala–D-γ-Glu-NH2) dipeptide was obtained from Calbiochem.
Since recombinant Pal, expressed with an E. coli pET vector, co-purified with endogenous E. coli UMDP 21, we tested whether this might also be the case for recombinant ArfA-c. To this end, we denatured purified ArfA-c in 8 M urea and subjected it to size exclusion chromatography in 8 M urea to remove any potentially associated molecules. We then refolded the eluted protein in urea-free buffer and examined its 1H/15N HSQC spectrum. As previously noted for ArfA-b 12, the native fold of ArfA-c could be readily restored after removal of denaturant by dialysis. No difference was detected between the spectra obtained before or after denaturation and refolding, indicating that ArfA-c, unlike Pal, does not co-purify with E. coli UMDP.
NMR Spectroscopy
NMR experiments were performed at 25°C or 40°C on a Bruker AVANCE 600 MHz spectrometer or a Bruker AVANCE 800 MHz spectrometer, each equipped with a cryoprobe. The pulse sequences are described in detail in the literature 35; 36; 37; 38; 39; 40; 41; 42; 43; 44. The NMR data were processed and analyzed using NMRPipe 45, NMRViewJ 46 and Sparky 47.
Resonance assignments for UMDP were obtained by means of 2D TOCSY with a 60 ms mixing time, and by comparison with previous published assignments for UMKP 48. The backbone 1H, 15N, and 13C chemical shifts were resolved using two-dimensional 1H/15N- and 1H/13C-HSQC experiments, and assigned using three-dimensional 1H/15N/13C HNCA and HNCACB experiment. Assignments of the side chain resonances were obtained with three-dimensional HCC(CO)NH, CC(CO)NH, HCCH-COSY/TOCSY and 13C-NOESY-HSQC experiments. Homonuclear 1H NOEs were measured from three-dimensional 15N- and 13C-edited NOESY-HSQC spectra, obtained with mixing times of 100 ms. 1H/15N Residual dipolar couplings (RDCs) were measured using in-phase/anti-phase 15N/1H HSQC experiments 41 with samples in magnetically oriented Pf1 phage (10 mg/ml). Heteronuclear 1H/15N NOE measurements were made using difference experiments with and without 5 s of saturation of the 1H resonances between scans 42. For saturation transfer difference experiments 43; 44, selective radiofrequency saturation was obtained by applying a train of frequency-selective 50 ms Gaussian pulses, separated by a 1 ms delay. Two experiments, each with on- or off-resonance saturation, were performed in an interleaved fashion, and the difference spectrum was obtained by subtracting the resulting free induction decays. Water suppression was achieved by excitation sculpting 49.
Structure calculations
Structure calculations were performed using XPLOR-NIH version 2.27 50. Structures were visualized with PyMol 51. Electrostatic potentials were calculated in units of kT/e (k=Boltzmann constant; T=absolute temperature; e=proton charge) using the programs PDB2PQR 52 and APBS 53. A total of 400 structures were generated from extended random coil coordinates, as described previously 12. Structures were calculated using restraints measured at pH7 and 25°C, including: backbone ϕ and ψ dihedral angles derived from analysis of HA, N, C, CA, and CB chemical shifts with the database program TALOS+ 54; distances derived from NOEs; and orientations derived from 1H/15N RDCs, analyzed with the XPLOR-NIH script CalcTensor and with REDCAT 55. The 40 structures with lowest energies were subjected to a final refinement in water using the explicit solvent refinement module available in XPLOR-NIH 56. Structure calculations for wild-type ArfA-c included two single long distance restraints (4.2 ± 2.4 Å) between the methyl protons of L232 and the amide protons of A224 and V281 to loosely constrain the β1-α2 loop, as suggested by the results of the L232G mutation.
