INTRODUCTION
Upon agonist stimulation, phospholipase C metabolizes phosphatidylinositol 4,5-bisphosphate into the intracellular second messengers 1,2-diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (IP3) (Berridge, 1993; Hokin, 1985; Majerus, 1992; Mikoshiba, 1997; Nishizuka, 1986). The PLC-generated second messenger DAG is well known as an activator of protein kinase C, while IP3 regulates cellular calcium flux by binding to IP3 receptors and subsequently induce the release of Ca2+ from internal stores. IP3 is metabolized to form a number of more polar inositol phosphates (IPs) and diphosphoryl inositol phosphates (PP-IPs), also commonly referred to as inositol pyrophosphates (Irvine and Schell, 2001; Majerus, 1992; Shears, 1998; York, 2006). In the last fifteen years, there has been a rapid accumulation of knowledge defining the metabolic pathways of these molecules, the enzymes involved in their production, and their biological roles (Figure 1). Four kinases have been identified as conserved in nearly all eukaryotes from budding yeast to man, and two additional enzymes have been found in selected metazoans and plants. These gene products may be categorized into two classes: 1) kinases that produce inositol phosphomonoesters and 2) those that produce inositol diphosphates. The cloning and cellular studies of these gene products has led to significant increases in our knowledge of the metabolic pathways of these molecules and their biological roles in eukaryotes from budding yeast Saccharomyces cerevisiae to man. Here we discuss a subset of recent advances that highlight roles for IP and PP-IP molecules in cellular processes and organism development.
FIGURE 1. INOSITOL PHOSPHATE AND PYROPHOSPHATE PATHWAYS.
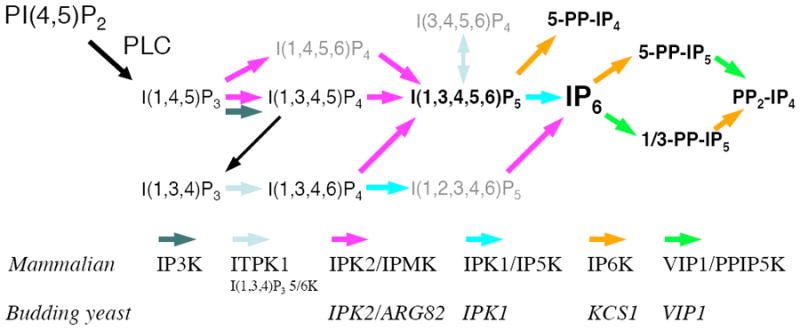
Activation of phosphoinositide-specific phospholipase C (PLC) triggers the conversion of phosphatidylinositol 4,5-bisphosphate, PI(4,5)P2, to the second messengers I(1,4,5)P3 and 1,2-diacylglycerol (not shown). Metabolism of I(1,4,5)P3 occurs to generate numerous inositol phosphate and inositol pyrophosphate chemical codes. There are several evolutionarily conserved inositol phosphate kinases that contribute to the production of these regulatory molecules: in mammals there are 6 distinct kinase activities, whereas in the budding yeast there are 4 gene products. The commonly used gene name abbreviations are listed. Inositol phosphatases are omitted for clarity except for the black arrow linking I(1,3,4,5)P4 and I(1,3,4)P3, which is encoded by a 5-phosphatase INPP5.
ENZYME REGULATION OF IP AND PP-IP SYNTHESIS IN BUDDING YEAST
For simplicity, we will begin our discussion of the routes of inositol phosphate metabolism in the single cell eukaryote, S. cerevisiae. The budding yeast genome encodes four kinases responsible for synthesizing an array of IP and PP-IP species (Figure 1) (Mulugu et al., 2007; Odom et al., 2000; Saiardi et al., 2000a; Saiardi et al., 2000b; York et al., 1999). In the budding yeast, Ipk2 is the first enzyme required for PLC dependent synthesis of IPs, and is a dual-specificity IP3/IP4 6-/3-kinase that converts inositol 1,4,5-trisphosphate [I(1,4,5)P3 or IP3] to inositol 1,3,4,5,6-pentakisphophate [I(1,3,4,5,6)P5 or IP5] via the intermediate inositol 1,4,5,6-tetrakisphosphate [I(1,4,5,6)P4 or IP4]. IP5 is subsequently metabolized by another kinase Ipk1, an IP5 2-kinases, to inositol 1,2,3,4,5,6-hexakisphosphate [I(1,2,3,4,5,6)P6 or IP6]. Inositol pyrophosphates [PP- IP4, PP-IP5 (IP7), PP2-IP3 and PP2- IP4 (IP8)] are generated by two kinases called Kcs1 and Vip1 in yeast. Kcs1 and Vip1 function as both inositol hexakisphosphate (IP6K) and inositol heptakisphosphate kinases (PPIP5K). They generate different IP7 isomers: Kcs1 make 5-IP7 and Vip1 makes 1/3-IP7 (Lin et al., 2009) – 1/3 designates enantiomers that have not yet been resolved and it is possible that Vip1 makes 1-, 3- or both isomers of IP7. Kcs1 and Vip1 collectively work together to generate IP8 from IP6.
