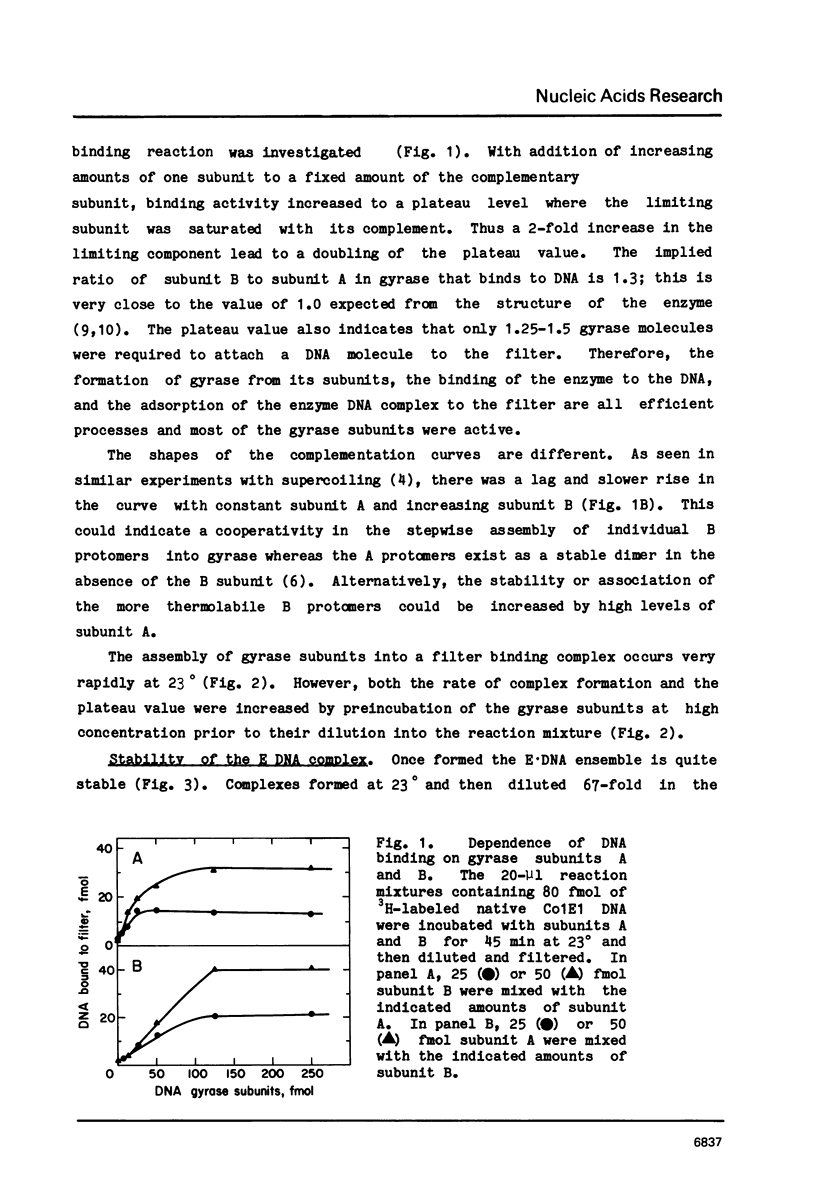
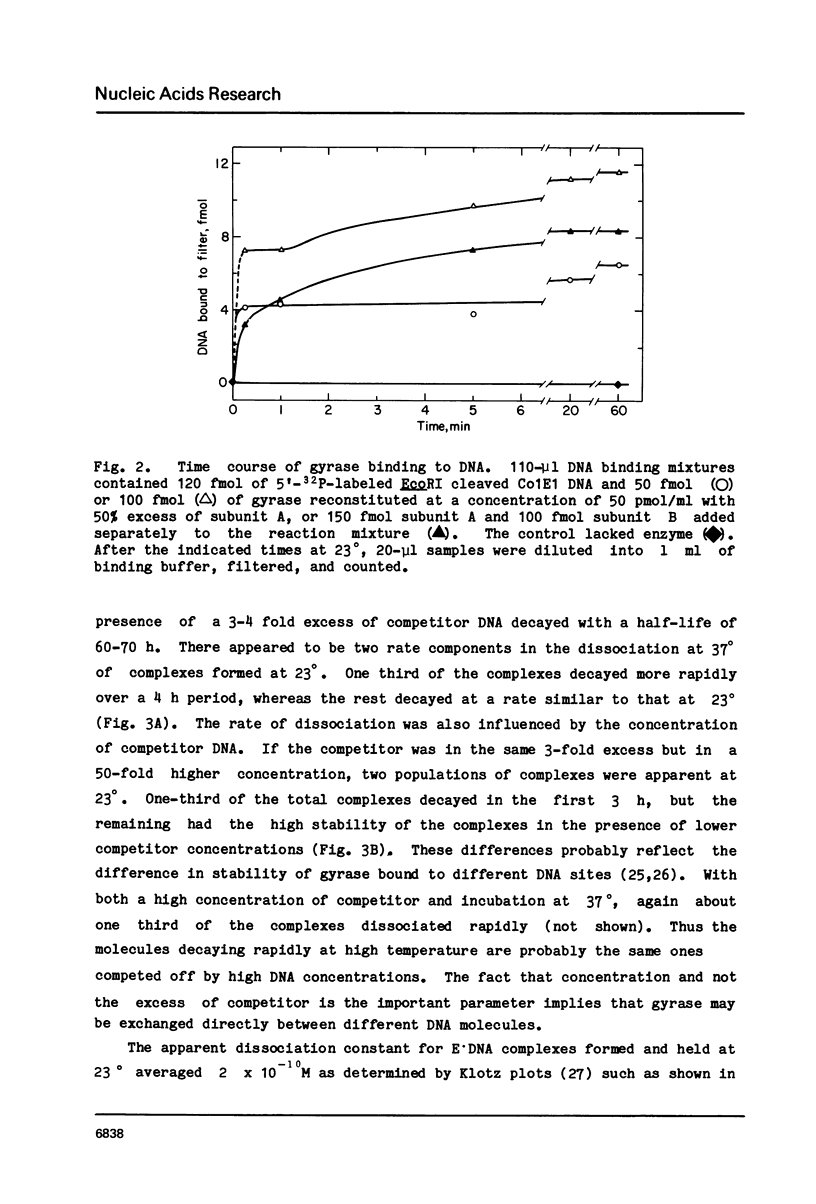
Abstract
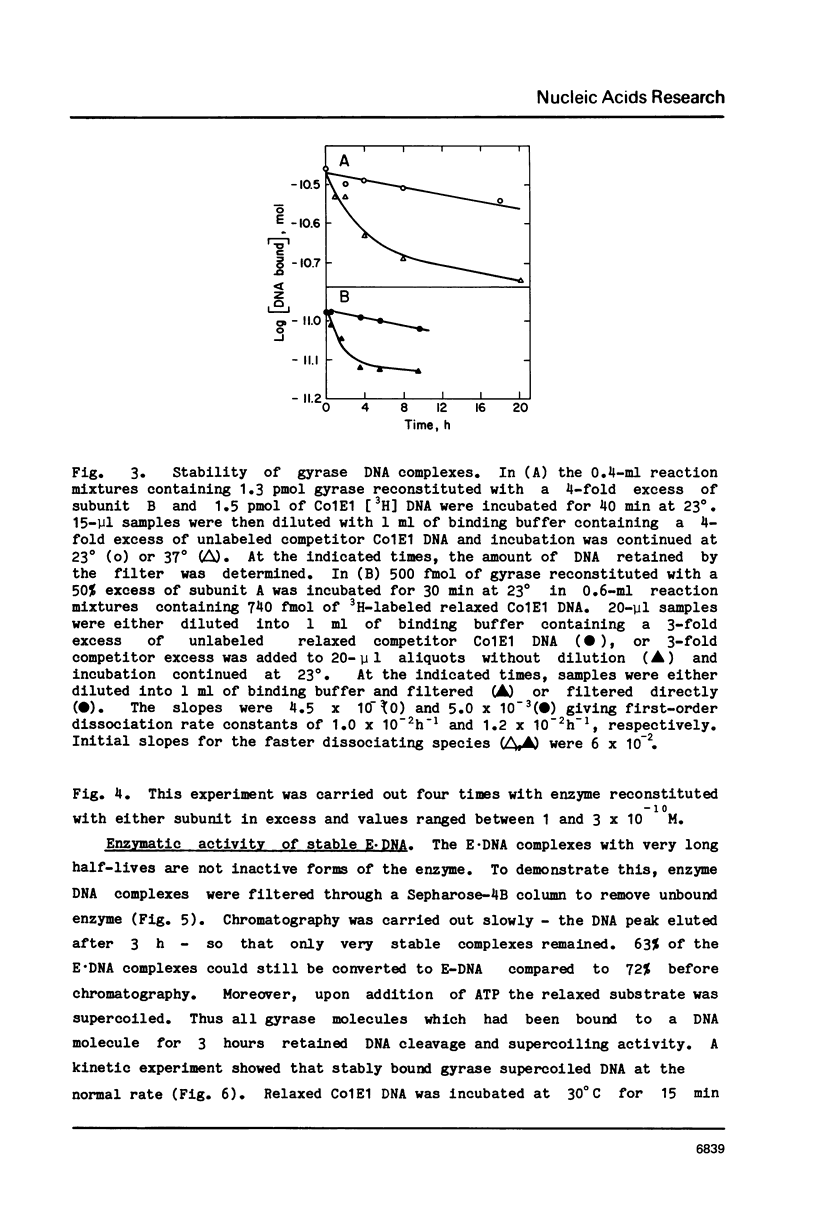
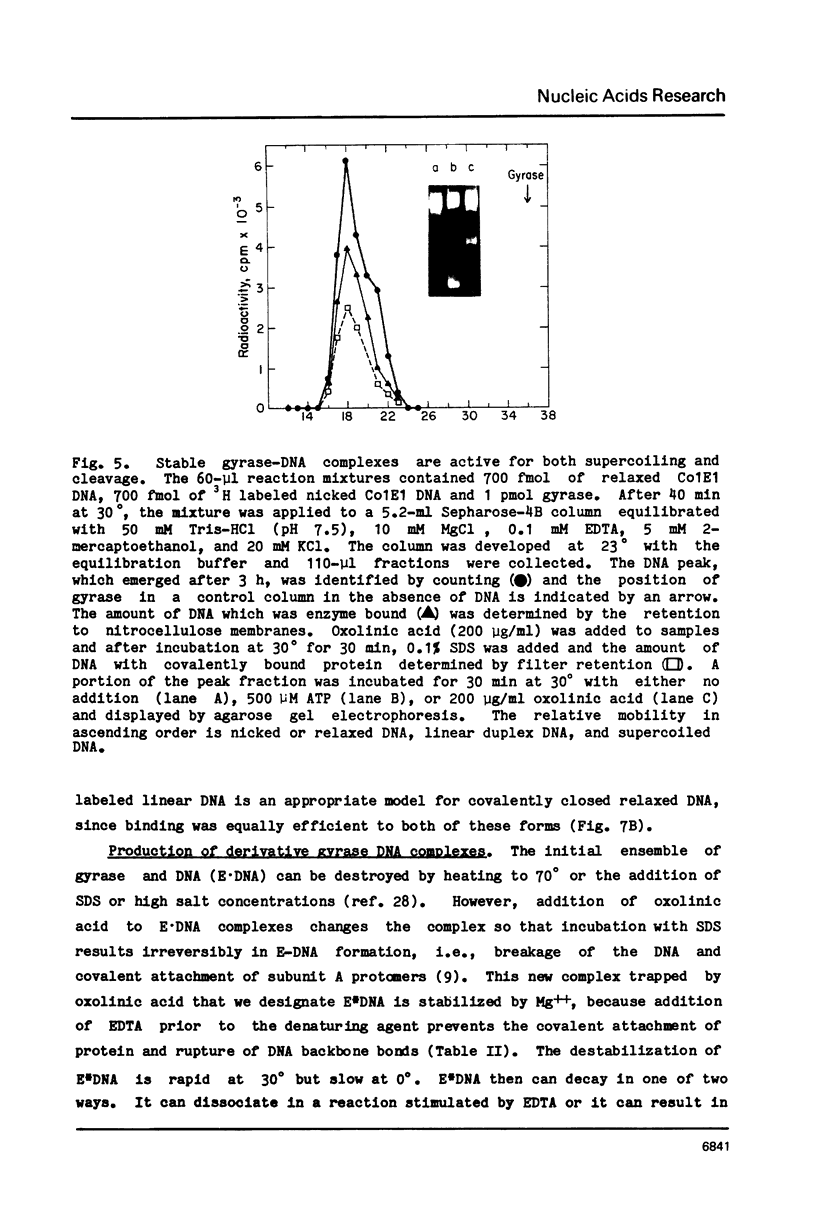
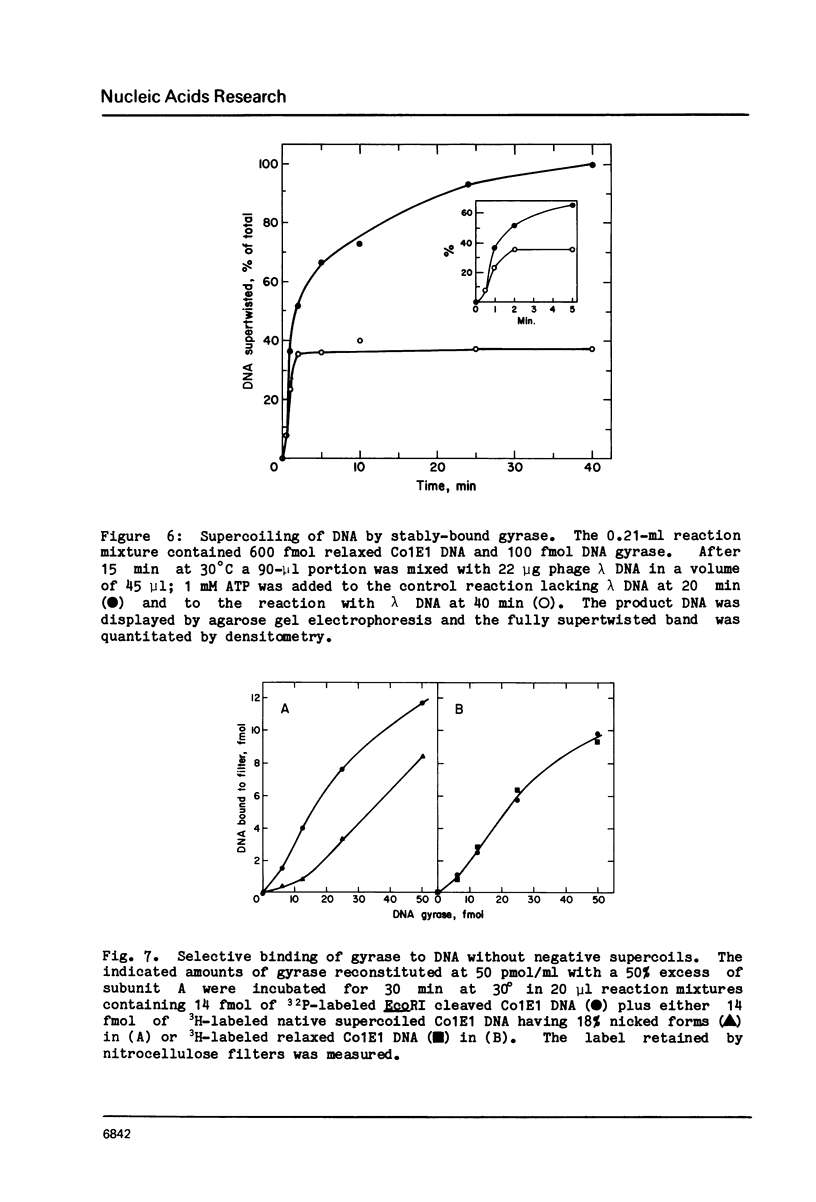
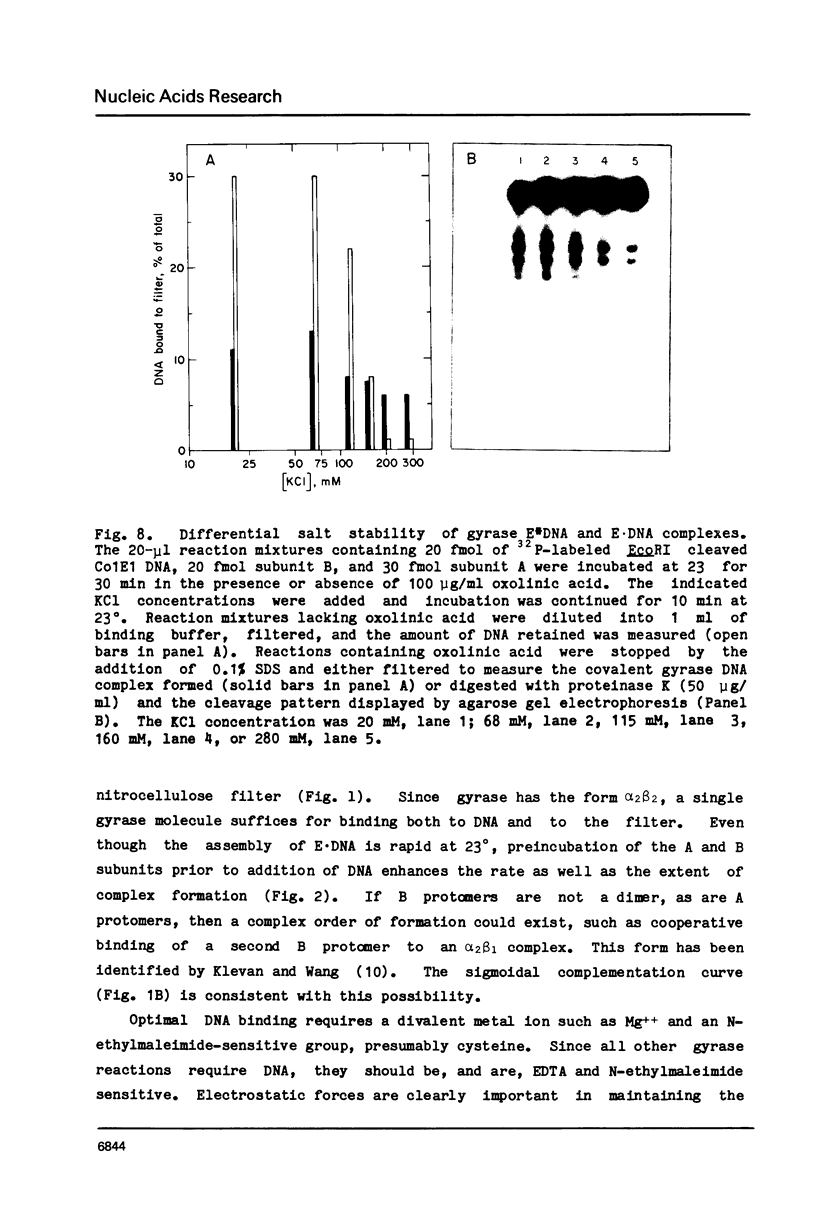
Three distinct Escherichia coli DNA gyrase complexes with DNA can be identified using a nitrocellulose filter-binding assay. One complex consists of an ensemble of two subunit A and two subunit B protomers bound noncovalently to specific sequences of DNA. High levels of each subunit alone are inactive but a single gyrase molecule binds DNA to a filter. At 23 degrees, the complex has a dissociation constant of approximately 10(-10) M and a half-time of decay of about 60 h. It is sufficiently stable that it can be purified by gel filtration and retain full supercoiling activity. Gyrase binds preferentially to relaxed DNA over supercoiled DNA by a factor of about 10. On addition of oxolinic acid, a second complex is formed that is distinguished by its stability in high ionic strength solutions and by efficient conversion to a