Abstract
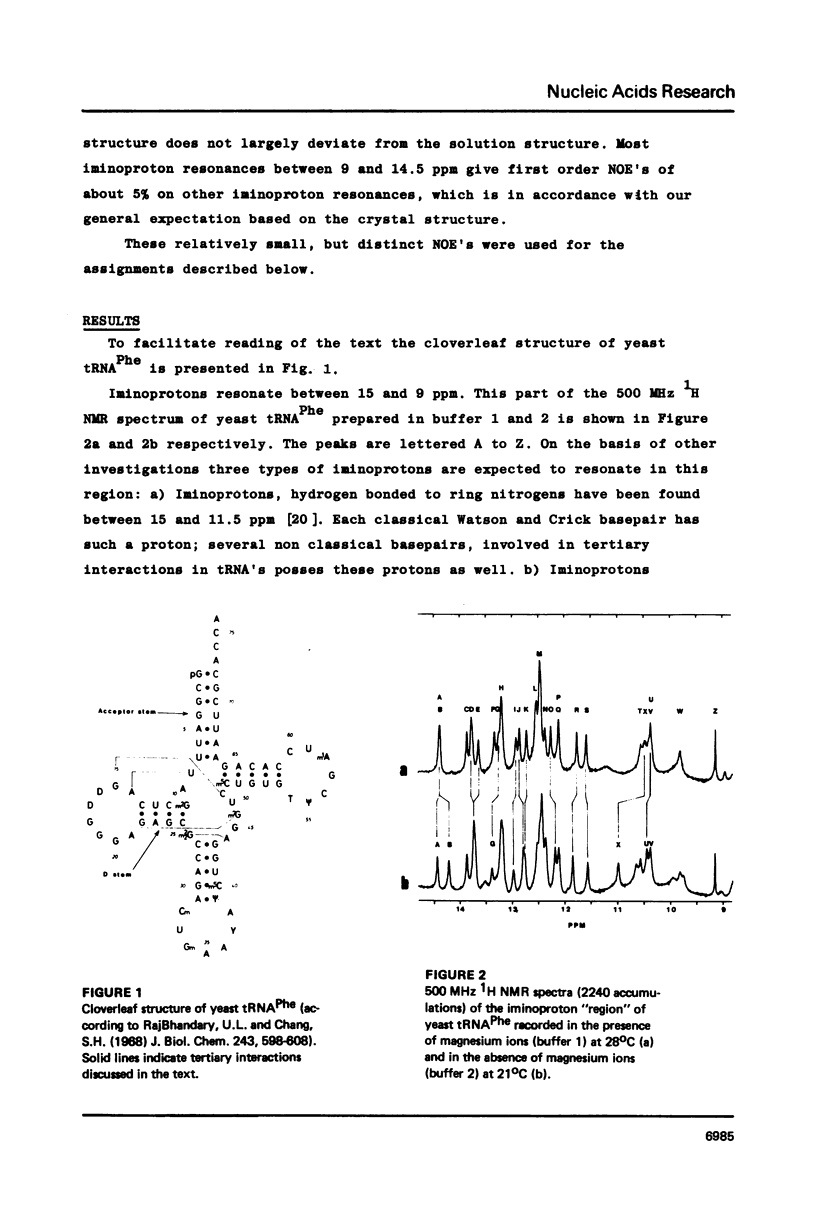
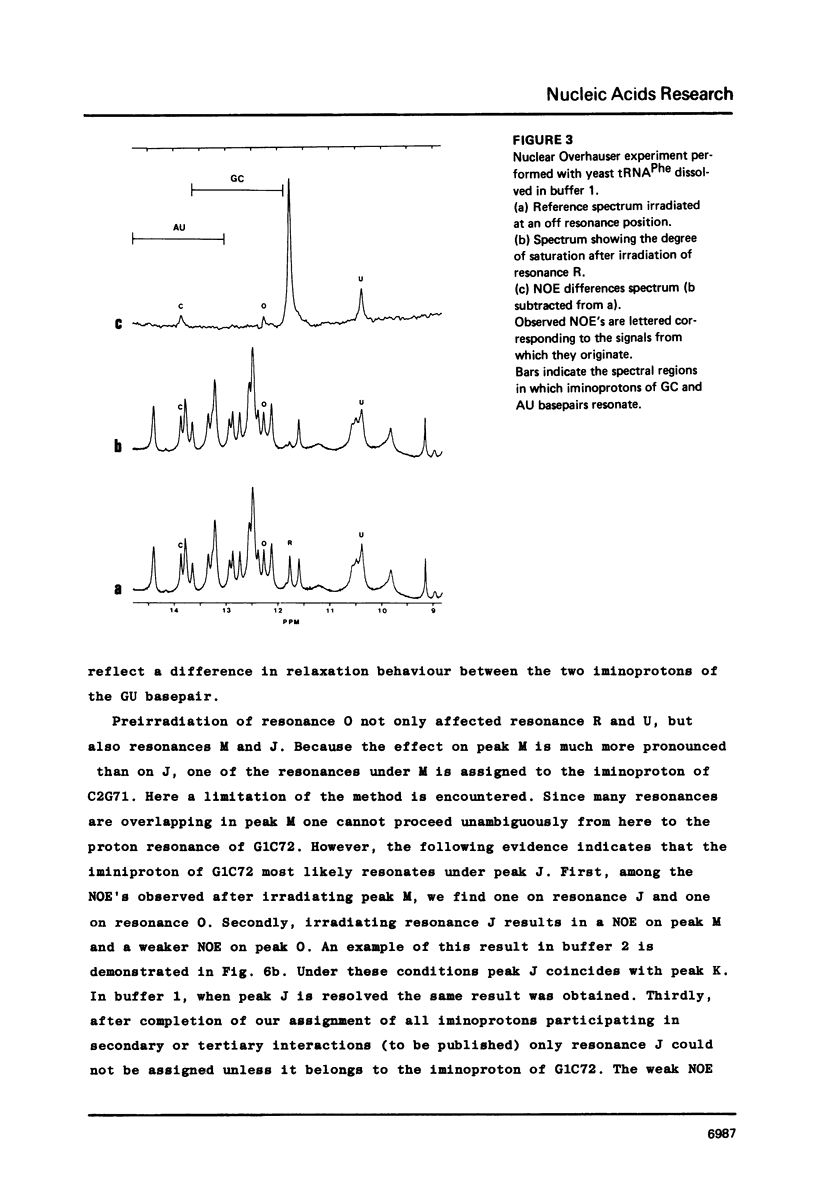
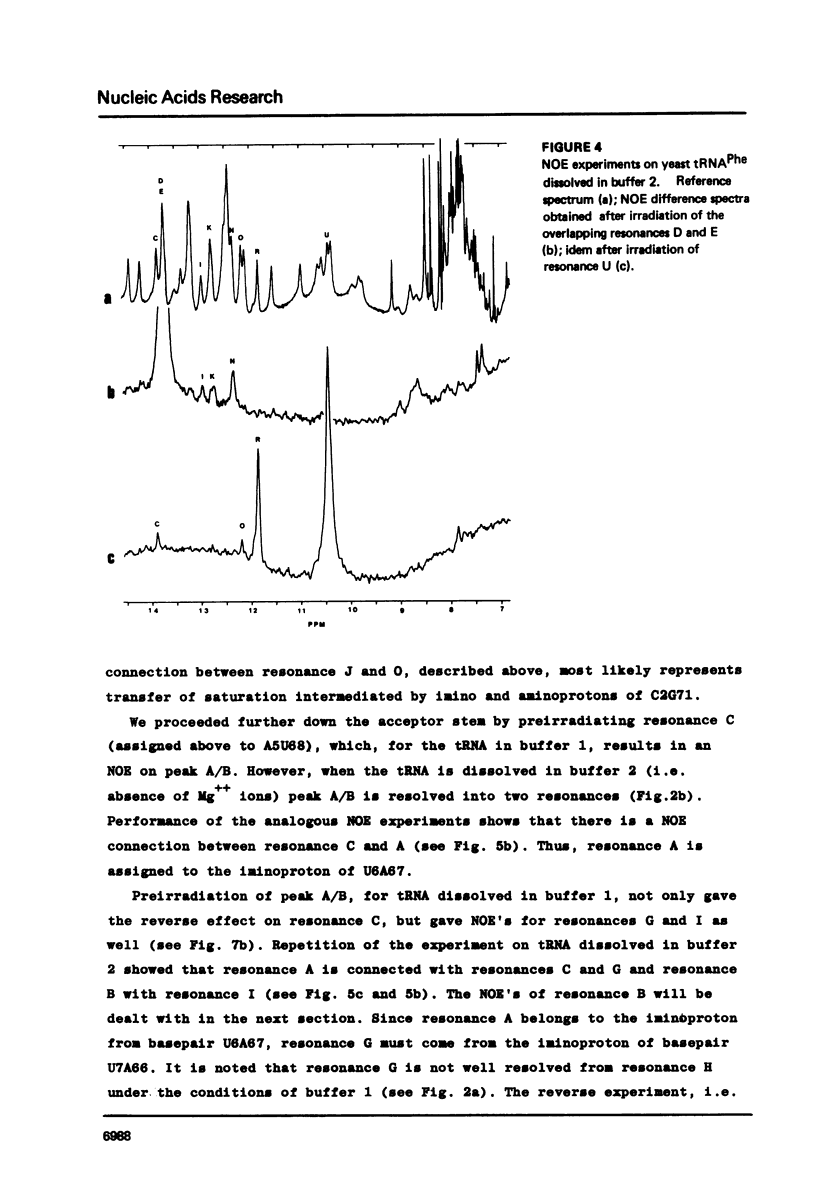
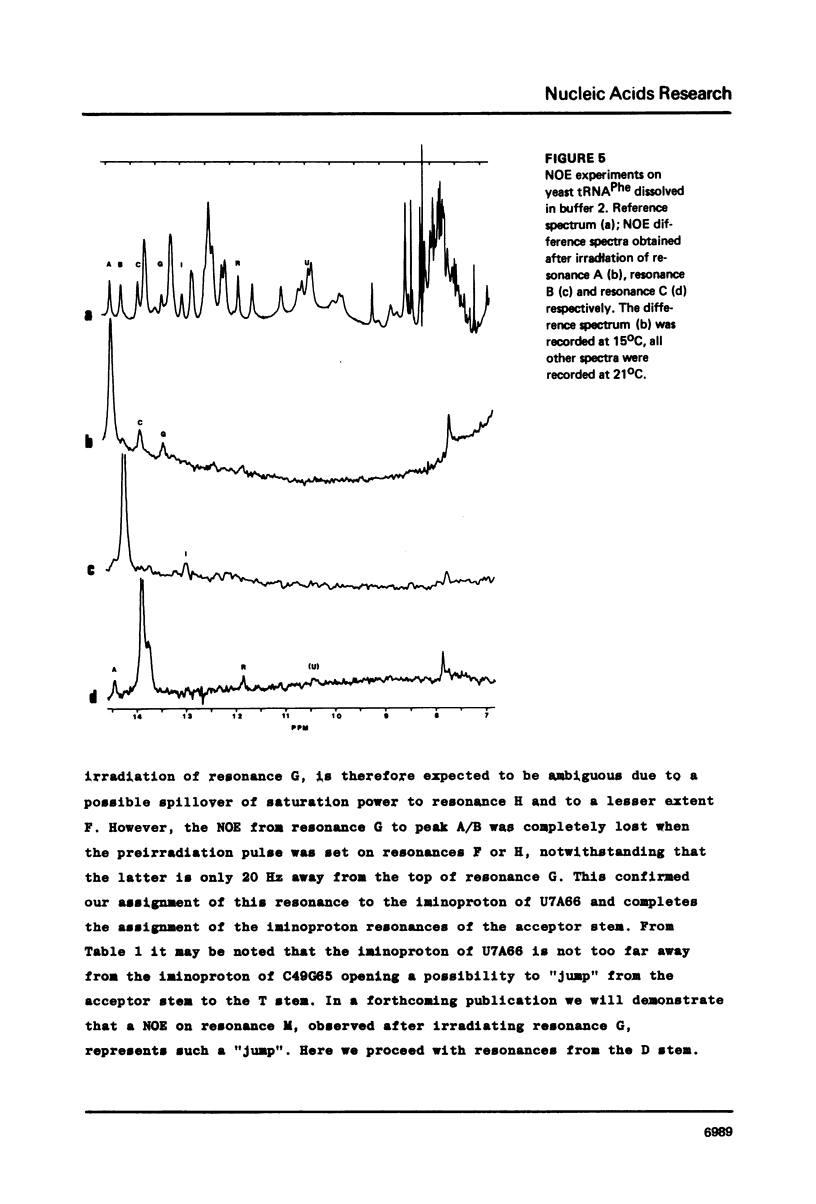
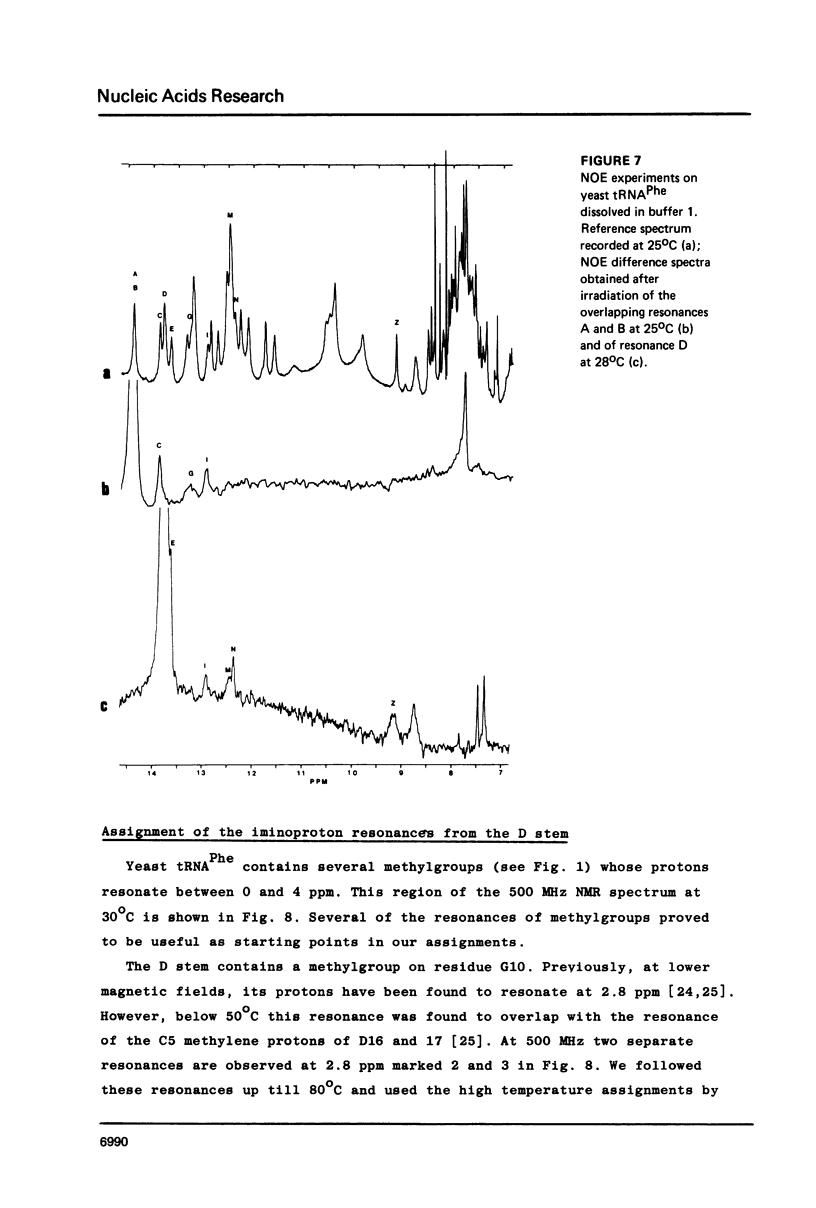
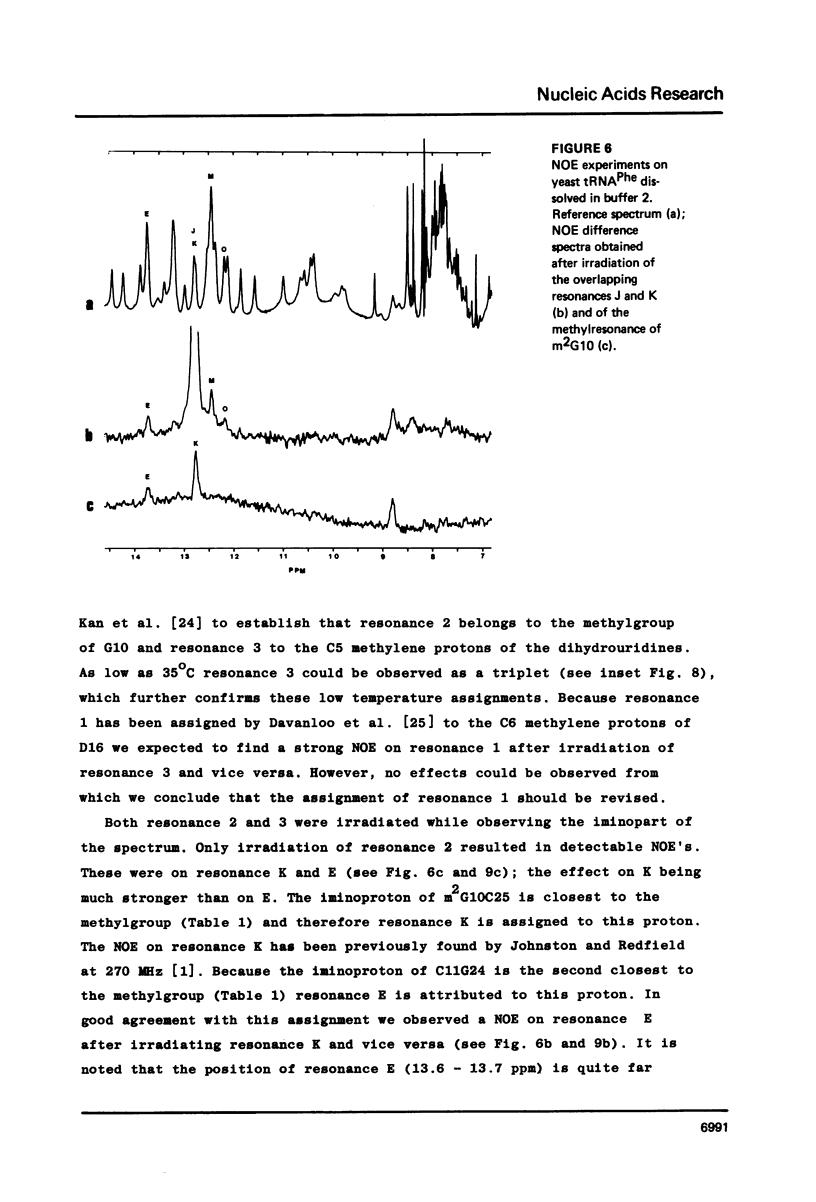
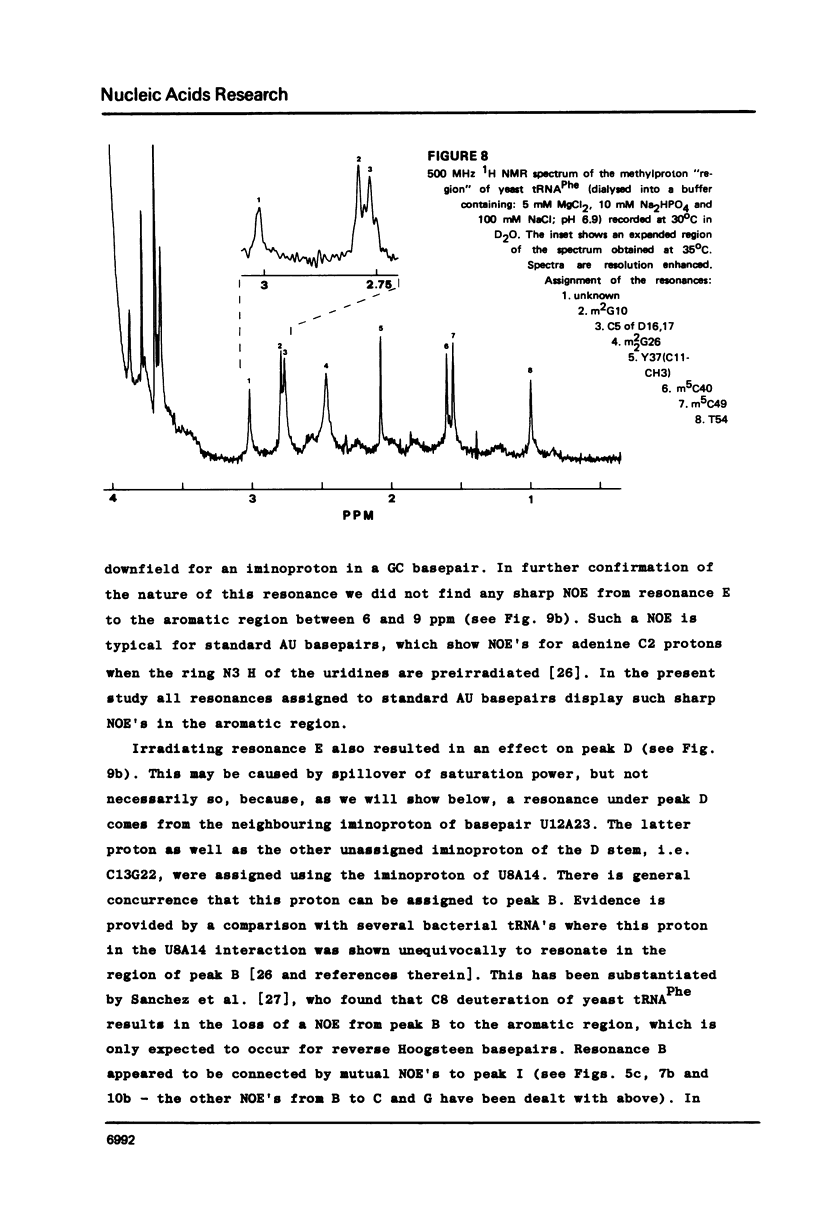
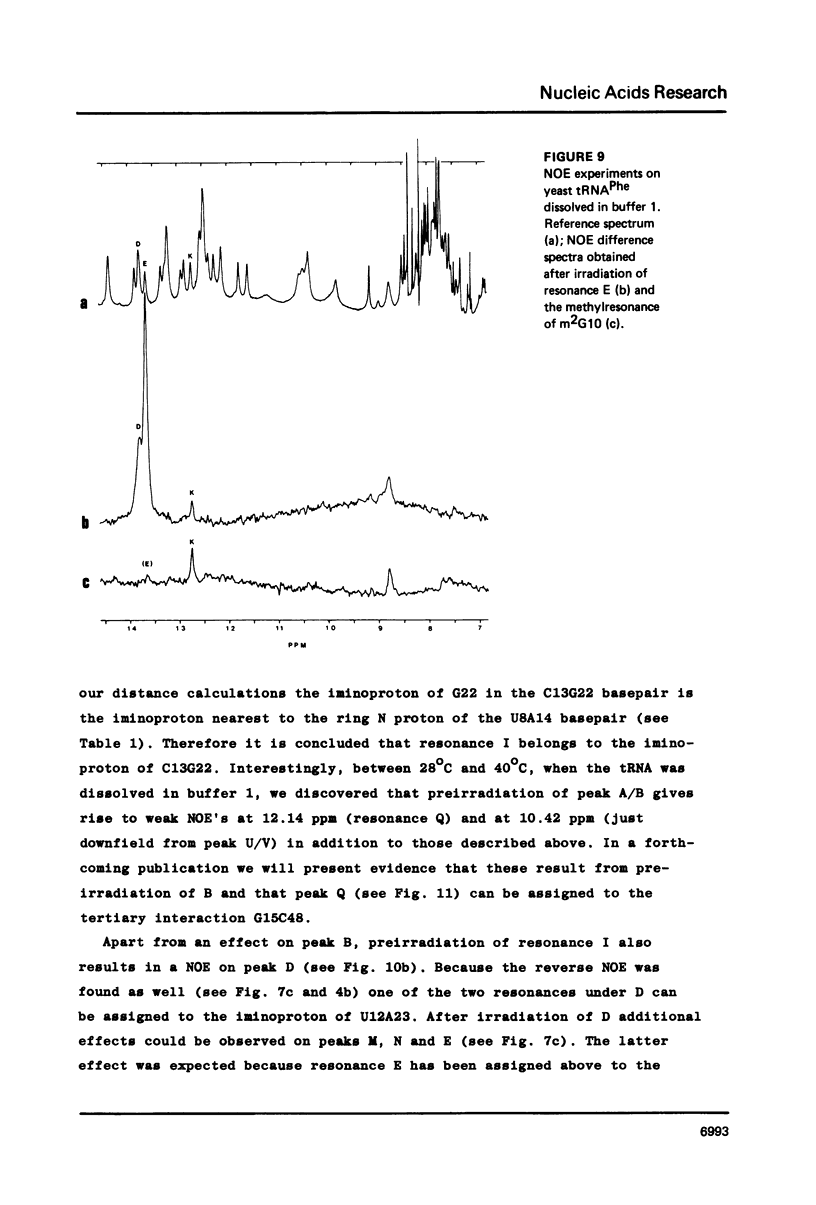
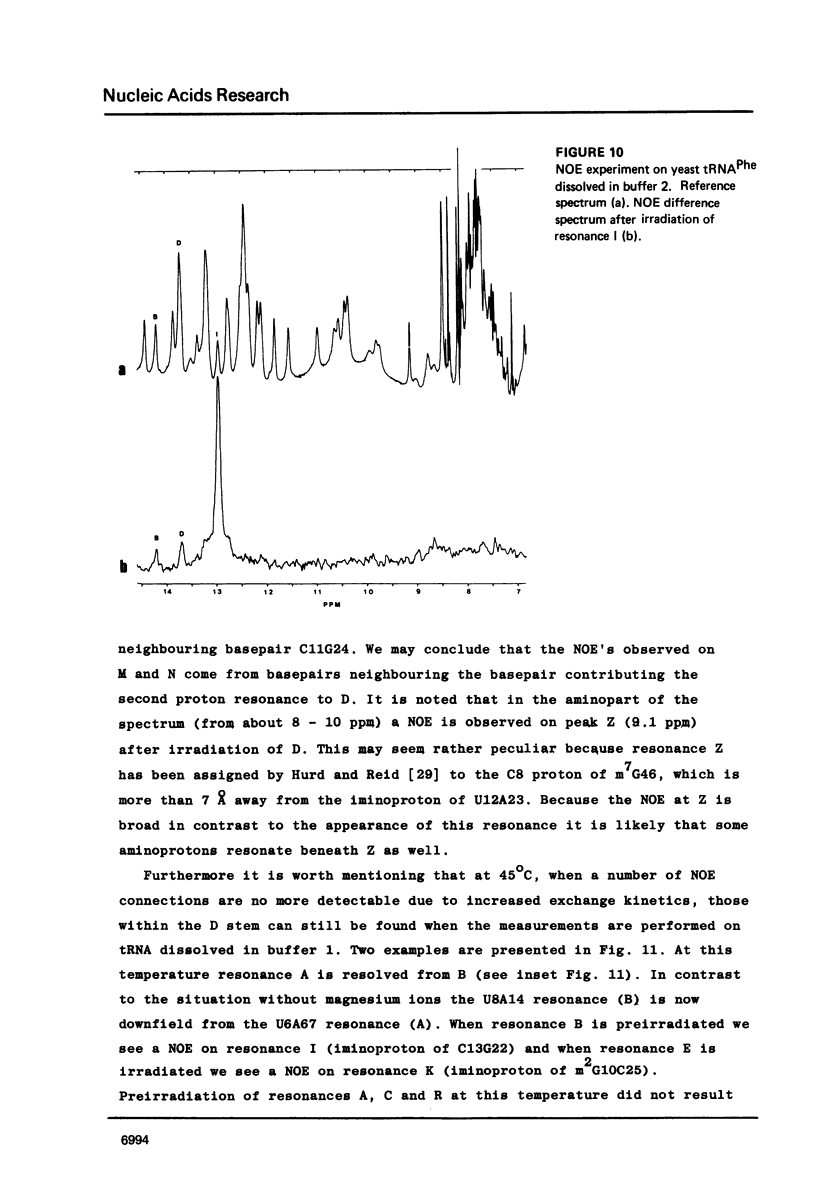
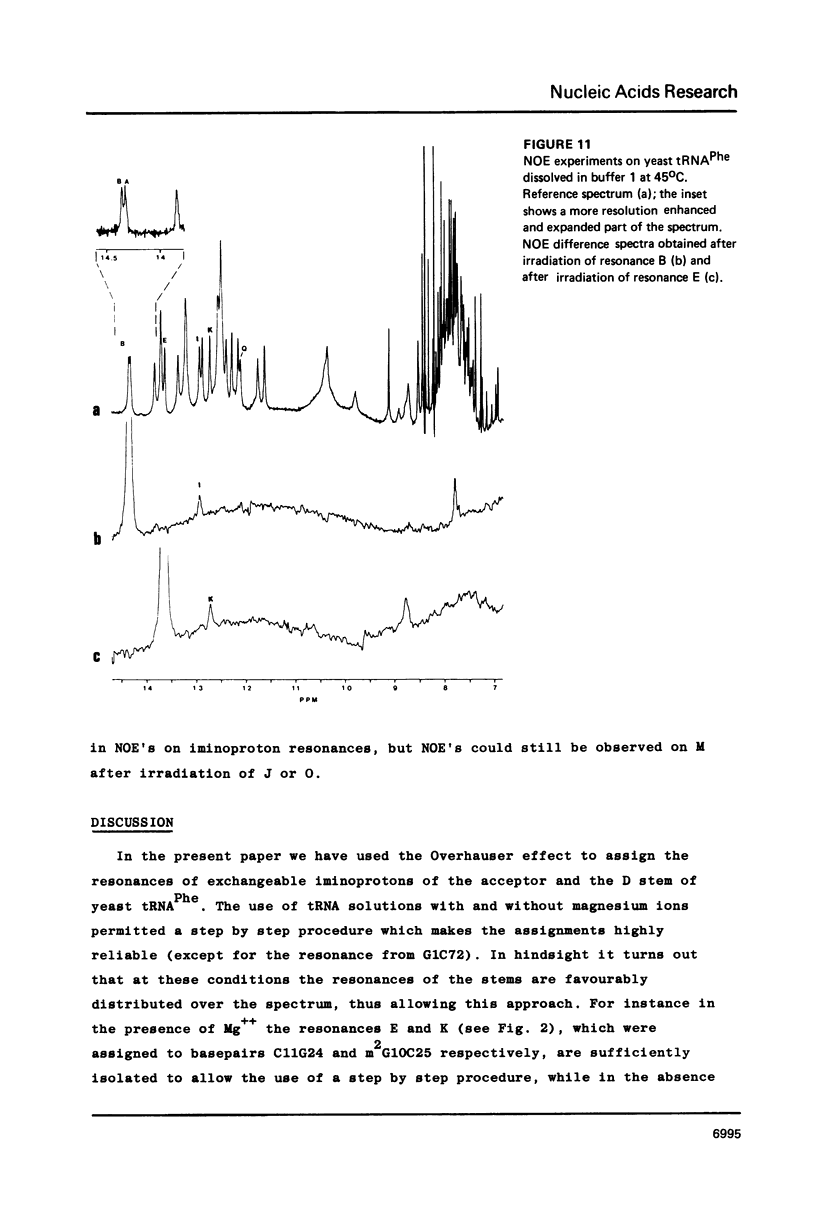
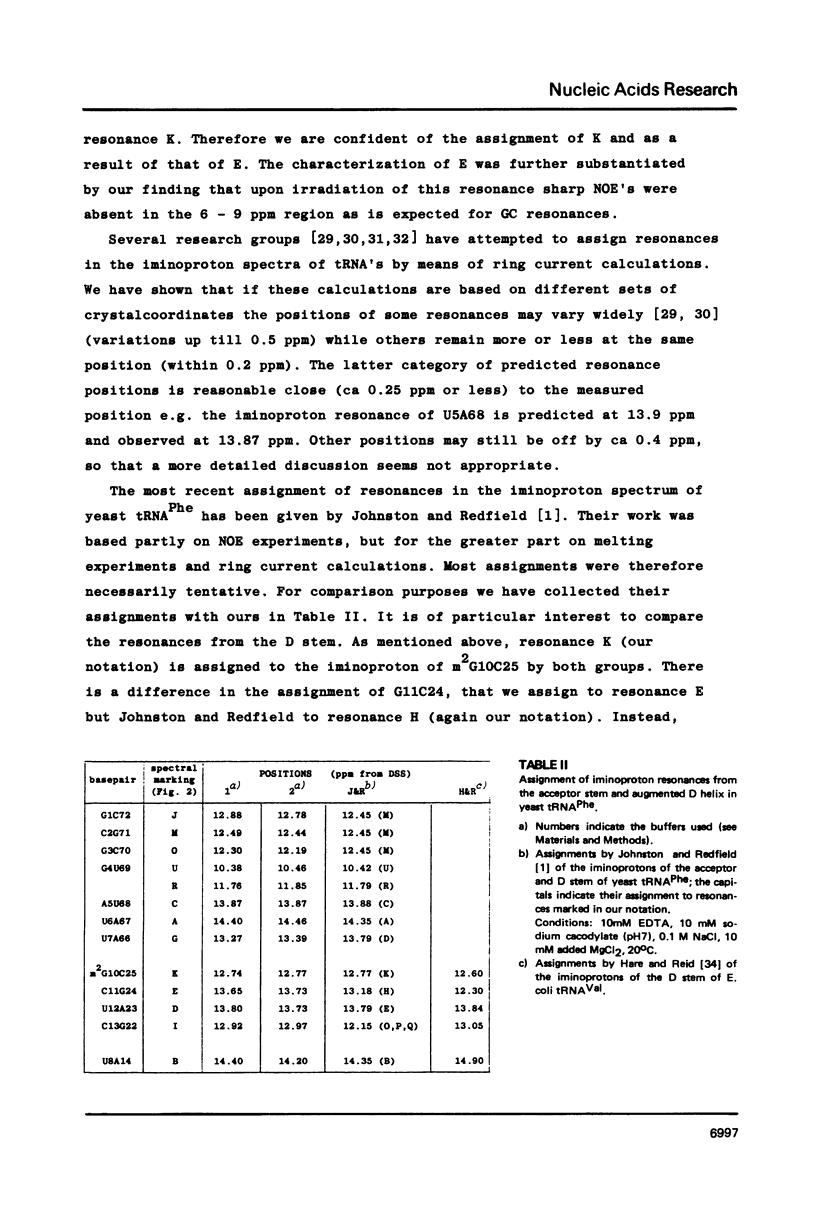
Resonances of the water exchangeable iminoprotons of the acceptor and D stem of yeast tRNAPhe have been assigned by means of Nuclear Overhauser Effects (NOE's). Assignments were made for spectra recorded from tRNA dialysed against a buffer with 110 mM sodium and 5 mM magnesium ions and against a buffer with 430 mM sodium and no magnesium ions. Remarkable is the assignment of a resonance at 13.6 - 13.7 ppm to the iminoproton of C11G24. This assignment as well as those of G1C72, G3C70, U7A66, U12A23 and C13G22 are different from those made previously on the basis of less direct evidence. NOE experiments