Except for Vip1, the enzymes involved in IPs and PP-IPs can be found in the nucleus. Yeast PLC1 has been shown to associate with the kinetochore, and nuclear PLC isoforms have been identified in mammals (Cocco et al., 1996; Lin et al., 2000; Manzoli et al., 1995; Maraldi et al., 1995). Ipk2 in yeast and other organisms is predominantly nuclear (Bercy et al., 1987; Fujii and York, 2005; Nalaskowski et al., 2002; Odom et al., 2000; Seeds et al., 2004; Xia et al., 2003). Yeast Ipk1 resides in the nuclear envelope (York et al., 1999). Kcs1 and its mammalian homologs IP6Ks have both nuclear and cytoplasmic distributions (Luo et al., 2002; Saiardi et al., 2000a; Saiardi et al., 2000b). The nuclear presence of these enzymes suggests IPs and PP-IPs have nuclear functions; indeed studies in yeast demonstrated they are involved in diverse nuclear processes such as transcriptional control, mRNA export and telomere length regulation (Alcazar-Roman and Wente, 2008; York, 2006; York et al., 2001). In recent years, much progress has been made in understanding the biology of IPs and PP-IPs in other organisms by employing a reverse genetics approach.
METABOLISM OF IP AND PP-IP SPECIES IN PLANTS, FLIES AND MAMMALS
The genes involved in IPs and PP-IPs synthesis are evolutionary conserved from yeast to man, however in mammals there are two additional classes of kinase as well as gene duplications for several of the six kinases (Figure 1). In eukaryotic species examined to-date, all appear to require a Ipk2 homolog (also known as inositol phosphate multi-kinase, IPMK) and a Ipk1 homolog to produce IP5 and IP6 (Chang et al., 2002; Frederick et al., 2005; Fujii and York, 2005; Nalaskowski et al., 2002; Saiardi et al., 2001; Seeds et al., 2004; Stevenson-Paulik et al., 2005; Stevenson-Paulik et al., 2002; Verbsky et al., 2005a; Verbsky et al., 2005b; Verbsky et al., 2002; Xia et al., 2003). However, additional IPKs are identified in higher organisms, which represent alternative biosynthetic routes to IP6. In plants and human cells, I(1,3,4)P3 5/6-kinases have been identified and disruption of these 5/6-kinases decreased IP6 levels in maize and human cells (Shi et al., 2003; Verbsky et al., 2005b; Wilson and Majerus, 1997). I(1,3,4)P3 5/6-kinase (alias ITPK1) has also been shown to regulate chloride channel activity via novel 1-kinase activity towards I(3,4,5,6)P4 and I(1,3,4,5,6)P5 1-phosphatase (Ho et al., 2002; Yang and Shears, 2000). The structures of I(1,3,4)P3 5/6-kinase/ITPK1 have illuminate the bases for these activities (Chamberlain et al., 2007; Miller et al., 2005). On the other hand, I(1,4,5)P3 3-kinases have been identified in mammals and Drosophila. There is no evidence that these 3-kinases are required for IP6 synthesis in mouse cells or Drosophila (Leyman et al., 2007; Seeds et al., 2004). However, an alternative route to generate IP6 involving 5/6-kinase and 3-kinase has been elucidated in human cells (Majerus, 1992; Menniti et al., 1993; Verbsky et al., 2005b). In this pathway, I(1,4,5)P3 is first phosphorylated by IP3 3-kinase to I(1,3,4,5)P4, which is then dephosphorylated by 5-Ptase to I(1,3,4)P3. I(1,3,4)P3 is subsequently metabolized to I(1,3,4,6)P4 by IP3 5/6-kinase. I(1,3,4,6)P4 is sequentially phosphorylated by Ipk2 and Ipk1 to make IP5 and finally IP6. In slime mold and duckweed, an inositol lipid-independent pathway of IP6 synthesis has been proposed (Biswas et al., 1978; Brearley and Hanke, 1996; Stephens and Irvine, 1990), in which I(3)P is generated from glucose 6-phosphate by inositol 3-phosphate synthase (MIPS). I(3)P is successively phosphorylated to I(3,4)P2, I(3,4,6)P3, I(3,4,5,6)P4, I(1,3,4,5,6)P5 and finally to IP6. The enzymology involved in this pathway is not fully understood at this point. The existence of multiple biosynthetic routes to IP6 may increase signaling versatility and redundancy in higher organisms.
