Abstract
The pattern of T lymphocyte homing is hypothesized to be controlled by combnations of chemokine receptors and complementary chemokines. Here, we use numerical simulation to explore the relationship among chemokine potency and concentration, signal transduction, and adhesion. We have developed a form of Adhesive Dynamics – a mechanically accurate stochastic simulation of adhesion – that incorporates stochastic signal transduction using the next subvolume method. We show that using measurable parameter estimates derived from a variety of sources, including signaling measurements that allow us to train parameter values, we can readily simulate approximate time scales for T lymphocyte arrest. We find that adhesion correlates with total chemokine receptor occupancy, not the frequency of occupation, when multiple chemokine receptors feed through a single G-protein. A general strategy for selective T-lymphocyte recruitment appears to require low affinity chemokine receptors. For a single chemokine receptor, increases in multiple cross-reactive chemokines can lead to an overwhelming increase in adhesion. Overall, the methods presented here provide a predictive framework for understanding chemokine control of T-lymphocyte recruitment.
Keywords: T Lymphocyte, Chemokine, Adhesive Dynamics
INTRODUCTION
T lymphocyte arrest within the microvasculature is an essential process in the development of adaptive immunity that directs naïve, effector, and memory cell populations to secondary lymphoid organs and peripheral tissues [1,2]. Cell arrest is triggered by chemokine-mediated ‘inside-out’ signaling, which activates passive, low-affinity surface integrins to active, high-affinity conformations as lymphocytes tether and roll along post-capillary venules [3]. High affinity binding between β2, β1, and β7 integrins and their endothelial ligands directly facilitates shear-resistant cell adhesion and subsequent extravasation into surrounding tissues [4]. Affinity regulation of resting integrins has been linked to small GTPases including Rap1 and RhoA, whose activities are in turn regulated by signals originating from G-proteins associated with active chemokine receptors [5,6]. Experimental studies of lymphocyte recruitment have suggested that T cells integrate inside-out signals very efficiently, allowing rolling cells to arrest within a narrow window (< 500 μm) downstream of the initial chemokine stimulus [7].
It is now firmly established that the combinatorial expression of rolling, activation, and arrest receptors acts as a molecular ‘zip-code’ that addresses specific cell lineages to appropriate tissues expressing a complimentary combination of ligands [8]. In this way, T lymphocytes carry with them molecular instructions to traffic between specific anatomic compartments selectively, rather than indiscriminately. Growing evidence suggests that individual T cell clones are responsive to multiple chemokine stimuli, which confounds the traditional view of non-overlapping trafficking cues [9]. Unlike the surface receptors that mediate rolling and arrest, numerous lymphocyte chemokine receptors recognize multiple chemokine ligands which may be expressed concomitantly within target tissues [10,11]. Furthermore, the majority of T lymphocyte lineages express two or more chemokine receptors which independently recognize distinct sub-families of chemokines [12–14].
Several hypotheses for multiple chemokine interactions have been advanced, including redundancy, antagonism, and synergism [11,15]. In the simplest scenario, redundant chemokine interactions imply that T cells may not discriminate among ligands that are ‘promiscuous’ for a common chemokine receptor. Conversely, antagonistic or cooperative chemokine signals could provide molecular cross-talk that either limits or enhances the cellular response, respectively. While each of these hypotheses are rooted in elementary concepts of receptor biology, the relevance and theoretical origin for such regulation in the recruitment cascade remains poorly defined.
In this study, we address possible roles for chemokine networks using computational simulations that capture the dynamics of T lymphocyte arrest. The simulations herein are built upon Adhesive Dynamics (AD) [42, 43], a stochastic simulation of receptor mediated cell adhesion which calculated the outcome of surface encounter from a mechanically accurate mechanical energy balance. We have recently developed a suite of AD methods that incorporates signal transduction, and used these methods that integrate mechanics and signaling to calculate the dynamics of neutrophil adhesion [45, 46]. Here, we describe a mathematical model of T lymphocyte arrest which includes integrin regulation that has been trained upon experimental measurements of inside-out signaling dynamics and is capable of predicting characteristic metrics of T cell arrest obtained from in vitro flow assays. This framework allows us to identify how changes in molecular componentry bring about qualitative and quantitative changes in lymphocyte recruitment, and to define the physical determinants of chemokine potency. Together, our calculations uncover a role for the additive integration of multiple chemokine cues, which suggest a previously unrecognized mechanism for generating diversity in the T lymphocyte trafficking program.
RESULTS
In vitro Characterization of T Lymphocyte Arrest
To identify characteristic dynamics of T lymphocyte arrest, we performed preliminary experiments with Jurkat T cells, a lymphoma cell line widely used in studies of T cell signaling and adhesion [3–5]. Prior studies have shown that Jurkat cells concomitantly express high levels of chemokine receptor CXCR4 [16] and integrin αLβ2 [17] which recognize the chemokine CXCL12 and endothelial ICAM-1, respectively. While resting low-affinity αLβ2/ICAM-1 interactions support transient cell tethering under flow, inducible high-affinity αLβ2/ICAM-1 complexes mediate stable cell arrest [8]. To recapitulate lymphocyte activation and arrest in vitro, cells were perfused into parallel plate flow chambers coated with 5 μg/mL ICAM-1, and 0–4 μg/mL CXCL12 at a characteristic shear rate of 100 s−1. This minimal combination of adhesion ligand, chemokine, and fluid kinematics closely mimics the environment encountered by naïve and memory T lymphocytes entering post-capillary high endothelial venules (HEV) of secondary lymphoid tissues [1].
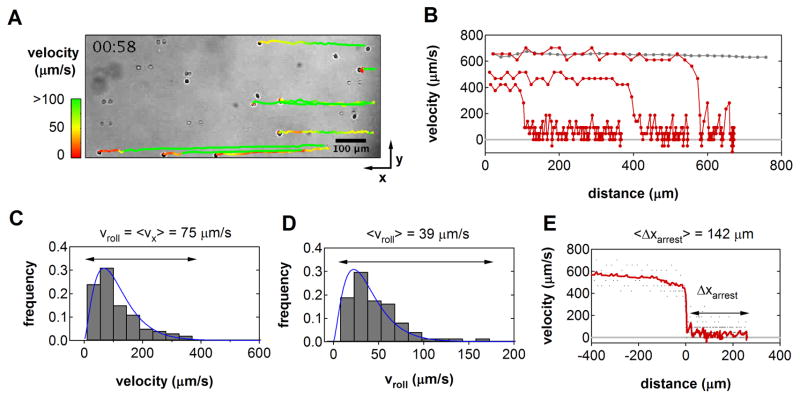
Cell motion was monitored by video microscopy and analyzed with particle tracking software to reconstruct precise trajectories during interactions with the substrate (Figure 1). Listed in Table 1, velocity and arrest statistics were tabulated for those cells that entered the field of view in a stream line just above the substrate, tethered, and subsequently transitioned from rolling to firm adhesion. Upon tethering, cells immediately exhibited a step-wise, stop-and-go motion characteristic of leukocyte rolling. Consistent with theory, the instantaneous translational velocities measured for single cells varied according to a gamma distribution during the rolling phase [18], with mean rolling velocities ranging from 13–180 μm/s (Fig. 1C–D). The relative frequency of average rolling velocities observed among all cells was approximately log-normally distributed about 39 μm/s, indicative of heterogeneity in the population-level response. After a variable rolling period, cells came to an abrupt arrest (Fig. 1E). The distance traveled between tethering and arrest, ranged between 28–320 μm, consistent with previous measurements of lymphocyte arrest in both in vitro flow assays and in vivo studies [19,20].
Figure 1.
In vitro Dynamics of T Lymphocyte Recruitment (A) Video micrograph of Jurkat T cells interacting with immobilized CXCL12/ICAM-1 in shear flow. Cells were perfused into parallel plate flow chambers at a shear rate of 100 s−1 (right to left). Trajectories for eight individual cells that ultimately arrested were reconstructed from video analysis. (B) Three characteristic velocity-distance profiles for cells that tethered, rolled, and arrested within the frame of view (red). The trajectory of a non-adherent cell is shown for comparison (gray). (C) Instantaneous velocity distribution for one individual cell prior to arrest. The macroscopic rolling velocity, vroll, is given by the mean of the distribution. (D) Population-level distribution of mean rolling velocities indicated significant heterogeneity among cells. In both (C) and (D), velocities varied according a gamma distribution (blue line), consistent with theory [18]. (E) Velocity-distance profiles from all cell trajectories were synchronized to the moment of tether formation to identify the range of arrest distances. The population average velocity as a function of distance is indicated (red).