performed at 45 degrees C support the view that the D stem together with the tertiary interaction U8A14 is one of the most stable parts of the molecule in the presence of magnesium ions. A comparison of the spectra recorded under the two different buffer conditions shows that an excess of 320 mM sodium ions is not capable to force the tRNA in the same conformation as 5 mM magnesium ions can do.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alma N. C., Harmsen B. J., Hull W. E., van der Marel G., van Boom J. H., Hilbers C. W. Double-resonance experiments at 500 MHz on gene-5 protein and its complex with octadeoxyriboadenylic acid. Biochemistry. 1981 Jul 21;20(15):4419–4428. doi: 10.1021/bi00518a029. [DOI] [PubMed] [Google Scholar]
- Cornelis A. G., Haasnoot J. H., den Hartog J. F., de Rooij M., van Boom J. H., Cornelis A. Local destabilisation of a DNA double helix by a T--T wobble pair. Nature. 1979 Sep 20;281(5728):235–236. doi: 10.1038/281235a0. [DOI] [PubMed] [Google Scholar]
- Davanloo P., Sprinzl M., Cramer F. Proton nuclear magnetic resonance of minor nucleosides in yeast phenylalanine transfer ribonucleic acid. Conformational changes as a consequence of aminoacylation, removal of the Y base, and codon--anticodon interaction. Biochemistry. 1979 Jul 24;18(15):3189–3199. doi: 10.1021/bi00582a001. [DOI] [PubMed] [Google Scholar]
- Dubs A., Wagner G., Wüthrich K. Individual assignments of amide proton resonances in the proton NMR spectrum of the basic pancreatic trypsin inhibitor. Biochim Biophys Acta. 1979 Mar 27;577(1):177–194. doi: 10.1016/0005-2795(79)90020-5. [DOI] [PubMed] [Google Scholar]
- Geerdes H. A., Hilbers C. W. Ring current shifts in GU base pairs. FEBS Lett. 1979 Nov 1;107(1):125–128. doi: 10.1016/0014-5793(79)80478-0. [DOI] [PubMed] [Google Scholar]
- Geerdes H. A., Hilbers C. W. The iminoproton NMR spectrum of yeast tRNA-Phe predicted from crystal coordinates. Nucleic Acids Res. 1977 Jan;4(1):207–221. doi: 10.1093/nar/4.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot C. A., den Hartog J. H., de Rooij J. F., van Boom J. H., Altona C. Loopstructures in synthetic oligodeoxynucleotides. Nucleic Acids Res. 1980 Jan 11;8(1):169–181. doi: 10.1093/nar/8.