BIOLOGICAL ROLES OF IP AND PP-IP MOLECULES IN YEAST
A major role of Ipk2 and its products, IP4 and IP5, is transcriptional regulation in response to changes in environmental and nutritional cues. IPK2 is allelic to ARG82, and is a component of the arginine responsive ArgR-Mcm1 complexes. Ipk2 and its kinase activity have been shown to be required for formation of the active ArgR-Mcm1 complexes which controls the transcription regulation of arginine responsive genes that are required for the utilization of arginine or ornithine as the only nitrogen source in budding yeast (Bechet et al., 1970; El Alami et al., 2003; Messenguy and Dubois, 1993; Odom et al., 2000). On the other hand, Ipk2 has been shown to be required for the induction of transcription of some phosphate responsive genes (e.g. Pho5) by modulation of the chromatin remodeling complexes SWI/SNF and INO80, under high phosphate conditions (El Alami et al., 2003; Steger et al., 2003). In vitro, IP4 and IP5 have been shown to stimulate nucleosome mobilization by the yeast SWI/SNF complex (Shen et al., 2003). Recently, Vancura’s group showed that Plc1 and IPs are involved in regulation of activity of the kinetochore by facilitating recruitment of another chromatin remodeling complex, RSC, which stands for Remodels Structure of Chromatin. RSC has been shown to be required for maintaining the structure of the centromere and for proper kinetochore function (Cairns et al., 1996; Desai et al., 2009).
The yeast Ipk1 was first identified as a regulator of mRNA export in a synthetic lethal screen with the gle1-2 mutant, where Gle1 is an essential factor associated with the nuclear pore complex (NPC) required for mRNA export (Murphy and Wente, 1996; York et al., 1999). It was subsequently shown that the product of IPK1, IP6, and Gle1 regulates mRNA export through the binding to and synergistic activation of the RNA-dependent ATPase activity of a nuclear pore-associated DEAD-box protein Dbp5, which is essential for RNA export (Alcazar-Roman et al., 2006; Snay-Hodge et al., 1998). Recently, it has also been proposed that IP6, Gle1 and DbP5 cooperate to control translation termination (Bolger et al., 2008).
KCS1 was initially identified as a suppressor of mutation of the protein kinase C gene (Huang and Symington, 1995). kcs1 null cells have small and fragmented vacuoles (Saiardi et al., 2000b) and is required for resistance to salt stress, cell wall integrity and vacuole morphogenesis (Dubois et al., 2002). These phenotypes maybe due to the defect in endocytosis observed in the kcs1 mutant (Saiardi et al., 2002). In addition, Kcs1 activity modulates telomere homeostasis such that loss and gain of PP-IP4 in cells increase and decrease average telomere length, respectively (Saiardi et al., 2005; York et al., 2005). The affect of PP-IP4 on telomere length requires the presence of Tel1 but does not depend on Ku70. It is not clear whether or not these pathways mechanistically overlap with a putative role of Kcs1 in regulating of DNA hyper-recombination (Luo et al., 2002).
Vip1 was identified by a biochemical approach to look for the enzyme responsible for the remaining IP6 kinase activity in the kcs1 ddp1 double mutant cells (Mulugu et al., 2007; York et al., 2005). In addition to a kinase domain, Vip1 harbors a C-terminal phosphatase domain of unidentified function (Fridy et al., 2007; Mulugu et al., 2007). In the budding yeast, Vip1-generated 1(3)-IP7, but not Kcs1-gerenated 5-IP7, acts as a signaling molecule regulating the yeast phosphate (Pi)-responsive (PHO) signaling pathway (Lee et al., 2007). 1(3)-IP7 binds to the CDI-Cyclin-CDK complex Pho81-Pho80-Pho85 and promote Pho81-dependent inactivation of Pho80-Pho85, leading to dephosphorylation and nuclear accumulation of the transcription factor of Pho4 and subsequent transcription of the PHO genes (Lee et al., 2008; York and Lew, 2008). On the other hand, in the fission yeast S. pombe, the homolog of Vip1, Asp1, has been suggested to play a role in regulating actin-related protein complexes (ARP) (Feoktistova et al., 1999). Over-expression of a kinase only mutant of Asp1 is toxic to the cells and the toxicity can be alleviated in an arp3 mutant background, strongly suggesting Vip1 generated 1(3)-IP7 functionally interact with the Arp complex (Mulugu et al., 2007).