A Kinetic Model for Chemokine-Triggered ‘Inside-Out’ Integrin Activation
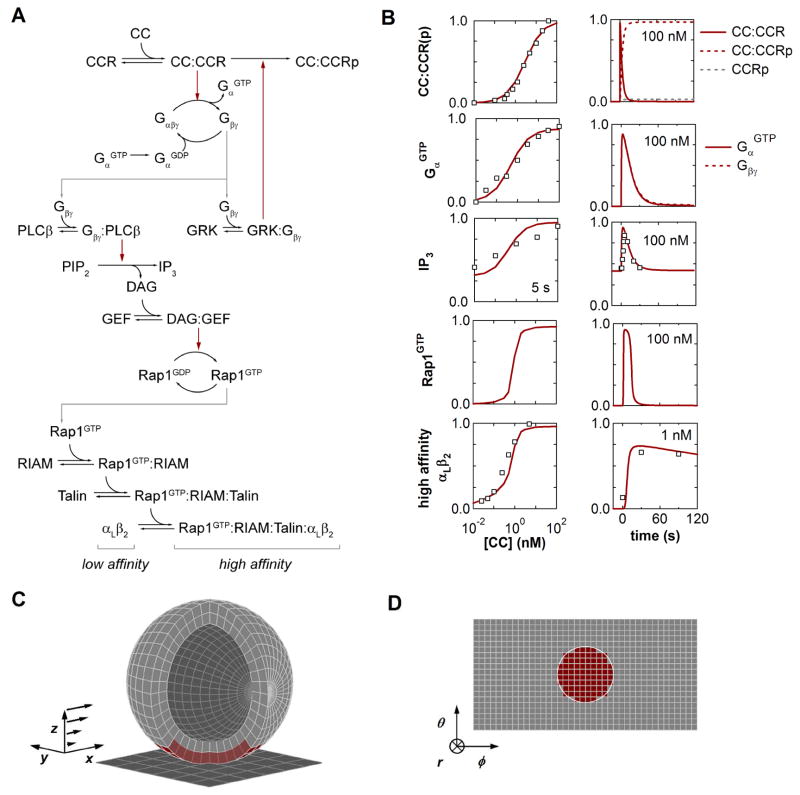
A mathematical model for chemokine-mediated signaling events implicated in the activation of leukocyte integrins was constructed according to current biochemical hypotheses (Figure 2). Elementary reaction dynamics were first modeled in a continuum representation by ordinary differential equations (ODEs) in order to identify characteristic reaction rate parameters that might be used in a stochastic, spatially resolved model of greater detail. Rate laws for constituent reactions between proteins/complexes were formulated according to standard mass-action formalisms describing bimolecular association and unimolecular dissociation (Table S1). Where appropriate, catalytic phosphorylation and guanine nucleotide exchange were modeled by Michaelis-Menten kinetics. Although this latter formalism invokes specific assumptions regarding the dynamics of reactive intermediates [21], a lumped kinetic representation seems most appropriate in consideration of the level of insight we wish to glean from simulated signal dynamics.
Figure 2.
Kinetic Model for Chemokine-Triggered Inside-Out Integrin Activation. (A) Inside-out reaction network linking chemokine recognition to integrin activation. Arrows indicate molecular association/dissociation (black) or catalytic phosphorylation/nucleotide transfer (red). (B) Experimental and simulated inside-out dynamics. The kinetic model (lines) was trained against a collection of experimental data (symbols) describing inside-out activation of αLβ2 integrins in human neutrophils responding to soluble CXCL8. (C) Model geometry for simulating T lymphocyte adhesion under flow. Membrane and cytosolic compartments were discretized into subvolumes based on a spherical lattice. (D). Projected cell surface from the reference frame of the cell. In (C) and (D), the extent of the cell-substrate contact zone is indicated by the subvolumes highlighted in red.
Biochemical reactions describing five cascading signaling modules were included in the kinetic model based on recent characterization of inside-out pathways in mammalian leukocytes [4]. The present model includes mechanisms representing: (i) ligation and phosphorylation of G-protein coupled chemokine receptors (CCR) [22,23]; (ii) collision-coupled G-protein activation and subsequent stimulation of phospholipase C beta (PLCβ) and G-protein coupled receptor kinase (GRK) activity by Gβγ subunits [24,25]; (iii) phosphatidylinositol biphosphate (PIP2) cleavage to diacylglycerol (DAG) and inositol (1,4,5) triphosphate (IP3) [26,27]; (iv) Rap1 GTPase nucleotide transfer by calcium and diacylglycerol-regulated guanine nucleotide exchange factor (GEF) [28]; and (v) adaptor-mediated assembly of Rap1GTP/RIAM/talin integrin-activation complexes [29]. The composite network consists of 25 distinct molecular species and complexes coupled by 28 reactions, which are in turn characterized by 29 kinetic rate parameters. This modular structure is intended to generalize canonical pathways that have been studied extensively in other cell types and have recently been implicated in hematopoietic lineages such as neutrophils, monocytes, and lymphocytes [27,30–32]. Therefore, we do not impose a strict interpretation upon the identity or isoform of species belonging to homologous families whose expression patterns may be cell-type-specific.
Additional signaling molecules that have well-established roles in regulating leukocyte integrin affinity were ultimately omitted from the model based on the inability to identify complete pathway dependencies. For example, both the small GTPase RhoA and protein kinase C (PKC) are known to differentially modulate integrin affinity and clustering downstream of chemokine receptors in the context of adhesion under flow [4,6]; however, intermediate effectors linking these regulators to integrin activation remain unidentified [33,34]. We are also aware of RAPL involvement in linking Rap1 to integrin activation, although this effector appears to be specific for integrin αLβ2 [29]. In this respect, the proposed model recapitulates but one effector cascade which likely operates in conjunction with parallel pathways. As additional data become available, the network topology presented here will be subject to further refinement.
In order to identify estimates for the 29 reaction rate parameters listed in Table S1, the ODE model was trained against a collection of legacy data from the literature describing the human neutrophil response to a soluble chemokine stimulus of CXCL8. We anticipate that further biochemical characterization of inside-out signaling will facilitate a similar exercise for T lymphocytes, but for now the choice of CXCL8-mediated integrin activation in neutrophils activation reflects the desire to rely on the most comprehensive data set for a single leukocyte lineage. Illustrated in Figure 2, these data include the CXCL8 dose response for (i) chemokine receptor binding as determined by Scatchard analysis [35]; (ii) G-protein activation as reported by GTPase activity assay [36]; (iii) IP3 elevation as determined by competitive radiobinding assay [37]; and (iv) αLβ2 affinity modulation as reported by epitope-specific antibody binding [38]. Also included in the calibration data set were time-series data from (v) IP3 release and (vi) αLβ2 activation experiments reported in [37,38].
To reduce the degrees of freedom within the model, resting concentrations for each molecular species were fixed at characteristic values identified from the literature (Table S2). Estimates for the remaining rate parameters in each module were determined using a Latin hypercube sampling strategy to randomly search parameter space for those combinations that best replicated the selected calibration data (Materials and Methods) [39]. In all but a few cases, the distribution of optimal parameter values were broad and lacked clearly defined optimum, indicating that the available experimental data poorly constrained parameter space. This uncertainty was not unanticipated, and therefore a single parameter vector from among the optimized family collection of parameters values was selected for all subsequent simulations (Table S1).
Integrated Spatio-Temporal Simulation of T Lymphocyte Signaling and Adhesion
A salient feature of leukocyte recruitment is the small number of adhesion complexes (< 100) known to mediate adhesion within the cell-substrate contact interface [40]. The deterministic kinetic model was therefore reformulated using a stochastic, spatio-temporal formalism – the Next Subvolume Method (NSM) – to capture the dynamics of single molecule interactions [41]. Illustrated in Figure 2C and D, the lymphocyte membrane and cytoplasmic compartments are idealized as the union of distinct area and volume elements which are assumed to be well-mixed. Each subvolume contains a discrete number of molecular species which may diffuse among neighboring elements and react according to the rates prescribed from the ODE model. As described in Materials and Methods, the novel extension in the present work is the generalization of the NSM to non-cubic subvolumes. This approach affords an accurate representation of reaction and diffusion events that take place exclusively within the cell contact zone.
To simulate the effects of localized activation of inside-out effectors upon T cell adhesion, the NSM model was integrated within Adhesive Dynamics (AD), a simulator of leukocyte adhesion [42]. We have used AD to extensively study the determinants of static and dynamic leukocyte adhesion [43,44], and have recently described methods for integrating adhesion and signaling calculations [40,45,46]. This strategy allows us to dissect the molecular events that ultimately trigger cell arrest. Here, we specifically consider the transition between L-selectin/sLex-mediated rolling and αLβ2/ICAM-1-mediated cell arrest that is stimulated by a generic chemokine, CC, and its receptor, CCR.