The 20 lowest energy structures were evaluated with PROCHECK 57. They have no dihedral angle violations greater than 5°, no bond angle violations greater than 5°, no NOE distance restraint violations greater than 0.5 Å, and no bond distance violations greater than 0.05 Å. Only 1.2% (wild type) or 0.9% (D236A mutant) of the residues had ϕ/ψ dihedral angles in generously allowed and/or disallowed regions of the Ramachandran plot; these residues are located in the termini outside the structured core of the protein or in connecting loops. Statistics for the resulting family of 20 structures are reported in Table S1.
To model the complex of ArfA-c with MDP (MurNAc–L-Ala–D-γ-Glu–m-DAP–D-Ala–D-Ala), we first generated an extended MDP peptide and subjected it to Cartesian coordinate Powell minimization, then we optimized the interface between ArfA-c(D236A) and MDP using semi-rigid body simulated annealing 58. In this step, the MDP peptide was flexible, ArfA-c side chain atoms (from CB onward) with NMR peaks affected by addition of UMDP were allowed to rotate and translate, and all other ArfA-c atoms were held fixed. The starting position of MDP relative to ArfA-c was guided by the chemical shift mapping and STD results of ArfA-c(D236A) with UMDP, and by the published complex of Pal with UMDP 21. The energy function included distance restraints derived from chemical shift mapping and STD (Table S2), as well as terms for covalent geometry, Van der Waals contacts, and the torsion angle database Rama potential 59. The 10 lowest energy structures out of 100 calculated structures were selected and analyzed.
Sequence analysis
Amino acid sequences were aligned with ClustalW 60, and manually edited based on the structure using Jalview 61.
Accession numbers
Atomic coordinates, resonance assignments and NMR restraints have been deposited in the Protein Data Bank (PDB) for wild-type ArfA-c (PDB: 2LCA) and ArfA-c(D236A) (PDB: 2LBT). NMR assignments have been deposited in the Biological Magnetic Resonance Bank (BMRB: 17575).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Stanley Opella for providing purified Pf1 bacteriophage, Jasmina Rakic and Yuan Yang for their assistance with protein preparation, and Houhui Song for preparing the original wild-type ArfA expression plasmid. This research was supported by a grant from the National Institutes of Health (AI074805). It utilized the NIH-supported Biodefense and Emerging Infections Research Resources Repository (NIAID), and NIH-supported NMR Facilities at the Sanford Burnham Medical Research Institute (CA030199) and at the University of California San Diego (S10RR23773).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: ArfA, ammonia release facilitator; BON, Bacterial OsmY and Nodulation; DFS, Differential Scanning Fluorimetry; DTT, dithiotheritol; m-DAP, meso-diaminopimelate; GSSG, glutathione disulfide; GlcNAc, N-acetyl-D-glucosamine; Mur, muramic acid; MurNAc, N-acetylmuramic acid; MurNGlyc, N-glycolylmuramic acid; MDP, MurNAc–L-Ala–D-γ-Glu–m-DAP–D-Ala–D-Ala; UMDP, uridine-5'-diphosphate–MurNAc–L-Ala–D-γGlu–m-DAP–DAla–D-Ala; UMKP, uridine-5'-diphosphate–MurNAc–L-Ala–D-γ-Glu–L-Lys–D-Ala–D-Ala; GMAG, GlcNAc–MurNAc–L-Ala–D-γ-Glu-NH2
Supplemental information related to this article can be found online.