PP-IPs have also been proposed to be phosphate donor in non-enzymatic phosphorylation of proteins in eukaryotic cells (Bhandari et al., 2007; Draskovic et al., 2008; Saiardi et al., 2004). It is thought that the beta-phosphate from 5-PP-IP5 is non-enzymatically donated to a “pre-primed” phosphoserine residue on the recipient protein thereby generating a protein-serine-pyrophosphate and IP6 as products (Bhandari et al., 2007). A wide range of proteins have been implicated as recipients (Azevedo et al., 2009). Elucidation of the biological roles of this pathway is an area of active research, although to our knowledge it has not yet been shown by physical methods that serine-pyrophosphate modified recipient proteins exist in cells.
ROLES OF IP MESSENGERS IN STRESS/HORMONE SIGNALING AND DEVELOPMENT IN PLANTS
In the plant Arabidopsis thaliana, two homologs of the yeast Ipk2 has been identified to-date, i.e. AtIpk2α and AtIpk2β (Stevenson-Paulik et al., 2005; Stevenson-Paulik et al., 2002; Xia et al., 2003). Different genes located on chromosome 5 encode AtIpk2α and AtIpk2β. Despite their low overall sequence identities to the yeast Ipk2 (12-18%), purified recombinant AtIPK2α and AtIpk2β are inositol phosphate 6/3/5-kinases, capable of generating I(1,3,4,5,6)P5 (IP5) from I(1,4,5)P3 (IP3) predominately through an I(1,4,5,6)P4 (IP4) intermediate. Expression of either AtIpk2 can restore IP4 and IP5 synthesis in the ipk2 null yeast strain and complement its temperature sensitivity, indicating AtIpk2α and AtIpk2β are true functional homologs of the yeast Ipk2 (Stevenson-Paulik et al., 2002).
Both AtIpk2α and AtIpk2β are ubiquitously expressed in the leaves, stem, roots, siliques and flowers in adult plants by semi-quantitative PCR (Stevenson-Paulik et al., 2002). In addition, using a reporter of AtIpk2α expression in which the 1.5kb 5’ upstream region of the gene is fused to the E. coli GUS reporter gene, the Xue lab found that AtIpk2α is also expressed in the emerging seedlings, seedlings at different developmental stages, roots, pollen grains and growing pollen tubes (Xia et al., 2003; Xu et al., 2005). Also, a strong expression is detected in anthers and the stigma in flowers using the GUS reporter. When expressed in onion epidermal cells, AtIpk2α-GFP localizes in the nucleus as well as the plasma membrane, suggesting nuclear roles for this protein. Transgenic plants expressing an AtIpk2α anti-sense using its own promoter were made which results in a strong reduction in expression of AtIpk2α while the transcript level of AtIpk2β is unaffected. Interestingly, the homozygous AtIpk2α anti-sense transgenic plants exhibit higher germination frequencies, increase in pollen tube lengths and enhanced root growth and early seedling growth, consistent with the expression of AtIpk2α in these tissues. These data indicated AtIpk2α impacts on plant development by acting as negative regulator of pollen germination and root growth. As the Xue group has shown that exogenous application of IP3 can enhance root growth, the phenotypic consequences of inhibition of AtIpk2α may in part due to an accumulation of IP3 in addition to depletion of IP4 and IP5.