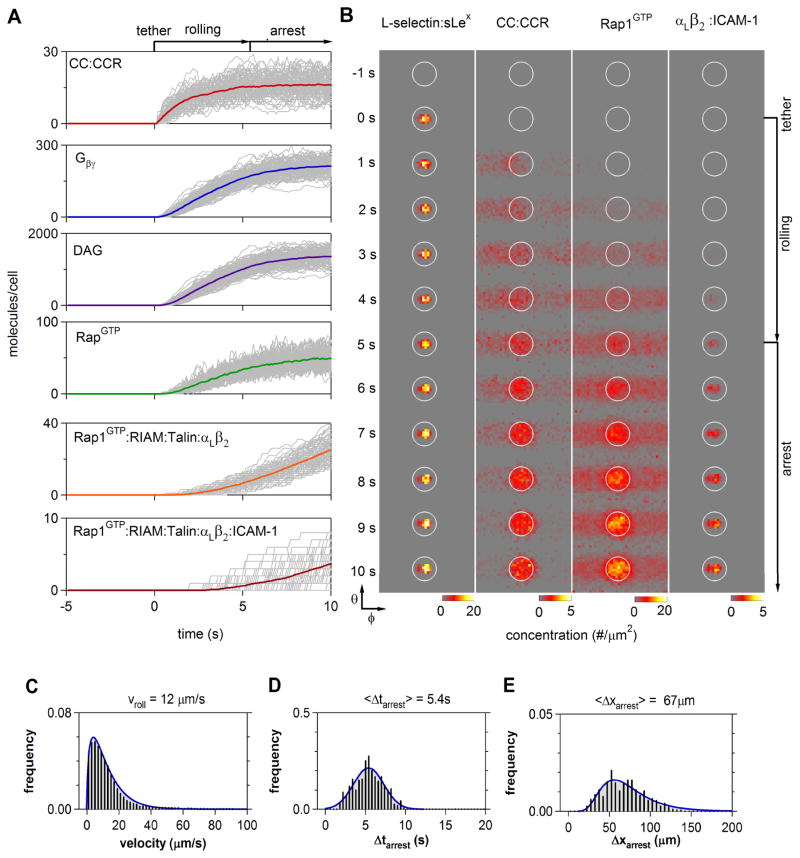
Figure 3 summarizes the collective information generated from integrated simulations of T cell arrest. These data include the temporal and spatial evolution of both intracellular signaling effectors and extracellular adhesion complexes (Fig. 3A and B), as well as the kinematics of cell adhesion (Fig. 3C–E). To emphasize the role of stochastic fluctuations in both signaling and adhesion events, 500 single cell trajectories were generated for a fixed chemokine stimulus of 103 CC sites/μm2. Upon tethering, the number of bound chemokine receptors rapidly approached a steady-state which remained approximately constant over the time scales for cell arrest (Fig. 3A). The effect of chemokine receptor occupancy propagated through the inside-out network and within several seconds, scaffold-mediated activation of resting integrins brought about an abrupt halt to cell rolling. Consistent with previous findings, simulated T cell arrest only required the formation of a small number of high-affinity integrin bonds with the cell substrate (< 10) [45,47].
Figure 3.
Integrated Simulation of T Lymphocyte Recruitment. Time course and statistics of signaling events during T cell rolling and arrest were compiled from five-hundred simulations. (A) Accumulation of the indicated species is shown for individual cells (gray); color trajectories denote population averages. All simulations were synchronized to the moment of initial tether formation (t = 0 s). (B) Spatio-temporal snapshots of select membrane-associated species from the cell reference frame. In each column, the membrane concentration of the indicated species was projected to a plane centered on the cell-substrate contact zone (encircled, cf. Fig. 1D). Cell rolling results in the convection of molecules activated in the contact zone from right to left through the reference frame, as seen in the early stages of CCR binding and Rap1GTP activation. Frames correspond to 1 s intervals (C–E) Predicted distributions of instantaneous cell velocities, (C), arrest times, Δtarrest, (D), and arrest distances, Δxarrest (E).
Integrated simulations also predicted complex spatial patterns of signal propagation during cell rolling and arrest (Fig. 3B). While selectin and integrin binding was naturally limited to the contact zone, chemokine receptors and downstream effectors were effectively activated across a wide swath of the plasma membrane due to the convective motion of the cell relative to the substrate. For example, the simulated accumulation of active Rap1GTP was symmetrical about the axis of cell rotation. Upon arrest, subsequent inside-out signaling was largely confined to the contact zone, which facilitated further integrin binding and adhesion maturation.
The corresponding ensemble of cell arrest dynamics likewise revealed a complex interrelationship between rolling velocity, arrest time, and arrest distance (Fig. 3C–E), which emerge from the concurrent realization of stochastic signal integration and transient adhesive interactions during rolling. The times required to mediate cell arrest were normally distributed around a mean value of 5 ± 2 s, which corresponded well with the 2–8 s range of arrest times observed experimentally (Fig. 3D; Table 1). The predicted rolling distances that preceded arrest (10–150 μm) were also similar in magnitude to in vitro trajectories. However, the distribution of arrest distances was significantly skewed downstream (Fig. 3E), suggesting that the occasional large translational velocities during rolling results in a natural asymmetry about the population average distance to arrest.
Table 1.
T Lymphocyte Recruitment Parameters
| statistic | velocity (μm/s) | Δvroll (μm/s) | Δtarrest (s) | Δxarrest (μm) |
|---|---|---|---|---|
| in vitroa | ||||
| range | 0–330 | 13–180 | 2.3–7.9 | 28–320 |
| mean | 75 | 39 | 4.7 | 140 |
| standard deviation | 87 | 22 | 2.4 | 120 |
| in vivob | ||||
| range | - | 12–72 | 2–5 | 25–125 |
| mean | - | 50 | 4 | 92 |
| standard deviation | - | - | - | - |
This study.
Murine lymphocytes [20]
Efficiency and Fidelity of Cell Arrest
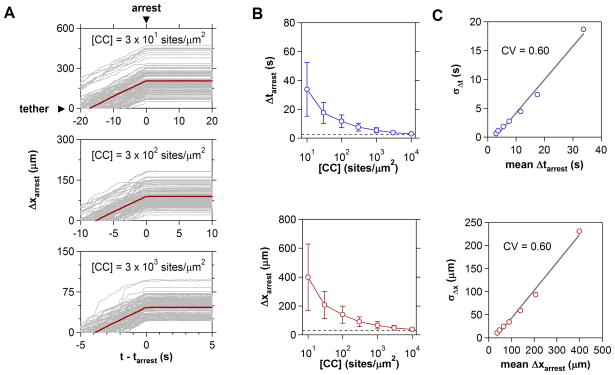
In resting lymphoid tissues, basal expression of homeostatic chemokines such as CCL19 and CCL21 promote constitutive recruitment of naïve and central memory T cell populations bearing CCR7. During an active immune response, expression of these ligands is downregulated ten- to one hundred-fold [48], whereas a second class of inflammatory chemokines specific for differentiated effector cells becomes upregulated. To address how changes in chemokine levels alter cell arrest properties, simulations were performed over five decades of chemokine concentrations spanning 100–105 sites/μm2 (Fig. 4). In all predicted trajectories, T cells did not exhibit any marked deceleration in the moments preceding arrest, confirming the observation that relatively few active integrins are required to mediate near-instantaneous arrest [47].
Figure 4.
Chemokine Regulation of T Cell Arrest. (A) Effect of increasing chemokine substrate concentration on time (Δtarrest) and distance (Δxarrest) to cell arrest. 500 trajectories synchronized to the moment of arrest and distance of tether formation are shown. The population mean is indicated in red. (B) Cell arrest metrics over four decades of chemokine concentration. Dashed lines indicate the minimum time and distance required to integrate inside-out stimuli. (C) Standard deviations of Δtarrest and Δxarrest distributions increase linearly with population mean. For both metrics, the coefficient of variation (CV) equals 0.60.
With increasing chemokine presentation, the distance to cell arrest asymptotically approached a threshold response distance of ~30 μm, indicating that there exists a limiting, albeit very rapid, response time, ~2.5 s, required to trigger cell arrest. The dispersion in cell arrest times/distances likewise decreased with increasing stimulus levels. Careful inspection revealed that the coefficient of variation (CV) describing arrest distributions was nearly constant (CV ~0.6). Thus, chemokine levels not only control the average distance to T cell arrest, but also the fidelity of recruitment from the fluid stream.