REFERENCES
- 1.Hett EC, Rubin EJ. Bacterial growth and cell division: a mycobacterial perspective. Microbiol Mol Biol Rev. 2008;72:126–156. doi: 10.1128/MMBR.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaur D, Guerin ME, Skovierova H, Brennan PJ, Jackson M. Chapter 2: Biogenesis of the cell wall and other glycoconjugates of Mycobacterium tuberculosis. Advances in Applied Microbiology. 2009;69:23–78. doi: 10.1016/S0065-2164(09)69002-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3963–3967. doi: 10.1073/pnas.0709530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zuber B, Chami M, Houssin C, Dubochet J, Griffiths G, Daffe M. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J Bacteriol. 2008;190:5672–5680. doi: 10.1128/JB.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sani M, Houben EN, Geurtsen J, Pierson J, de Punder K, van Zon M, Wever B, Piersma SR, Jimenez CR, Daffe M, Appelmelk BJ, Bitter W, van der Wel N, Peters PJ. Direct visualization by cryo-EM of the mycobacterial capsular layer: a labile structure containing ESX-1-secreted proteins. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000794. e1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen L, Thompson CJ. Foundations of antibiotic resistance in bacterial physiology: the mycobacterial paradigm. Trends Microbiol. 2006;14:304–312. doi: 10.1016/j.tim.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Senaratne RH, Mobasheri H, Papavinasasundaram KG, Jenner P, Lea EJ, Draper P. Expression of a gene for a porin-like protein of the OmpA family from Mycobacterium tuberculosis H37Rv. J Bacteriol. 1998;180:3541–3547. doi: 10.1128/jb.180.14.3541-3547.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raynaud C, Papavinasasundaram KG, Speight RA, Springer B, Sander P, Bottger EC, Colston MJ, Draper P. The functions of OmpATb, a pore-forming protein of Mycobacterium tuberculosis. Mol Microbiol. 2002;46:191–201. doi: 10.1046/j.1365-2958.2002.03152.x. [DOI] [PubMed] [Google Scholar]
- 9.Molle V, Saint N, Campagna S, Kremer L, Lea E, Draper P, Molle G. pH-dependent pore-forming activity of OmpATb from Mycobacterium tuberculosis and characterization of the channel by peptidic dissection. Mol Microbiol. 2006;61:826–837. doi: 10.1111/j.1365-2958.2006.05277.x. [DOI] [PubMed] [Google Scholar]
- 10.Alahari A, Saint N, Campagna S, Molle V, Molle G, Kremer L. The N-Terminal Domain of OmpATb Is Required for Membrane Translocation and Pore-Forming Activity in Mycobacteria. J Bacteriol. 2007;189:6351–6358. doi: 10.1128/JB.00509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y, Auguin D, Delbecq S, Dumas E, Molle G, Molle V, Roumestand C, Saint N. Structure of the Mycobacterium tuberculosis OmpATb protein: a model of an oligomeric channel in the mycobacterial cell wall. Proteins. 2011;79:645–661. doi: 10.1002/prot.22912. [DOI] [PubMed] [Google Scholar]
- 12.Teriete P, Yao Y, Kolodzik A, Yu J, Song H, Niederweis M, Marassi FM. Mycobacterium tuberculosis Rv0899 adopts a mixed alpha/beta-structure and does not form a transmembrane beta-barrel. Biochemistry. 2010;49:2768–2777. doi: 10.1021/bi100158s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song H, Huff J, Janik K, Walter K, Keller C, Ehlers S, Bossmann SH, Niederweis M. Expression of the ompATb operon accelerates ammonia secretion and adaptation of Mycobacterium tuberculosis to acidic environments. Mol Microbiol. 2011;80:900–918. doi: 10.1111/j.1365-2958.2011.07619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veyron-Churlet R, Brust B, Kremer L, Blanc-Potard AB. Expression of OmpATb is dependent on small membrane proteins in Mycobacterium bovis BCG. Tuberculosis (Edinb) 2011 doi: 10.1016/j.tube.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Marassi FM. Mycobacterium tuberculosis Rv0899 defines a family of membrane proteins widespread in nitrogen-fixing bacteria. Proteins. 2011;79:2946–2955. doi: 10.1002/prot.23151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeats C, Bateman A. The BON domain: a putative membrane-binding domain. Trends in Biochemical Sciences. 2003;28:352–355. doi: 10.1016/S0968-0004(03)00115-4. [DOI] [PubMed] [Google Scholar]