The expression of the other plant Ipk2 homolog, AtIpk2β, can be induced by abiotic stress (Yang et al., 2008). Transcript levels of AtIpk2β increase when subject to cold or drought stress, and decrease upon high salt or ABA addition, and AtIpk2β protein levels increase in response to mannitol. In order to investigate the role of AtIpk2β in stress signaling in plants, the Zhang’s lab generated transgenic tobacco plants constitutively expressing the AtIpk2β gene. Expression of AtIpk2β does not cause any discernable change in plant morphology. However, the AtIpk2β -expressing plants exhibit an enhanced tolerance to various kinds of stress: they are more resistant to root and seedlings growth retardation caused by high salt concentrations, osmotic stress induced by PEG, drought conditions, H2O2-induced damage and short term -20°C treatment. The observed increase tolerance can partly be explained by an increase in proline levels in the transgenic plant. Proline was reported to be involved in free radical scavenger (Hong et al., 2000) and in protection of macromolecules from dehydration (Yancey, 2005). Also, elevation of catalase activity in the transgenic plant can offer addition protection from stress-induced reactive oxygen species. More importantly, the constitutive expression of AtIpk2β results in the constitutive or increased transcription of stress responsive genes such as lipid transfer protein, fructose-bisphosphate aldolase and raffinose synthase family protein/ seed inhibition protein. The induction of stress-response genes is considered as an important step in stress adaptation in plants (Thomashow, 1999). Transgenic plants expressing some of the stress responsive genes result in enhanced tolerance in stress (Smart et al., 2000; Yamada et al., 2000). Taken together, the results suggest a nuclear role of AtIpk2β in transducing abiotic stress-induced signals by transcriptional regulation of stress responsive genes. Whether AtIpk2β impacts on transcriptional regulation of stress-inducible genes in plant, by acting as a component of a transcriptional complex, or as modulator of chromatin structure just like the roles of yeast Ipk2 in arginine responsive and phosphate arginine gene expression (El Alami et al., 2003; Odom et al., 2000; Shen et al., 2003; Steger et al., 2003), or by an entirely novel mechanism, is currently unclear.
Surprisingly, over-expression of AtIpk2β does not confer stress tolerance in Arabidopsis (Yang et al., 2008), suggesting a different role AtIpk2β play in this organism. In fact, the Xia group observed that over-expression of AtIpk2β in Arabidopsis results in increase in branching of axillary shoots and hence increased axillary branches in mature plants (Zhang et al., 2007). Axillary shoot branching requires the formation of axillary meristems and the outgrowth of axillary buds, and in the AtIpk2β over-expressing lines both developmental processes are speeded up. Auxin is an important plant hormone that modulates growth in response to a plethora of developmental and environmental cues. Also, auxin has been shown to be important regulators of axillary shoot branching (Teale et al., 2006; Woodward and Bartel, 2005). The major natural occurring auxin, indole-3-acetic acid (IAA), has been shown to inhibit axillary bud growth and hence axillary shoot branching (Chatfield et al., 2000; Napoli et al., 1999). Indeed, Xia group found that AtIpk2β regulates axillary shoot branching via auxin signaling. The expression patterns of AtIpk2β and auxin transcriptional reporters are similar. Moreover, expression of AtIpk2β can be induced with addition of exogenous IAA in a dose-dependent manner, indicating AtIPK2β is an IAA responsive gene. IAA does not affect expression of AtIpk2α. Over-expression of AtIpk2β can alleviate the inhibition of elongation of primary roots by IAA, demonstrating AtIpk2β can negatively regulate IAA signaling. This regulation appears to be at least partly transcriptional, as over-expression of AtIpk2β results in decreased expression of one of the auxin biosynthetic gene CYP83B1, increase in expression of an auxin-transport gene PIN4 and, more importantly, decrease in transcription of the auxin inducible gene MAX4, which are required for auxin-mediated bud inhibition, and a cytochrome P450 homolog SPS which is required for suppression of axillary meristem initiation (Bainbridge et al., 2005; Tantikanjana et al., 2001). This study demonstrated another nuclear role for AtIpk2β in negative regulation of auxin hormone signaling by transcriptional control of some of the auxin responsive genes.
A recent structural study of the Arabidopsis ubiquitin ligase complex TIR1-ASK1 (Tan et al., 2007) provides a hint of the mechanism by which IPs may regulate auxin signaling transcriptionally. Transduction of auxin signaling is regulated by ubiquitin dependent proteolytic system. TIR1, which stands for transport inhibitor response protein 1, is a F-box protein and a subunit of the Skp1-Cullin-F-box protein (SCFTIR1) ubiquitin ligase complex. ASK1 (Arabidopsis SKP1) is the adaptor subunit of the SCFTIR1 complex and links TIR1 to the E3 ubiquitin ligase complex. Components of the SCFTIR1 complex, including TIR1, have been shown to localize in the nucleus in Arabidopsis tonoplast culture (Tao et al., 2005). TIR1 has been shown to be a bona fide auxin receptor (Dharmasiri et al., 2005; Kepinski and Leyser, 2004; Kepinski and Leyser, 2005). Upon binding to the SCFTIR1 complex, auxin promotes the interaction between the TIR1 subunit and the transcription repressor Aux/IAA proteins. Subsequently Aux/IAA proteins are poly-ubiquitinated by SCFTIR1 and then degraded via the 26S proteasome pathway. Proteolysis of Aux/IAA proteins relieves the inhibition of the auxin response factor (ARF) family of transcription factor by Aux/IAA proteins. The activated ARFs then turn on the transcription of the auxin responsive genes (Hagen and Guilfoyle, 2002; Liscum and Reed, 2002; Reed, 2001; Tiwari et al., 2001; Zenser et al., 2001). A surprising finding in Tan et al. is that an IP6 molecule was found to bind to the leucine-rich repeat domain of TIR1, adjacent to the auxin-binding site. IP6 appears to be a structural co-factor for TIR1 and its role in TIR1 function remain to be elucidated. However, one attractive hypothesis is that IP6 (or presumably other IPs), by binding to TIR1, regulates either auxin binding to the TIR1 or the ubiquitin ligase activity of the complex, and modulates the subsequent binding and degradation of Aux/IAA proteins, hence regulating the strength of the auxin signaling output.