third form upon addition of protein denaturants. The first and second complexes require Mg++ for optimal formation. The third form has been shown previously to contain denatured A protomers covalently linked to DNA that is broken at the site of attachment.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown P. O., Cozzarelli N. R. A sign inversion mechanism for enzymatic supercoiling of DNA. Science. 1979 Nov 30;206(4422):1081–1083. doi: 10.1126/science.227059. [DOI] [PubMed] [Google Scholar]
- Champoux J. J., McConaughy B. L. Purification and characterization of the DNA untwisting enzyme from rat liver. Biochemistry. 1976 Oct 19;15(21):4638–4642. doi: 10.1021/bi00666a014. [DOI] [PubMed] [Google Scholar]
- Cozzarelli N. R. DNA gyrase and the supercoiling of DNA. Science. 1980 Feb 29;207(4434):953–960. doi: 10.1126/science.6243420. [DOI] [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher L. M., Mizuuchi K., O'Dea M. H., Ohmori H., Gellert M. Site-specific interaction of DNA gyrase with DNA. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4165–4169. doi: 10.1073/pnas.78.7.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Itoh T., Tomizawa J. I. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., O'Dea M. H., Itoh T., Tomizawa J. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4474–4478. doi: 10.1073/pnas.73.12.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn I. M., Chappell J. B. A simple method for the preparation of 32-P-labelled adenosine triphosphate of high specific activity. Biochem J. 1964 Jan;90(1):147–149. doi: 10.1042/bj0900147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins N. P., Peebles C. L., Sugino A., Cozzarelli N. R. Purification of subunits of Escherichia coli DNA gyrase and reconstitution of enzymatic activity. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1773–1777. doi: 10.1073/pnas.75.4.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinkle D. C., Chamberlin M. J. Studies of the binding of Escherichia coli RNA polymerase to DNA. I. The role of sigma subunit in site selection. J Mol Biol. 1972 Sep 28;70(2):157–185. doi: 10.1016/0022-2836(72)90531-1. [DOI] [PubMed] [Google Scholar]