- 17.De Mot R, Vanderleyden J. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol Microbiol. 1994;12:333–334. doi: 10.1111/j.1365-2958.1994.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 18.Vandal OH, Nathan CF, Ehrt S. Acid resistance in Mycobacterium tuberculosis. Journal of Bacteriology. 2009;191:4714–4721. doi: 10.1128/JB.00305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brennan PJ, Nikaido H. The envelope of mycobacteria. Annu. Rev. Biochem. 1995;64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- 21.Parsons LM, Lin F, Orban J. Peptidoglycan recognition by Pal, an outer membrane lipoprotein. Biochemistry. 2006;45:2122–2128. doi: 10.1021/bi052227i. [DOI] [PubMed] [Google Scholar]
- 22.Roujeinikova A. Crystal structure of the cell wall anchor domain of MotB, a stator component of the bacterial flagellar motor: implications for peptidoglycan recognition. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10348–10353. doi: 10.1073/pnas.0803039105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grizot S, Buchanan SK. Structure of the OmpA-like domain of RmpM from Neisseria meningitidis. Molecular Microbiology. 2004;51:1027–1037. doi: 10.1111/j.1365-2958.2003.03903.x. [DOI] [PubMed] [Google Scholar]
- 24.Blair DF, Kim DY, Berg HC. Mutant MotB proteins in Escherichia coli. J Bacteriol. 1991;173:4049–4055. doi: 10.1128/jb.173.13.4049-4055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim JH, Kim MS, Kim HE, Yano T, Oshima Y, Aggarwal K, Goldman WE, Silverman N, Kurata S, Oh BH. Structural basis for preferential recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan recognition proteins. J Biol Chem. 2006;281:8286–8295. doi: 10.1074/jbc.M513030200. [DOI] [PubMed] [Google Scholar]
- 26.Swaminathan CP, Brown PH, Roychowdhury A, Wang Q, Guan R, Silverman N, Goldman WE, Boons GJ, Mariuzza RA. Dual strategies for peptidoglycan discrimination by peptidoglycan recognition proteins (PGRPs) Proc Natl Acad Sci U S A. 2006;103:684–689. doi: 10.1073/pnas.0507656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cascales E, Bernadac A, Gavioli M, Lazzaroni JC, Lloubes R. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J Bacteriol. 2002;184:754–759. doi: 10.1128/JB.184.3.754-759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordon AH, Hart PD, Young MR. Ammonia inhibits phagosome-lysosome fusion in macrophages. Nature. 1980;286:79–80. doi: 10.1038/286079a0. [DOI] [PubMed] [Google Scholar]
- 29.Rezwan M, Laneelle MA, Sander P, Daffe M. Breaking down the wall: fractionation of mycobacteria. Journal of Microbiological Methods. 2007;68:32–39. doi: 10.1016/j.mimet.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 30.Song H, Sandie R, Wang Y, Andrade-Navarro MA, Niederweis M. Identification of outer membrane proteins of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2008;88:526–544. doi: 10.1016/j.tube.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akins DR, Robinson E, Shevchenko D, Elkins C, Cox DL, Radolf JD. Tromp1, a putative rare outer membrane protein, is anchored by an uncleaved signal sequence to the Treponema pallidum cytoplasmic membrane. Journal of Bacteriology. 1997;179:5076–5086. doi: 10.1128/jb.179.16.5076-5086.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cowles CE, Li Y, Semmelhack MF, Cristea IM, Silhavy TJ. The free and bound forms of Lpp occupy distinct subcellular locations in Escherichia coli. Mol Microbiol. 2011;79:1168–1181. doi: 10.1111/j.1365-2958.2011.07539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Persson C, Oldenvi S, Steiner H. Peptidoglycan recognition protein LF: a negative regulator of Drosophila immunity. Insect Biochem Mol Biol. 2007;37:1309–1316. doi: 10.1016/j.ibmb.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Fesik SW, Zuiderweg ER. Heteronuclear three-dimensional NMR spectroscopy of isotopically labelled biological macromolecules. Q Rev Biophys. 1990;23:97–131. doi: 10.1017/s0033583500005515. [DOI] [PubMed] [Google Scholar]