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare D. R., Reid B. R. Direct assignment of the dihydrouridine-helix imino proton resonances in transfer ribonucleic acid nuclear magnetic resonance spectra by means of the nuclear Overhauser effect. Biochemistry. 1982 Apr 13;21(8):1835–1842. doi: 10.1021/bi00537a020. [DOI] [PubMed] [Google Scholar]
- Hilbers C. W., Shulman R. G., Kim S. H. High resolution NMR study of the melting of yeast tRNA Phe. Biochem Biophys Res Commun. 1973 Dec 10;55(3):953–960. doi: 10.1016/0006-291x(73)91235-7. [DOI] [PubMed] [Google Scholar]
- Hingerty B., Brown R. S., Jack A. Further refinement of the structure of yeast tRNAPhe. J Mol Biol. 1978 Sep 25;124(3):523–534. doi: 10.1016/0022-2836(78)90185-7. [DOI] [PubMed] [Google Scholar]
- Hurd R. E., Reid B. R. Nuclear magnetic resonance studies on the tertiary folding of transfer ribonucleic acid: assignment of the 7-methylguanosine resonance. Biochemistry. 1979 Sep 4;18(18):4017–4024. doi: 10.1021/bi00585a027. [DOI] [PubMed] [Google Scholar]
- Hurd R. E., Reid B. R. Nuclear magnetic resonance studies on transfer ribonucleic acid: assignment of AU tertiary resonances. Biochemistry. 1979 Sep 4;18(18):4005–4011. doi: 10.1021/bi00585a025. [DOI] [PubMed] [Google Scholar]
- Johnston P. D., Figueroa N., Redfield A. G. Real-time solvent exchange studies of the imino and amino protons of yeast phenylalanine transfer RNA by Fourier transform NMR. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3130–3134. doi: 10.1073/pnas.76.7.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston P. D., Redfield A. G. An NMR study of the exchange rates for protons involved in the secondary and tertiary structure of yeast tRNA Phe. Nucleic Acids Res. 1977 Oct;4(10):3599–3615. doi: 10.1093/nar/4.10.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston P. D., Redfield A. G. Nuclear magnetic resonance and nuclear Overhauser effect study of yeast phenylalanine transfer ribonucleic acid imino protons. Biochemistry. 1981 Mar 3;20(5):1147–1156. doi: 10.1021/bi00508a016. [DOI] [PubMed] [Google Scholar]