- Hinkle D. C., Chamberlin M. J. Studies of the binding of Escherichia coli RNA polymerase to DNA. II. The kinetics of the binding reaction. J Mol Biol. 1972 Sep 28;70(2):187–195. doi: 10.1016/0022-2836(72)90532-3. [DOI] [PubMed] [Google Scholar]
- Jones O. W., Berg P. Studies on the binding of RNA polymerase to polynucleotides. J Mol Biol. 1966 Dec 28;22(2):199–209. doi: 10.1016/0022-2836(66)90126-4. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K., Wang J. C. Mapping the topography of DNA wrapped around gyrase by nucleolytic and chemical probing of complexes of unique DNA sequences. Cell. 1981 Mar;23(3):721–729. doi: 10.1016/0092-8674(81)90435-9. [DOI] [PubMed] [Google Scholar]
- Klevan L., Wang J. C. Deoxyribonucleic acid gyrase-deoxyribonucleic acid complex containing 140 base pairs of deoxyribonucleic acid and an alpha 2 beta 2 protein core. Biochemistry. 1980 Nov 11;19(23):5229–5234. doi: 10.1021/bi00564a012. [DOI] [PubMed] [Google Scholar]
- Kreuzer K. N., McEntee K., Geballe A. P., Cozzarelli N. R. Lambda transducing phages for the nalA gene of Escherichia coli and conditional lethal nalA mutations. Mol Gen Genet. 1978 Nov 29;167(2):129–137. doi: 10.1007/BF00266906. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. DNA-DNA gyrase complex: the wrapping of the DNA duplex outside the enzyme. Cell. 1978 Nov;15(3):979–984. doi: 10.1016/0092-8674(78)90281-7. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Micrococcus luteus DNA gyrase: active components and a model for its supercoiling of DNA. Proc Natl Acad Sci U S A. 1978 May;75(5):2098–2102. doi: 10.1073/pnas.75.5.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K., O'Dea M. H., Gellert M. DNA gyrase: subunit structure and ATPase activity of the purified enzyme. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5960–5963. doi: 10.1073/pnas.75.12.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A., Cozzarelli N. R. Contacts between DNA gyrase and its binding site on DNA: features of symmetry and asymmetry revealed by protection from nucleases. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1416–1420. doi: 10.1073/pnas.78.3.