Sensitivity To Homing Receptor Expression
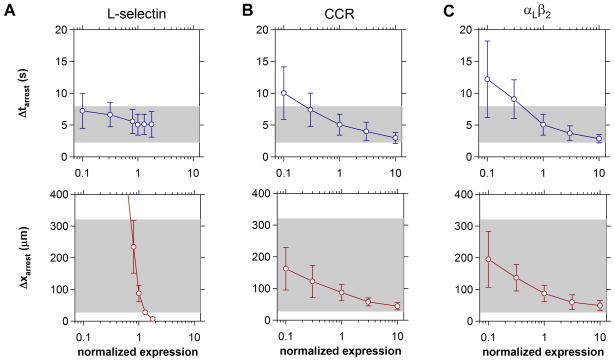
According to prevailing models for lymphocyte homing, tissue tropism is encoded by complimentary receptor phenotypes acquired during T cell maturation and differentiation [49]. To address whether tissues discriminate cells on the basis of qualitative or quantitative differences in receptor expression, arrest metrics were predicted for cell populations expressing variable levels selectins, chemokine receptors, or integrins (Fig. 5). Because the presence of tethering receptors principally affects the speed of lymphocyte rolling, the timing of cell arrest was largely unaffected by variations in L-selectin expression. Arrest distances however, were exquisitely sensitive to a reduction below optimal L-selectin levels (Fig. 5A). By comparison, variations in chemokine and integrin receptor expression had similar, quantitative influence upon both the timing and distance of cell arrest (Fig. 5B and C). Cells characterized by ten-fold lower levels of CCR or αLβ2 arrested just outside the window observed in flow chamber assays. Tenfold higher CCR and αLβ2 levels accelerated cell arrest, but only to a point, suggesting that there exists a threshold rate at which molecular diffusion and collision allow cells to integrate inside-out signals. Together, these predictions imply that vascular tissues recruit select T cell populations in a binary, all-or-none fashion via selectin- mediated rolling, and subsequently discriminate subpopulations based upon quantitative differences in chemokine and integrin expression.
Figure 5.
Impact of Surface Receptor Expression on T Cell Recruitment. Arrest statistics were compiled for cells expressing variable levels of (A) L-selectin, (B) chemokine receptor, and (C) integrin αLβ2. Receptor expression was varied one-at-a-time and normalized to those levels that yielded arrest metrics similar to those observed in adhesion assays (1.25×104 L-selectin/cell, 5×104 CCR/cell, and 104 αLβ2/cell). Shaded regions indicate the range of cell behaviors observed in vitro (cf. Table 1). The concentration of surface ligands were 102 sLex/μm2, 103 CC/μm2, and 103 ICAM-1/μm2. Data reflect the mean ± s.d. of twenty simulations. In (A), cells cease to roll at ~3× above normalized L-selectin levels.
Additive Integration of Multiple Chemokine Stimuli
It is an open question whether the presentation of multiple chemokines serves a functional role in T lymphocyte trafficking. Both secondary lymphoid organs and inflamed tissues have been shown to produce a rich chemokine environment [50]. Recent reports have also described apparent cross-talk between multiple chemokines during T cell migration [9,51]. It therefore seems reasonable to anticipate that multiple chemokines may cooperate at the level of inside-out activation during lymphocyte recruitment.
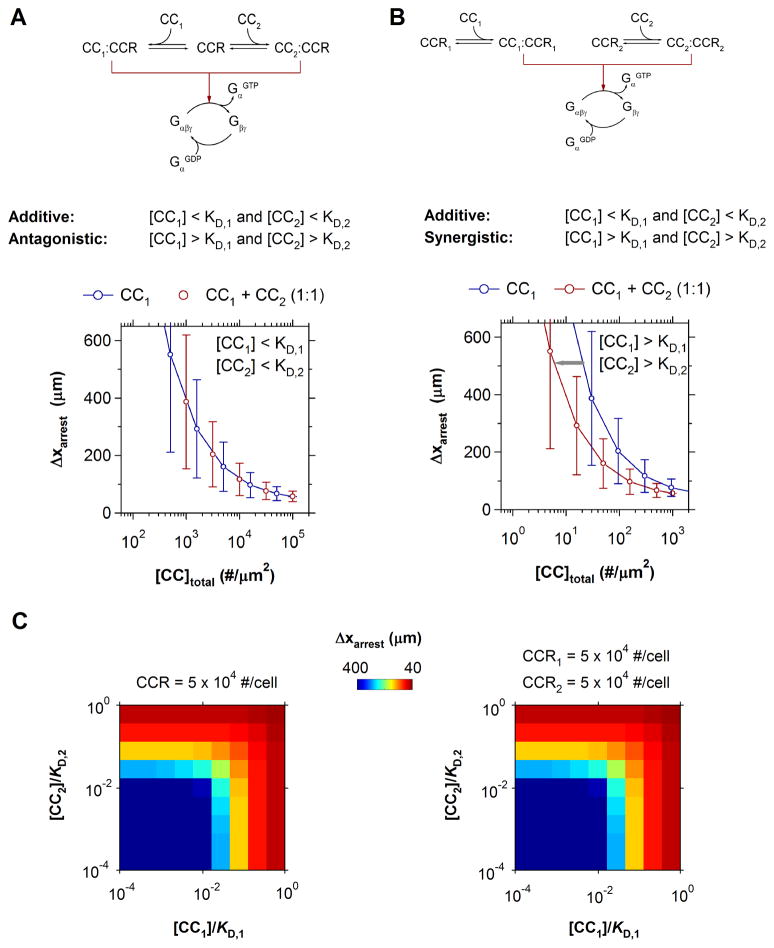
Shown in Fig. 6, two common chemokine-receptor networks were simulated to identify the quantitative effects of multiple chemokine recognition. The first encompasses simultaneous recognition of two chemokines by one receptor. Relevant examples of this motif include CCL19/21 binding by CCR7 in naïve T cells, CXCL9/11 binding by CXCR3 in Th1 cells, and CCL17/22 binding by CCR4 in Th2 cells [9, 10]. The second motif considers the expression of multiple chemokine receptors. Notable examples of double positive cells include CCR7+CXCR4+ naïve cells, CXCR3+CCR5+ Th1 cells, and CCR3+CCR4+ Th2 cells [12, 13]. In simulating each motif, the expression of each chemokine receptor was maintained at fixed, characteristic levels (5×104 CCR/cell).
Figure 6.
Additive Integration of Multiple Chemokine Stimuli. (A–B) Receptor signaling motifs for two chemokines. Predicted T cell arrest distances for the corresponding motifs under additive conditions (left) or synergistic conditions (right) are shown. (C) Predicted arrest distances for increasing substrate concentrations of two chemokines at physiological affinities: KD.1 = KD.2 = 5×105 μm−2. For either motif, the net effect of multiple chemokines on T cell arrest is identical to an equivalent concentration of one chemokine. In all simulations, CCR = CCR1 = CCR2 = 5×104 #/cell.
For cells expressing only one chemokine receptor, receptor competition will be minimal and the predicted receptor occupancy effectively additive so long as the two chemokines are present in sub-saturating amounts; i.e. [CC1] < KD,1 and [CC2] < KD,2 (Fig. 6A). However, a single chemokine may maximally activate all available receptors if present at saturating concentrations and a second chemokine stimulus will therefore result in receptor competition and ligand antagonism. For cells expressing two distinct receptors, stimulation by two chemokines will also be additive at low concentrations (Fig. 6B). At higher concentrations, the availability of a second receptor increases the saturation threshold, giving rise to a synergistic response. In the context of T cell arrest, such synergism is manifest in a leftward shift in the dose-response for cell arrest distance to lower total chemokine concentrations.
Which regime will these chemokine-receptor motifs operate within under physiological conditions? Using an average two-dimensional affinity for CXCL8 - CXCR1/2 binding (KD = 105 μm2) [52] and a maximal chemokine concentrations of 10,000 sites/μm2, scaling considerations indicate that the relative low affinity prevents either network from maximally activating the available number of surface receptors (i.e. CC/KD ≤ 100). Instead, both networks should operate in a regime where the depletion of available receptors is negligible and receptor engagement by chemokines is approximately additive [52].
This response similarity between motifs was indeed observed in the corresponding simulations of arrest dynamics. For all possible chemokine combinations, there was a negligible difference between the arrest responses triggered by one or two chemokine receptors (Fig. 6C). Furthermore, the combined effect of multiple chemokines was closely mimicked by an equivalent dose of either chemokine in isolation (Fig. 6C). Thus, the efficacy of cell arrest is determined by the total concentration of complimentary chemokines that a given cell may recognize.
Determinants and Consequences of Variable Chemokine Potency
An alternate hypothesis for multiple chemokine recognition suggests that chemokines exhibit varying levels of agonistic potency in their ability to trigger inside-out signals [10]. In this scenario, a strong chemokine agonist is predicted to stimulate robust T cell arrest of multiple cell populations despite wide variations in surface receptor expression. In contrast, a weakly potent chemokine agonist would stimulate only those populations expressing high receptor levels. While disparate agonist potencies have been reported for chemokines that share a common receptor, the physical determinants of chemokine potency in the context of lymphocyte recruitment remain poorly defined [23,53].
An intriguing proposal purports that the overall frequency of chemokine receptor engagement may be as important in determining agonist potency as receptor binding affinity [10]. This mode of ligand discrimination has been implicated in a number of biological processes, including the serial engagement of T cell receptors by low affinity ligands and enhancing G-protein receptor activation [54,55].