- Johnston P. D., Redfield A. G. Pulsed FT-NMR double resonance studies of yeast tRNAPhe: specific nuclear Overhauser effects and reinterpretation of low temperature relaxation data. Nucleic Acids Res. 1978 Oct;5(10):3913–3927. doi: 10.1093/nar/5.10.3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston P. D., Redfield A. G. Study of transfer ribonucleic acid unfolding by dynamic nuclear magnetic resonance. Biochemistry. 1981 Jul 7;20(14):3996–4006. doi: 10.1021/bi00517a008. [DOI] [PubMed] [Google Scholar]
- Kan L. S., Ts'o P. O. 1H NMR studies of transfer RNA III: the observed and the computed spectra of the hydrogen-bonded NH resonances of baker's yeast transfer-RNA Phe. Nucleic Acids Res. 1977;4(5):1633–1647. doi: 10.1093/nar/4.5.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan L. S., Ts'o P. O., Sprinzl M., vd Harr F., Cramer F. 1H nuclear magnetic resonance studies of transfer RNA: the methyl and methylene resonances of baker's yeast phenylalanine transfer RNA and its fragments. Biochemistry. 1977 Jul 12;16(14):3143–3154. doi: 10.1021/bi00633a017. [DOI] [PubMed] [Google Scholar]