AtIpk1 is the Arabidopsis Ipk1 homolog, identified by homology search using several fungal Ipk1 protein sequences as queries (Stevenson-Paulik et al., 2005). Recombinant AtIpk1 can phosphorylate I(1,3,4,5,6)P5 to IP6. It also functions as a 2-kinase towards I(1,3,4,6)P4 and I(1,4,5,6)P4 enabling the production of I(1,2,3,4,6)P5 and I(1,2,4,5,6)P5. Expression of AtIpk1 in the yeast ipk1 null restores IP6 synthesis and rescues the temperature sensitivity of the mutant, indicating AtIpk1 is able to make IP6 in vivo (Sweetman et al., 2006). An AtIPK1 mutant, named atipk1-1 was identified in the Salk T-DNA collection. atipk1-1 contains a T-DNA insertion 77 nucleotides upstream of the stop codon in the last exon of the ORF. atipk1-1 mutant exhibits more then 70% drops in the AtIPK1 transcript as judged by Northern blot, and the seed phytate (IP6) levels in the atipk1-1 mutant are reduced by 83%, demonstrating AtIpk1 plays a critical role in synthesis of IP6 in plant. atipk1-1 is hypersensitive to inorganic phosphate levels in the growth medium, which are optimal for the wild type plant. The atipk1-1 mutant leaves, but not the wild type leaves, become abaxially curled (epinastic) when 1mM Pi is present in the medium. In addition, the atipk1-1 mutant exhibits a higher intracellular Pi levels, indicating atipk1-1is unable to maintain a normal phosphate homeostasis. Taken together, it seems that AtIpk1 and its IP products are required for phosphate sensing and homeostasis in plants.
Surprisingly, atipk1-1 mutant plants do not appear to have defects in auxin signaling pathways (Stevenson-Paulik et al., 2005). This is puzzling in light of the binding of IP6 to the auxin receptor, TIR1 as described above. It is possible that the 5-10% of IP6 produced in the atipk1-1 hypomorphic mutants is sufficient to generate enough functional TIR1, thus a complete null in IP6 production may indeed exhibit an auxin phenotype. Alternatively, loss of Ipk1 results in an accumulation of IP5 to levels nearly equivalent to IP6 in wild-type plants, thus it is plausible that IP5 is capable of compensating for the loss of IP6. To this end it is not known whether or not IP5 binds TIR1 with equal affinity. If IP5 serves a compensatory role, then it may be that loss of both AtIpk2α and AtIpk2β would result in a loss of auxin signaling.
IP6 is also implicated in ABA signaling in guard cells (Lemtiri-Chlieh et al., 2000; Lemtiri-Chlieh et al., 2003). Studies by Brearley group showed that IP6 level is rapidly induced by the stress hormone abscisic acid (ABA) in intact guard cells of Solanum tuberosum. Addition of submicromolar amount of IP6 into guard cell protoplasts through patch clamp phenocopies the inhibitory effect of ABA or Ca2+ on the inward rectifying K+ current. Co-administration of a Ca2+ chelator EGTA can alleviate this inhibitory effect by IP6, suggesting it is Ca2+ dependent. Indeed, Brearley group showed that IP6 does induce an increase in intracellular Ca2+, not by Ca2+ influx, but by triggering Ca2+ release from endomembrane store particularly the guard cell vacuole. These data strongly indicate that IP6 mediate ABA signaling in guard cells by mobilizing intracellular Ca2+ store. Similarly, IP4 and IP6 have been shown to regulate Ca2+ channel activity and store-operated Ca2+ influx by modulation of activities of IP3 5-phosphatase, protein phosphatase and adenyl cyclase (Hermosura et al., 2000; Larsson et al., 1997; Yang et al., 2001).