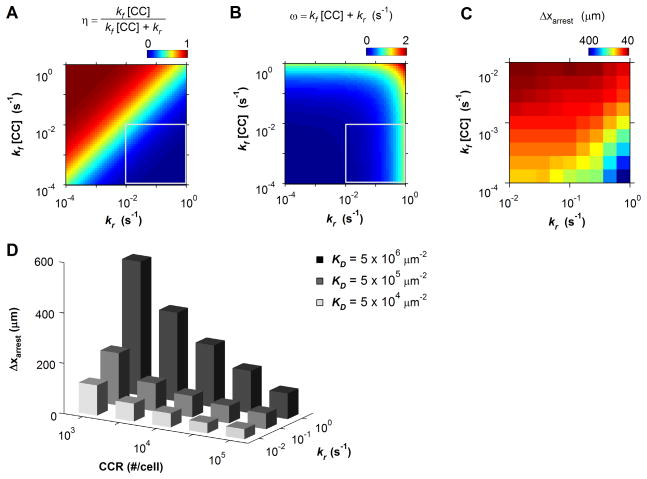
By definition, the binding affinity, kf/kr, and chemokine concentration, [CC], determines the fraction of receptors occupied at equilibrium, η:
| (1) |
The frequency of receptor engagement, ω, is similarly related to the forward, kf, and reverse, kr, association rates and chemokine concentration by:
| (2) |
To assess whether T cells discriminate chemokines on the basis of chemokine affinity or binding frequency, the efficiency of cell arrest was simulated over a range of chemokine-receptor kinetics: kf ≤ 10−3 μm2/s, [CC] ≤ 105 μm−2, kr ≥10−2 s−1 (Fig. 7). The predicted pattern in cell arrest efficiency most strongly correlated with the variations in fractional receptor occupancy, η, and only weakly correlated with binding frequency, ω (Fig. 7C). Therefore, the apparent potency with which a given chemokine triggers inside-out activation is primarily dictated by the overall binding affinity. Because two-dimensional binding kinetics are generally diffusion-limited [52], variations in chemokine affinity (and hence potency) should likely hinge upon differences in the molecular dissociation rate, kr, rather than significant variations in kf.
Figure 7.
Recruitment Sensitivity to Chemokine-Receptor Kinetics. (A) Fractional receptor occupancy as predicted by Eq. 6.1 and (B) frequency of receptor occupancy as predicted by Eq. 6.2. The enclosed regions in (A) and (B) indicate physiological parameter space. (C) Predicted arrest distance as a function chemokine-receptor kinetics. (D) Mean arrest distance as a function of chemokine affinity and chemokine receptor expression. In all simulations [CC] = 103 μm−2.
This prediction substantiates the claim that tissues might effectively modulate the strength of the inside-out signal by reciprocally expressing high- or low-affinity chemokines. Simulations confirmed that this form of regulation is likely useful in specifying population-level recruitment patterns (Fig. 7D). A ten-fold difference in chemokine dissociation rates between 10−1 s−1 and 100 s−1 was particularly effective at discriminating cells expressing low levels of chemokine receptor (< 105 #/cell). Whereas a high affinity interaction (kr ~10−2 s−1) effectively recruited a broad range of cells (Δxarrest < 200 μm), low affinity interactions (kr ~ 00 s−1) only triggered robust cell arrest in cells characterized by high chemokine receptor expression (105 receptors/cell). Thus, weak and strong chemokine-receptor affinities likely represent a general mechanism for generating selective and non-selective T cell recruitment patterns, respectively.
DISCUSSION
Dynamic inside-out regulation of lymphocyte integrins has been the subject of intense study, but quantitative descriptions of the molecular events involved have remained elusive. In this study, we developed a kinetic model of one inside-out pathway directly linking chemokine receptor binding to integrin affinity activation and cell adhesion in T lymphocytes [4,56]. Previously published models of integrin activation in leukocytes have been limited in molecular detail largely due to uncertainty in the identity of molecules involved in the signaling cascade [40,45]. While a good deal of uncertainty in pathway topology remains, the present model offers a reasonable starting point for future revisions. In particular, we fully anticipate that ongoing work to identify molecular effectors downstream of PKC and RhoA activation will facilitate the incorporation of parallel activation pathways [57,58]. A number of studies also hint at the role for positive feedback via ‘outside-in’ signalosome activation once a threshold number of integrins have been ligated [59,60]. Incorporating these and potentially other signaling effectors will be critical for assessing the role of intracellular cross-talk and phenotypic variability among lymphocyte subpopulations.
One challenge in developing such network models of biochemical signaling is identifying and measuring appropriate systems-level dynamics that appropriately constrain the kinetic parameter space. Our efforts to identify unambiguous parameter values revealed that available measurements of inside-out dynamics still leave some uncertainty in parameter estimates. This will necessarily be an area for future pathway-level studies, and efforts should focus on the design of cellular stimuli that uniquely constrain individual parameters [61]. However, we emphasize that the parameters have been trained on a wide variety of signaling experiments separate from adhesion experiments, and these parameters are consistent with known signaling phenomena. However, since these data are taken from different types of cells, there is some uncertainty in the values that apply for Jurkat cells or primary T-lymphocytes. These parameters, and the model itself, need to be validated using direct measurements of signaling and adhesion. This validation awaits further careful experimentation. One attractive approach that should be immediately applicable is RNAi-mediated knock-down of key signaling proteins. Signaling measurements made in knock-down cell lines reflect perturbed protein levels while leaving intrinsic rate parameters unchanged. In this way, simulations of knock-down cell dynamics can further constrain numerical estimates for reaction rate parameters. This method has already been utilized to successfully identify rate parameter distributions in a number of model cell lines, including murine B lymphocytes [22,62]. However, one key advance of the work we present here, compared to previous models in which signaling is course grained [40,45,46], is that, for the first time, we can make predictions of the effects of changes of individual proteins – modified through molecular biology – and then simulate how these individual changes affect the dynamics of adhesion. Furthermore, the incorporation of stochastic signaling elements seems appropriate, since in many cases small numbers of events are required rolling or arrest.
A detailed comparison to Caputo and Hammer [45] is warranted to explain the contribution of the current work. In Caputo and Hammer, a coarse grained signaling network for a single chemokine receptor was incorporated within AD; here, in contrast, a detailed signaling network that articulates each signaling molecule has been included In Caputo and Hammer, only changes in signaling dynamics within the interior of micovilli were considered; in this paper, changes in the concentration throughout the cytoplasm were included using the NSM. Furthermore, we have been able to estimate real reaction parameters for each participant in the signaling pathway, and we have extended the model to include multiple chemokine receptors which often are used in lymphocyte systems.
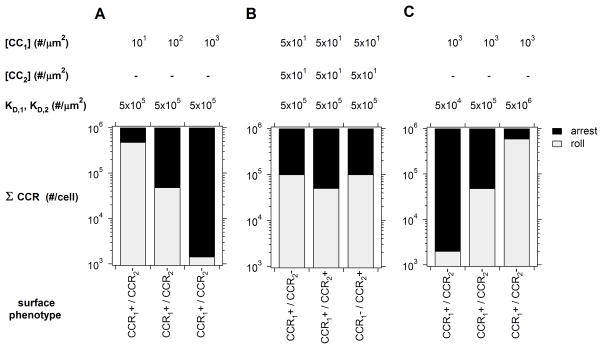
Several hypotheses have emerged from experimental studies of chemokine-triggered integrin activation in arresting T cells, but the theoretical basis for many of these have remain unexplored. Our primary approach to answer these questions has been the integration of signaling and adhesion models to predict the dynamics and efficacy of T cell recruitment. Beyond accurately replicating in vitro T lymphocyte arrest dynamics, our integrated model predictions provide theoretical evidence for three additional regulatory axes at the level of chemokine-chemokine receptor networks. Summarized in Fig. 8, these include natural regulation of chemokine presentation (density), additive integration of two or more chemokine stimuli, and selective presentation of low- and high-affinity chemokines.
Figure 8.
Three Modes of Selective T Lymphocyte Recruitment. The predicted range of T cell populations expressing one or two chemokine receptors that arrested within 100 μm of tethering are indicated. Regulated expression of one (A) or (B) two chemokines or chemokine affinity (C) resulted in population specific recruitment patterns.
In the face of vast diversity in surface receptor phenotypes, it appears that the immune system may modulate any one of these axes alone or in combination to further alter the identity of cells recruited to a specific tissue. Simply increasing chemokine expression levels may dramatically decrease the selective recruitment of T cell populations expressing variable levels of a single complementary receptor (Fig. 8A). Expressing two or more chemokines may represent a slight preference to recruit only double-positive cell subsets over single-positive populations. (Fig. 8B). And finally, expression of low to moderate affinity chemokines may represent a general strategy to only recruit cells with high receptor expression (Fig. 8C). A collection of experimental studies have in fact hinted at the existence of the mechanisms predicted here [9,10,63], but rigorous quantitative experiments monitoring individual cell trajectories should prove most definitive.