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A., Cozzarelli N. R. Site-specific cleavage of DNA by E. coli DNA gyrase. Cell. 1979 May;17(1):175–184. doi: 10.1016/0092-8674(79)90305-2. [DOI] [PubMed] [Google Scholar]
- Morrison A., Higgins N. P., Cozzarelli N. R. Interaction between DNA gyrase and its cleavage site on DNA. J Biol Chem. 1980 Mar 10;255(5):2211–2219. [PubMed] [Google Scholar]
- Peebles C. L., Higgins N. P., Kreuzer K. N., Morrison A., Brown P. O., Sugino A., Cozzarelli N. R. Structure and activities of Escherichia coli DNA gyrase. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):41–52. doi: 10.1101/sqb.1979.043.01.008. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Suzuki H., Bourgeois S. Lac repressor-operator interaction. I. Equilibrium studies. J Mol Biol. 1970 Feb 28;48(1):67–83. doi: 10.1016/0022-2836(70)90219-6. [DOI] [PubMed] [Google Scholar]
- Sugino A., Cozzarelli N. R. The intrinsic ATPase of DNA gyrase. J Biol Chem. 1980 Jul 10;255(13):6299–6306. [PubMed] [Google Scholar]
- Sugino A., Higgins N. P., Brown P. O., Peebles C. L., Cozzarelli N. R. Energy coupling in DNA gyrase and the mechanism of action of novobiocin. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4838–4842. doi: 10.1073/pnas.75.10.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino A., Higgins N. P., Cozzarelli N. R. DNA gyrase subunit stoichiometry and the covalent attachment of subunit A to DNA during DNA cleavage. Nucleic Acids Res. 1980 Sep 11;8(17):3865–3874. doi: 10.1093/nar/8.17.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino A., Peebles C. L., Kreuzer K. N., Cozzarelli N. R. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4767–4771. doi: 10.1073/pnas.74.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarus M., Berg P. On the properties and utility of a membrane filter assay in the study of isoleucyl-tRNA synthetase. Anal Biochem. 1970 Jun;35(2):450–465. doi: 10.1016/0003-2697(70)90207-1. [DOI] [PubMed] [Google Scholar]
- van de Sande J. H., Kleppe K., Khorana H. G. Reversal of bacteriophage T4 induced polynucleotide kinase action. Biochemistry. 1973 Dec 4;12(25):5050–5055. doi: 10.1021/bi00749a004. [DOI] [PubMed] [Google Scholar]