In Arabidopsis, there are four I(1,3,4)P3 5/6-kinase homologs identified to-date, namely AtITPK-1, AtITPK-2, AtITPK-3, and AtITPK-4 (Qin et al., 2005; Shi et al., 2003; Sweetman et al., 2007; Wilson and Majerus, 1997). Only the in vivo functions of AtItpk-1 have been explored (Qin et al., 2005). AtItpk-1 can phosphorylate I(1,3,4)P3 to either I(1,3,4,5)P4 or I(1,3,4,6,)P4 in vitro. In plants, a AtItpk1-GFP fusion protein is localized in the nucleus, where AtItpk1 binds to the COP9 signalosome (CSN), similar to what was observed for the human 5/6- kinase (Sun et al., 2002). The transcription and protein levels of AtItpk-1 are induced by red light (RL). Two T-DNA insertion mutants of AtITPK1, atitpk-1-1 and atitpk-1-2, have been isolated. AtITPK-1 transcript levels are drastically reduced in these mutants as judged by RT-PCR. When grown under pure RL, atitpk-1-1 and atitpk-1-2 exhibit shortened hypocotyls compared to WT, while transgenic plants over-expressing AtItpk-1 show increases in hypocotyls length. This suggests AtItpk-1 modulates photomorphogeneis under red light, possibly through association with the CSN. On the other hand, over-expression of a rice I(1,3,4)P3 5/6-kinase, OsITL1, in tobacco decreases tolerance to high salt during germination and seedling development, indicating OsItl1 may function as a negative regulator of osmotic stress signaling (Niu et al., 2008).
ROLES OF IP AND PP-IP MOLECULES IN EMBRYONIC DEVELOPMENT, FERTILITY AND INSULIN SIGNALING
Mouse Ipk2, alternatively known as inositol phosphate multikinase, has been identified on Chromosome 10 (Frederick et al., 2005). During embryogenesis, the mouse Ipk2 was found to express in the head fold, neural tube, bronchial arches, somites and hind limb bud. The mouse Ipk2 gene was disrupted by homologous recombination, which removed the exon 4 of the gene resulting in a complete loss of function. By metabolic labeling, the homozygous Ipk2 mutant embryo shows no detectable IP6 synthesis, while the levels of inositol lipids phosphatidylinositol, phosphatidylinositol phosphate and phosphatidylinositol 4,5-bisphosphate remain unchanged. Moreover, ES lines from the homozygous Ipk2 mutant exhibits drastic loss in both IP5 and IP6 levels, while the levels of IP3 [I(1,3,4)P3 and I(1,4,5)P3] and IP4 are similar to the WT ES lines. This confirms that the mouse Ipk2 is required for IPs synthesis in vivo. On the other hand, agonist-induced changes in IPs and Ca2+ levels are comparable in the ipk2 mutant ES cells and WT controls. Although Ipk2 is not required for the viability of ES cells, the homozygous Ipk2 mutant is embryonic lethal. Gastrulation seems to be unaffected in the absence of Ipk2. However, starting from embryonic day E8.5, the mutant embryos are developmentally delayed, and become smaller and shorter along the anterior-posterior axis compared to the WT littermate, suggesting a proliferation defect. Addition defects include separation of the allantois and the chorion, lack of somite formation and massive accumulation and folding of neurectoderm in the mid-hind region of the mutant embryos. This study highlights the important roles of Ipk2, and presumably production of IP5 and IP6, in the proper embryonic development in mice.
The mouse Ipk1 homolog, also known as I(1,3,4,5,6)P5 2-kinase, was studied by the Majerus group (Verbsky et al., 2005a). In these studies, Ipk1 was disrupted by a gene trap insertion where exon 1 of Ipk1 is fused to the β-galactosidase gene. While no cell lines have been derived from homozygous null embryos, mouse embryonic fibroblasts (MEF) lines derived from embryos heterozygous for the Ipk1 gene trap exhibit an accumulation IP5 and PP-IP4, indicating a decrease in conversion from IP5 to IP6 when one copy of Ipk1 is disrupted. Homozygous Ipk1 mutant is early embryonic lethal: the mutant embryos died and were reabsorbed before E8.5, demonstrating Ipk1 is essential for early embryonic development. Ipk1 is expressed in neural tube, notochord, somites, yolk sac, heart, cardiac vein, aortic, digestive tract and pharyngeal arches. Although the exact mechanism by which Ipk1 regulates embryonic development is unclear at present, the Majerus group suggested that strong expression of Ipk1 in the yolk sac are consistent with its role in nutrient absorption and delivery to the embryo and production of factors important for developmental patterning. Also, since increased levels of IP6 has been shown to protect HEK293 cells from TNFα- and Fas-induced apoptosis (Verbsky and Majerus, 2005), it may be possible that the lack of IP6 in the Ipk1 mutant leads to uncontrolled apoptosis and hence the embryonic phenotype.