An intriguing possibility is that all three axes are employed simultaneously and that lymphocyte recruitment represents the integration of multiple, variable chemokine inputs. In this context, certain combinations of chemokine stimuli - number, combination, and potency - should generate ‘master codes’ that promote indiscriminant recruitment of multiple lymphocyte clones; other combinations of molecular trafficking codes will be cell selective. Our work here provides theoretical evidence for such integration and signal diversity, and should motivate future studies of the biophysical parameters regulating T lymphocyte recruitment.
MATERIALS AND METHODS
Parallel plate flow chamber assay with Jurkat T Lymphocytes
The T lymphoma cell line Jurkat was maintained in RPMI1640 supplemented with 10% FCS, 2 mM L-glutamine and 25 μg/ml gentamicin in a 5% humidified CO2 incubator. A circular parallel-plate flow chamber (GlycoTech) with a gasket of width 0.25 cm, thickness 127 mm, and length 2 cm was used for flow experiments. Polystyrene Petri dishes (diameter: 35 mm; Falcon 1008) enclosed using a single well flexiPERM (Sigma) were coated with a mixed solution of 10 μg/ml Purified Recomb Protein G (Thermo Scientific) and 1 μg/ml recombinant mouse CXCL12 (R&D Systems) at room temperature for 2 hours. Surfaces were then washed with 1% BSA in PBS. 5 μg/ml of recombinant mouse ICAM-1/Fc (R&D Systems) was applied to the surfaces and allowed to bind the Protein G coated surfaces at room temperature for 2 hours. Surfaces were then blocked for nonspecific adhesion with 2% BSA in PBS at 4°C for overnight. Before performing experiments, dishes were washed with prewarmed Hanks’ balanced salt solution (HBSS) containing 0.5% bovine serum albumin, 10 mM HEPES, 2 mM Ca2+ and 2 mM Mg2+ (37°C). The flow chamber was mounted on an inverted microscope, Axiovert 200 M (Carl Zeiss, Gottingen, Germany) and enclosed by a microscope incubator, XL-3 (Carl Zeiss) to perform the flow chamber experiments at 37°C. The T cell solution in prewarmed HBSS+ (37°C), at a concentration of 106/ml, was perfused into the chamber using a syringe pump (Harvard apparatus) at a flow rate corresponding to a calculated wall shear rate of 100 s−1.
Data acquisition and cell tracking
T cell adhesion was observed in the field of view of 980×735 mm2 using bright field microscopy under a 10× objective (NA = 0.20; Type: A-Plan; Carl Zeiss). Adhesion was visualized for 10 minutes at the upstream boundary of substrate ligands to ensure adherent cells had not previously interacted with CXCL12, or ICAM-1. A microscope-linked CCD camera was used to record T cell adhesion events, and videos were subsequently redigitalized to 640×480 pixels at 29.97 fps with ffmpegX software. Cell tracking was performed in Matlab (Mathworks) using freely available routines developed by Daniel Blair and Eric Dufresne (http://physics.georgetown.edu/matlab/).
ODE model and parameter calibration
Biochemical reaction rate laws were selected for the chemokine-triggered ‘inside-out’ integrin activation network described in the text. Listed explicitly in Table S1, rates for molecular association and dissociation for the reaction A + B ↔ A:B were defined as
| (3) |
Rates for catalytic phosphorylation and guanine nucleotide exchange reactions E + S ↔ ES → E + P were assumed to follow Michaelis-Menten kinetics
| (4) |
For simplicity, assembly of multimolecular complexes (e.g Rap1GTP:RIAM:Talin:αLβ2) was allowed to occur sequentially, thereby avoiding explicit enumeration of three additional bi- and tri-molecular intermediates.
Parameter values for individual reactions were identified by first defining characteristic lower and upper limits for the relevant kinetic rate constants [64]. Unique parameter combinations within these limits were selected at random with uniform probability using Latin hypercube sampling [39]. For each candidate parameter set, dose response and time series profiles (calculated at times/doses appropriate to the respective experimental protocols) were generated by numerically integrating the system of coupled ODEs. To reduce additional degrees of freedom, resting species concentrations were fixed according to literature estimates (Table S2). Model predictions were compared against a collection of n measurements in m assays from [35–38] using a residual, R, defined as
| (5) |
where discrepancies between the reported and simulated molecular concentrations xj are scaled by the maximum observed value so as to normalize residual contributions across all experimental assays.
NSM model formulation
The NSM algorithm for simulating the stochastic evolution of reaction-diffusion networks has been described extensively elsewhere [41,65]. The algorithm is implemented here as originally described, but molecular diffusion rates were modified to simulate diffusion among subvolumes defined by orthogonal spherical coordinates (Fig. 1C). For cubic subvolumes, the diffusion rate, Tr→Δr, between one subvolume centered at r and its neighbor at r+Δr is simply determined by the diffusivity, D, and lattice spacing, dx; i.e. Tr→Δr = D/dx2. In spherical coordinates, it may be shown that the corresponding diffusion rates are given by:
| (6) |
In the limit of small spatial discretization Eqs. satisfy (i) detailed balance and (ii) the Einstein diffusion equation for orthogonal spherical coordinates [66]. To improve simulation time, rates for all possible diffusion events were pre-calculated and stored in an indexed array. In all simulations, the cytoplasm and membrane were discretized into an array of 40×20×2 subvolumes
Integrating NSM and Adhesive Dynamics
Inside-out activation of resting surface integrins during cell rolling was simulated by executing the NSM formulation of the kinetic model and Adhesive Dynamics (AD) in tandem. The two calculations interface via shared data structures accessible at any time during a simulation. Whereas AD exclusively determines the sequence of extracellular events, NSM simulates the evolution of intracellular processes. AD iterations begin by assessing the number of active integrins in the contact zone as determined from the previous iteration of NSM. Changes in integrin affinity will affect the likelihood of bond formation and ultimately the cell trajectory at the end of the AD time increment. In a similar manner, every NSM iteration queries the number of chemokine receptors bound within the contact zone and updates the propensity for intracellular reactions accordingly. In this manner, each component of the integrated model sequentially exchanges chemical information required by its counterpart.
Synchronizing the components of the integrated model is achieved by systematically switching between NSM and AD iterations as the simulation clock advances. The order of calculation is determined by the extracellular or intracellular event with the earliest execution time, which is systematically identified from a composite list of all possible events. This list - the event queue - consists of a fixed time increment for executing AD and n variable time increments associated with NSM, where n is the number of subvolumes. From this list, the next event selected for updating is that with the shortest execution time from the event queue. As in the original NSM, the composite event queue is implemented as an indexed binary heap for efficient temporal sampling.
Requisite parameters for AD are given in Tables S3 and S4. In addition to cell geometric properties, these include rheological constants describing microvilli deformation [67], and mechanochemical binding parameters for L-selectin/PNAd and αLβ2/ICAM interactions. Forward and reverse association rate parameters for L-selectin/PNAd, as well as high- and low-affinity αLβ2/ICAM binding (Table S4) were selected from previously reported measurements of catch-slip and Bell model kinetics, respectively [44,68].