- 36.Bax A, Grzesiek S. Methodological advances in protein NMR. Accounts of Chemical Research. 1993;26:131–138. [Google Scholar]
- 37.Cavanagh J, Fairbrother WJ, Palmer AG, Skelton NJ. Protein NMR spectroscopy : principles and practice. San Diego: Academic Press; 1996. [Google Scholar]
- 38.Clore GM, Gronenborn AM. NMR structure determination of proteins and protein complexes larger than 20 kDa. Current Opinion in Chemical Biology. 1998;2:564–570. doi: 10.1016/s1367-5931(98)80084-7. [DOI] [PubMed] [Google Scholar]
- 39.Ferentz AE, Wagner G. NMR spectroscopy: a multifaceted approach to macromolecular structure. Q Rev Biophys. 2000;33:29–65. doi: 10.1017/s0033583500003589. [DOI] [PubMed] [Google Scholar]
- 40.Kay LE. Nuclear magnetic resonance methods for high molecular weight proteins: a study involving a complex of maltose binding protein and beta-cyclodextrin. Methods in Enzymology. 2001;339:174–203. doi: 10.1016/s0076-6879(01)39314-x. [DOI] [PubMed] [Google Scholar]
- 41.Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J Magn Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 42.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J Biomol NMR. 1994;4:727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 43.Mayer M, Meyer B. Characterization of Ligand Binding by Saturation Transfer Difference NMR Spectroscopy. Angewandte Chemie International Edition. 1999;38:1784–1788. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 44.Mayer M, Meyer B. Group Epitope Mapping by Saturation Transfer Difference NMR To Identify Segments of a Ligand in Direct Contact with a Protein Receptor. Journal of the American Chemical Society. 2001;123:6108–6117. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 45.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 46.Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. Journal of Biomolcular NMR. 1995;5:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 47.Goddard TD, Kneller DG. SPARKY 3. San Francisco: University of California; 2004. [Google Scholar]
- 48.Kodani S, Ishida K, Murakami M. Occurrence and identification of UDP-N-acetylmuramyl-pentapeptide from the cyanobacterium Anabaena cylindrica. FEMS Microbiology Letters. 1999;176:321–325. doi: 10.1111/j.1574-6968.1999.tb13678.x. [DOI] [PubMed] [Google Scholar]
- 49.Stott K, Stonehouse J, Keeler J, Hwang TL, Shaka AJ. Excitation Sculpting in High-Resolution Nuclear Magnetic Resonance Spectroscopy: Application to Selective NOE Experiments. Journal of the American Chemical Society. 1995;117:4199–4200. [Google Scholar]
- 50.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. Journal of Magnetic Resonance. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 51.DeLano WL. PyMol. 2005 www.pymol.org. [Google Scholar]
- 52.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. Journal of Biomolecular NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valafar H, Prestegard JH. REDCAT: a residual dipolar coupling analysis tool. J Magn Reson. 2004;167:228–241. doi: 10.1016/j.jmr.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 56.Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 57.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283. [Google Scholar]
- 58.Clore GM. Accurate and rapid docking of protein-protein complexes on the basis of intermolecular nuclear overhauser enhancement data and dipolar couplings by rigid body minimization. Proc Natl Acad Sci U S A. 2000;97:9021–9025. doi: 10.1073/pnas.97.16.9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuszewski J, Gronenborn AM, Clore GM. Improvements and extensions in the conformational database potential for the refinement of NMR and X-ray structures of proteins and nucleic acids. J Magn Reson. 1997;125:171–177. doi: 10.1006/jmre.1997.1116. [DOI] [PubMed] [Google Scholar]
- 60.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.