- Komoroski R. A., Allerhand A. Natural-abundance carbon-13 Fourier-transform nuclear magnetic resonance spectra and spin lattice relaxation times of unfractionated yeast transfer-FNA. Proc Natl Acad Sci U S A. 1972 Jul;69(7):1804–1808. doi: 10.1073/pnas.69.7.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen F. M., Hoch J. C., Dobson C. M. A structural study of the hydrophobic box region of lysozyme in solution using nuclear Overhauser effects. Biochemistry. 1980 Jun 10;19(12):2597–2607. doi: 10.1021/bi00553a011. [DOI] [PubMed] [Google Scholar]
- Robillard G. T., Tarr C. E., Vosman F., Berendsen H. J. Similarity of the crystal and solution structure of yeast tRNAPhe. Nature. 1976 Jul 29;262(5567):363–369. doi: 10.1038/262363a0. [DOI] [PubMed] [Google Scholar]
- Roy S., Redfield A. G. Nuclear Overhauser effect study and assignment of D stem and reverse-Hoogsteen base pair proton resonances in yeast tRNAAsp. Nucleic Acids Res. 1981 Dec 21;9(24):7073–7083. doi: 10.1093/nar/9.24.7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt P. G., Tompson J. G., Agris P. F. Transfer RNA structure by carbon NMR: C2 of adenine, uracil and cytosine. Nucleic Acids Res. 1980 Feb 11;8(3):643–656. doi: 10.1093/nar/8.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman J. L., Holbrook S. R., Warrant R. W., Church G. M., Kim S. H. Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement. J Mol Biol. 1978 Aug 25;123(4):607–630. doi: 10.1016/0022-2836(78)90209-7. [DOI] [PubMed] [Google Scholar]
- Sánchez V., Redfield A. G., Johnston P. D., Tropp J. Nuclear Overhauser effect in specifically deuterated macromolecules: NMR assay for unusual base pairing in transfer RNA. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5659–5662. doi: 10.1073/pnas.77.10.5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropp J., Redfield A. G. Environment of ribothymidine in transfer ribonucleic acid studied by means of nuclear Overhauser effect. Biochemistry. 1981 Apr 14;20(8):2133–2140. doi: 10.1021/bi00511a010. [DOI] [PubMed] [Google Scholar]