Supplementary Material
Acknowledgments
This work was supported National Institutes of Health Grants HL087353 and AI082292. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supporting Information Available. Supporting information, including the tables of parameters used for these simulations, is included. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Butcher EC, Picker LJ. Lymphocyte homing and homeostasis. Science. 1996;272:60–66. doi: 10.1126/science.272.5258.60. [DOI] [PubMed] [Google Scholar]
- 2.Butcher EC. Leukocyte-endothelial cell recognition: three (or more) steps to specificity and diversity. Cell. 1991;67:1033–1036. doi: 10.1016/0092-8674(91)90279-8. [DOI] [PubMed] [Google Scholar]
- 3.Laudanna C, Kim JY, Constantin G, Butcher E. Rapid leukocyte integrin activation by chemokines. Immunol Rev. 2002;186:37–46. doi: 10.1034/j.1600-065x.2002.18604.x. [DOI] [PubMed] [Google Scholar]
- 4.Kinashi T. Intracellular signalling controlling integrin activation in lymphocytes. Nat Rev Immunol. 2005;5:546–559. doi: 10.1038/nri1646. [DOI] [PubMed] [Google Scholar]
- 5.Katagiri K, Hattori M, Minato N, Irie S, Takatsu K, Kinashi T. Rap1 is a potent activation signal for leukocyte function-associated antigen 1 distinct from protein kinase C and phosphatidylinositol-3-OH kinase. Mol Cell Biol. 2000;20:1956–1969. doi: 10.1128/mcb.20.6.1956-1969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laudanna C, Campbell JJ, Butcher EC. Role of Rho in chemoattractant-activated leukocyte adhesion through integrins. Science. 1996;271:981–983. doi: 10.1126/science.271.5251.981. [DOI] [PubMed] [Google Scholar]
- 7.Pachynski RK, Wu SW, Gunn MD, Erle DJ. Secondary lymphoid-tissue chemokine (SLC) stimulates integrin alpha 4 beta 7-mediated adhesion of lymphocytes to mucosal addressin cell adhesion molecule-1 (MAdCAM-1) under flow. J Immunol. 1998;161:952–956. [PubMed] [Google Scholar]
- 8.Springer TA. Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell. 1994;76:301–314. doi: 10.1016/0092-8674(94)90337-9. [DOI] [PubMed] [Google Scholar]
- 9.Bai Z, Hayasaka H, Kobayashi M, Li W, Guo Z, Jang MH, Kondo A, Choi B, Iwakura Y, Miyasaka M. CXC chemokine ligand 12 promotes CCR7-dependent naive T cell trafficking to lymph nodes and Peyer’s patches. J Immunol. 2009;182:1287–1295. doi: 10.4049/jimmunol.182.3.1287. [DOI] [PubMed] [Google Scholar]
- 10.D’Ambrosio D, Albanesi C, Lang R, Girolomoni G, Sinigaglia F, Laudanna C. Quantitative differences in chemokine receptor engagement generate diversity in integrin-dependent lymphocyte adhesion. J Immunol. 2002;169:2303–2312. doi: 10.4049/jimmunol.169.5.2303. [DOI] [PubMed] [Google Scholar]
- 11.Weber C, Koenen RR. Fine-tuning leukocyte responses: towards a chemokine ‘interactome’. Trends Immunol. 2006;27:268–273. doi: 10.1016/j.it.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Sallusto F, Lenig D, Mackay CR, Lanzavecchia A. Flexible programs of chemokine receptor expression on human polarized T helper 1 and 2 lymphocytes. J Exp Med. 1998;187:875–883. doi: 10.1084/jem.187.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell DJ, Debes GF, Johnston B, Wilson E, Butcher EC. Targeting T cell responses by selective chemokine receptor expression. Semin Immunol. 2003;15:277–286. doi: 10.1016/j.smim.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 14.Lee B, Sharron M, Montaner LJ, Weissman D, Doms RW. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci U S A. 1999;96:5215–5220. doi: 10.1073/pnas.96.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mantovani A. The chemokine system: redundancy for robust outputs. Immunol Today. 1999;20:254–257. doi: 10.1016/s0167-5699(99)01469-3. [DOI] [PubMed] [Google Scholar]
- 16.Ding Z, Issekutz TB, Downey GP, Waddell TK. L-selectin stimulation enhances functional expression of surface CXCR4 in lymphocytes: implications for cellular activation during adhesion and migration. Blood. 2003;101:4245–4252. doi: 10.1182/blood-2002-06-1782. [DOI] [PubMed] [Google Scholar]
- 17.Weber KS, York MR, Springer TA, Klickstein LB. Characterization of lymphocyte function-associated antigen 1 (LFA-1)-deficient T cell lines: the alphaL and beta2 subunits are interdependent for cell surface expression. J Immunol. 1997;158:273–279. [PubMed] [Google Scholar]
- 18.Zhao Y, Chien S, Skalak R. A stochastic model of leukocyte rolling. Biophys J. 1995;69:1309–1320. doi: 10.1016/S0006-3495(95)79998-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DiVietro JA, Brown DC, Sklar LA, Larson RS, Lawrence MB. Immobilized stromal cell-derived factor-1alpha triggers rapid VLA-4 affinity increases to stabilize lymphocyte tethers on VCAM-1 and subsequently initiate firm adhesion. J Immunol. 2007;178:3903–3911. doi: 10.4049/jimmunol.178.6.3903. [DOI] [PubMed] [Google Scholar]
- 20.Bargatze RF, Butcher EC. Rapid G protein-regulated activation event involved in lymphocyte binding to high endothelial venules. J Exp Med. 1993;178:367–372. doi: 10.1084/jem.178.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christopher VR, Adam PA. Stochastic chemical kinetics and the quasi-steady-state assumption: Application to the Gillespie algorithm. The Journal of Chemical Physics. 2003;118:4999–5010. [Google Scholar]
- 22.Maurya MR, Subramaniam S. A kinetic model for calcium dynamics in RAW 264.7 cells: 1. Mechanisms, parameters, and subpopulational variability. Biophys J. 2007;93:709–728. doi: 10.1529/biophysj.106.097469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohout TA, Nicholas SL, Perry SJ, Reinhart G, Junger S, et al. Differential desensitization, receptor phosphorylation, beta-arrestin recruitment, and ERK1/2 activation by the two endogenous ligands for the CC chemokine receptor 7. J Biol Chem. 2004;279:23214–23222. doi: 10.1074/jbc.M402125200. [DOI] [PubMed] [Google Scholar]
- 24.Mahama PA, Linderman JJ. Calcium Signaling in Individual Bc(3)H1 Cells - Speed of Calcium Mobilization and Heterogeneity. Biotechnology Progress. 1994;10:45–54. [Google Scholar]
- 25.Daaka Y, Pitcher JA, Richardson M, Stoffel RH, Robishaw JD, et al. Receptor and G betagamma isoform-specific interactions with G protein-coupled receptor kinases. Proc Natl Acad Sci U S A. 1997;94:2180–2185. doi: 10.1073/pnas.94.6.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu D, Huang CK, Jiang H. Roles of phospholipid signaling in chemoattractant-induced responses. J Cell Sci. 2000;113 (Pt 17):2935–2940. doi: 10.1242/jcs.113.17.2935. [DOI] [PubMed] [Google Scholar]
- 27.Hyduk SJ, Chan JR, Duffy ST, Chen M, Peterson MD, Waddell TK, Digby GC, Szaszi K, Kapus A, Cybulsky MI. Phospholipase C, calcium, and calmodulin are critical for alpha4beta1 integrin affinity up-regulation and monocyte arrest triggered by chemoattractants. Blood. 2007;109:176–184. doi: 10.1182/blood-2006-01-029199. [DOI] [PubMed] [Google Scholar]
- 28.Ghandour H, Cullere X, Alvarez A, Luscinskas FW, Mayadas TN. Essential role for Rap1 GTPase and its guanine exchange factor CalDAG-GEFI in LFA-1 but not VLA-4 integrin mediated human T-cell adhesion. Blood. 2007;110:3682–3690. doi: 10.1182/blood-2007-03-077628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rose DM, Alon R, Ginsberg MH. Integrin modulation and signaling in leukocyte adhesion and migration. Immunol Rev. 2007;218:126–134. doi: 10.1111/j.1600-065X.2007.00536.x. [DOI] [PubMed] [Google Scholar]
- 30.M’Rabet L, Coffer PJ, Wolthuis RM, Zwartkruis F, Koenderman L, Bos JL. Differential fMet-Leu-Phe- and platelet-activating factor-induced signaling toward Ral activation in primary human neutrophils. J Biol Chem. 1999;274:21847–21852. doi: 10.1074/jbc.274.31.21847. [DOI] [PubMed] [Google Scholar]
- 31.Caron E, Self AJ, Hall A. The GTPase Rap1 controls functional activation of macrophage integrin alphaMbeta2 by LPS and other inflammatory mediators. Curr Biol. 2000;10:974–978. doi: 10.1016/s0960-9822(00)00641-2. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Yu M, Podd A, Wen R, Chrzanowska-Wodnicka M, White GC, Wang D. A critical role of Rap1b in B-cell trafficking and marginal zone B-cell development. Blood. 2008;111:4627–4636. doi: 10.1182/blood-2007-12-128140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laudanna C, Campbell JJ, Butcher EC. Elevation of intracellular cAMP inhibits RhoA activation and integrin-dependent leukocyte adhesion induced by chemoattractants. J Biol Chem. 1997;272:24141–24144. doi: 10.1074/jbc.272.39.24141. [DOI] [PubMed] [Google Scholar]
- 34.Laudanna C, Mochly-Rosen D, Liron T, Constantin G, Butcher EC. Evidence of zeta protein kinase C involvement in polymorphonuclear neutrophil integrin-dependent adhesion and chemotaxis. J Biol Chem. 1998;273:30306–30315. doi: 10.1074/jbc.273.46.30306. [DOI] [PubMed] [Google Scholar]
- 35.Petersen F, Flad HD, Brandt E. Neutrophil-activating peptides NAP-2 and IL-8 bind to the same sites on neutrophils but interact in different ways. Discrepancies in binding affinities, receptor densities, and biologic effects. J Immunol. 1994;152:2467–2478. [PubMed] [Google Scholar]
- 36.Kupper RW, Dewald B, Jakobs KH, Baggiolini M, Gierschik P. G-protein activation by interleukin 8 and related cytokines in human neutrophil plasma membranes. Biochem J. 1992;282 (Pt 2):429–434. doi: 10.1042/bj2820429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith RJ, Sam LM, Leach KL, Justen JM. Postreceptor events associated with human neutrophil activation by interleukin-8. J Leukoc Biol. 1992;52:17–26. doi: 10.1002/jlb.52.1.17. [DOI] [PubMed] [Google Scholar]
- 38.Lum AFH, Green CE, Lee GR, Staunton DE, Simon SI. Dynamic regulation of LFA-1 activation and neutrophil arrest on intercellular adhesion molecule 1 (ICAM-1) in shear flow. Journal of Biological Chemistry. 2002;277:20660–20670. doi: 10.1074/jbc.M202223200. [DOI] [PubMed] [Google Scholar]
- 39.Kinzer-Ursem TL, Linderman JJ. Both ligand- and cell-specific parameters control ligand agonism in a kinetic model of g protein-coupled receptor signaling. PLoS Comput Biol. 2007;3:e6. doi: 10.1371/journal.pcbi.0030006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krasik EF, Caputo KE, Hammer DA. Adhesive dynamics simulation of neutrophil arrest with stochastic activation. Biophysical Journal. 2008;95:1716–1728. doi: 10.1529/biophysj.107.119677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elf J, Ehrenberg M. Spontaneous separation of bi-stable biochemical systems into spatial domains of opposite phases. Systems Biology, IEE Proceedings. 2004;1:230–236. doi: 10.1049/sb:20045021. [DOI] [PubMed] [Google Scholar]
- 42.Hammer DA, Apte SM. Simulation of Cell Rolling and Adhesion on Surfaces in Shear-Flow - General Results and Analysis of Selectin-Mediated Neutrophil Adhesion. Biophysical Journal. 1992;63:35–57. doi: 10.1016/S0006-3495(92)81577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beste MT, Hammer DA. Steady-State Multiplicity in Receptor-Mediated Colloidal Adhesion. The Journal of Physical Chemistry C. 2007;111:2008–2016. [Google Scholar]
- 44.Beste MT, Hammer DA. Selectin catch-slip kinetics encode shear threshold adhesive behavior of rolling leukocytes. Proc Natl Acad Sci U S A. 2008;105:20716–20721. doi: 10.1073/pnas.0808213105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caputo KE, Hammer DA. Adhesion Dynamics Simulations of G-protein-mediated Chemokine-activated Neutrophil Adhesion. Biophysical Journal. 2009;96:2989–3004. doi: 10.1016/j.bpj.2008.12.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krasik EF, Yee KL, Hammer DA. Adhesive dynamics simulation of neutrophil arrest with deterministic activation. Biophysical Journal. 2006;91:1145–1155. doi: 10.1529/biophysj.105.070706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shamri R, Grabovsky V, Gauguet JM, Feigelson S, Manevich E, Kolanus W, Robinson MK, Staunton DE, von Andrian UH, Alon R. Lymphocyte arrest requires instantaneous induction of an extended LFA-1 conformation mediated by endothelium-bound chemokines. Nat Immunol. 2005;6:497–506. doi: 10.1038/ni1194. [DOI] [PubMed] [Google Scholar]
- 48.Mueller SN, Hosiawa-Meagher KA, Konieczny BT, Sullivan BM, Bachmann MF, Locksley RM, Ahmed R, Matloubian M. Regulation of homeostatic chemokine expression and cell trafficking during immune responses. Science. 2007;317:670–674. doi: 10.1126/science.1144830. [DOI] [PubMed] [Google Scholar]
- 49.Mora JR, Bono MR, Manjunath N, Weninger W, Cavanagh LL, Rosenblatt M, von Andrian UH. Selective imprinting of gut-homing T cells by Peyer’s patch dendritic cells. Nature. 2003;424:88–93. doi: 10.1038/nature01726. [DOI] [PubMed] [Google Scholar]
- 50.Bromley SK, Mempel TR, Luster AD. Orchestrating the orchestrators: chemokines in control of T cell traffic. Nat Immunol. 2008;9:970–980. doi: 10.1038/ni.f.213. [DOI] [PubMed] [Google Scholar]
- 51.Sebastiani S, Danelon G, Gerber B, Uguccioni M. CCL22-induced responses are powerfully enhanced by synergy inducing chemokines via CCR4: evidence for the involvement of first beta-strand of chemokine. Eur J Immunol. 2005;35:746–756. doi: 10.1002/eji.200525800. [DOI] [PubMed] [Google Scholar]
- 52.Lauffenburger DA, Linderman JJ. Receptors: models for binding, trafficking, and signalling. x. New York: Oxford University Press; 1993. p. 365. [Google Scholar]
- 53.Mariani M, Lang R, Binda E, Panina-Bordignon P, D’Ambrosio D. Dominance of CCL22 over CCL17 in induction of chemokine receptor CCR4 desensitization and internalization on human Th2 cells. Eur J Immunol. 2004;34:231–240. doi: 10.1002/eji.200324429. [DOI] [PubMed] [Google Scholar]
- 54.Mahama PA, Linderman JJ. A Monte Carlo study of the dynamics of G-protein activation. Biophys J. 1994;67:1345–1357. doi: 10.1016/S0006-3495(94)80606-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalergis AM, Boucheron N, Doucey MA, Palmieri E, Goyarts EC, Vegh Z, Luescher IF, Nathenson SG. Efficient T cell activation requires an optimal dwell-time of interaction between the TCR and the pMHC complex. Nat Immunol. 2001;2:229–234. doi: 10.1038/85286. [DOI] [PubMed] [Google Scholar]
- 56.Banno A, Ginsberg MH. Integrin activation. Biochem Soc Trans. 2008;36:229–234. doi: 10.1042/BST0360229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giagulli C, Scarpini E, Ottoboni L, Narumiya S, Butcher EC, Constatin G, Laudanna C. RhoA and zeta PKC control distinct modalities of LFA-1 activation by chemokines: critical role of LFA-1 affinity triggering in lymphocyte in vivo homing. Immunity. 2004;20:25–35. doi: 10.1016/s1074-7613(03)00350-9. [DOI] [PubMed] [Google Scholar]
- 58.Pasvolsky R, Grabovsky V, Giagulli C, Shulman Z, Shamri R, Feigelson SW, Laudanna C, Alon R. RhoA is involved in LFA-1 extension triggered by CXCL12 but not in a novel outside-in LFA-1 activation facilitated by CXCL9. J Immunol. 2008;180:2815–2823. doi: 10.4049/jimmunol.180.5.2815. [DOI] [PubMed] [Google Scholar]
- 59.Laudanna C, Alon R. Right on the spot. Chemokine triggering of integrin-mediated arrest of rolling leukocytes. Thromb Haemost. 2006;95:5–11. [PubMed] [Google Scholar]
- 60.Kremer KN, Humphreys TD, Kumar A, Qian NX, Hedin KE. Distinct role of ZAP-70 and Src homology 2 domain-containing leukocyte protein of 76 kDa in the prolonged activation of extracellular signal-regulated protein kinase by the stromal cell-derived factor-1 alpha/CXCL12 chemokine. J Immunol. 2003;171:360–367. doi: 10.4049/jimmunol.171.1.360. [DOI] [PubMed] [Google Scholar]
- 61.Apgar JF, Toettcher JE, Endy D, White FM, Tidor B. Stimulus design for model selection and validation in cell signaling. PLoS Comput Biol. 2008;4:e30. doi: 10.1371/journal.pcbi.0040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Albeck JG, MacBeath G, White FM, Sorger PK, Lauffenburger DA, Gaudet S. Collecting and organizing systematic sets of protein data. Nature Reviews Molecular Cell Biology. 2006;7:803–812. doi: 10.1038/nrm2042. [DOI] [PubMed] [Google Scholar]
- 63.DiVietro JA, Smith MJ, Smith BRE, Petruzzelli L, Larson RS, Lawrence MB. Immobilized IL-8 triggers progressive activation of neutrophils rolling in vitro on P-selectin and intercellular adhesion molecule-1. Journal of Immunology. 2001;167:4017–4025. doi: 10.4049/jimmunol.167.7.4017. [DOI] [PubMed] [Google Scholar]
- 64.Aldridge BB, Burke JM, Lauffenburger DA, Sorger PK. Physicochemical modelling of cell signalling pathways. Nat Cell Biol. 2006;8:1195–1203. doi: 10.1038/ncb1497. [DOI] [PubMed] [Google Scholar]
- 65.Hattne J, Fange D, Elf J. Stochastic reaction-diffusion simulation with MesoRD. Bioinformatics. 2005;21:2923–2924. doi: 10.1093/bioinformatics/bti431. [DOI] [PubMed] [Google Scholar]
- 66.Chen LG, Deem MW. Two-dimensional diffusion in the presence of topological disorder. Physical Review E. 2003;68 doi: 10.1103/PhysRevE.68.021107. [DOI] [PubMed] [Google Scholar]
- 67.Xu G, Shao JY. Double tether extraction from human neutrophils and its comparison with CD4+ T-lymphocytes. Biophys J. 2005;88:661–669. doi: 10.1529/biophysj.104.048363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wojcikiewicz EP, Abdulreda MH, Zhang X, Moy VT. Force spectroscopy of LFA-1 and its ligands, ICAM-1 and ICAM-2. Biomacromolecules. 2006;7:3188–3195. doi: 10.1021/bm060559c. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.