In mammalian cells, inositol hexakisphosphate kinases (IP6Ks, also known as IHPKs) are enzymes that convert IP6 to PP-IPs. In mice, there are three IP6K homologs, IP6K1, 2 and 3. The Snyder group has generated an ip6k1 knockout mice by targeted deletion of exon 6, which encodes the sub-domain required for catalytic activity (Bhandari et al., 2008). IP7 and IP8 productions from IP6 are virtually undetectable in whole cell extract and cytosolic fraction from MEFs derived from homozygous ipk6k1 mutant. Also, there is a drastic reduction in IP7 levels in the mutant MEFs, indicating IP6K1 is the major IP6K1 activity in mice. Homozygous ip6k1 mutant go through embryonic development and reach adulthood. However, males of homozygous mutant are sterile; there is a reduction in numbers of advanced spermatids in the seminiferous tubules and no sperm in the epididymis, demonstrating IP6K1 plays an important role in spermatogenesis. Male mutant mice are smaller than WT and there are reductions in weight in both males and females. Plasma levels of growth factor in the ip6k1 mutant are similar to those of the wild type. On the other hand, there is a 65-70% reduction in plasma insulin levels in the mutant mice. Since it has been shown that IP7 is required for efficient exocytosis of insulin-containing secretory granules in pancreatic β cells (Illies et al., 2007), the drop in insulin level is presumably due to a decrease in insulin secretion. This study demonstrates IP6K1 and IP7 are required for spermatogeneis and regulation of body size and weight through regulating insulin secretion. A study by the Berggren group demonstrated that in pancreatic β cells glucose can stimulate a transient increase in IP6 levels which induce Ca2+ influx by inhibition of protein phosphatase activity (Larsson et al., 1997). In light of data from Illes et al., the increase of IP6 may also lead to an increase of IP7 production and hence an increase in insulin secretion. So IP6K1 may also play a role in glucose sensing and stimuli-secretion coupling in pancreatic β cells. Of note, mutations in IHPK1, the human homolog of IP6K1, were identified in families having type II diabetes (Kamimura et al., 2004), thus implicating a role for IP6K1 in diabetes pathogenesis. Recently, the mouse knockout of IP6K2, referred to as IHPK2, was reported and found to develop normally (Morrison et al., 2009). Interestingly, chronic treatment of IP6K2 mutant mice with a UV-mimetic 4-nitroquinoline 1-oxide (4-NQO) results in a significant increase in the incidence of invasive squamous cell carcinoma (SCC), while also exhibiting resistance to ionizing radiation (Morrison et al., 2009). Thus the roles of IP6Ks remain an active and intriguing area of study.
ZEBRAFISH IPK1 REGULATES LEFT-RIGHT ASYMMETRY
The zebrafish Ipk1 is 57% identical to the human Ipk1, and can restore the IP6 synthesis in the yeast ipk2 null and complemented the synthetic lethality of the yeast gle1-2 ipk1-4 double mutant, indicating it is a functional IP5 2-kinase (Sarmah et al., 2005). Wente group then carefully examined the phenotype in zebrafish embryos in which Ipk1 is effectively knockdown by antisense morpholinos, and found that Ipk1 is involved in left-right (LR) asymmetry in zebrafish. In vertebrate embryos, internal organs show a conserved LR asymmetry, which is critical for proper development (McGrath et al., 2003; Webb and Miller, 2003). In zebrafish, a structure called Kupffer’s vesicle (KV) has been shown to play a role in generating LR asymmetry by rotational movement of its motile cilia (Essner et al., 2005; Essner et al., 2002). Among other factors, Ca2+ has also been implicated in LR asymmetry generation in mice and chicken (McGrath et al., 2003; Raya and Izpisua Belmonte, 2004). In control embryos, >95% of heart tubes are asymmetrically positioned in the left side, while in Ipk1-morpholino treated embryos, around half of the heart tubes are on the right side and the other half on the left, indicating loss of Ipk1 results in randomization of heart asymmetry in zebrafish embryos. Furthermore, the asymmetrical positioning of other visceral organs (gut, pancreas, liver) and brain is also compromised by Ipk1 knockdown. Using a genetically encoded Ca2+ sensor Flash-pericam, Wente group then showed that there is a transient left oriented intracellular Ca2+ flux near KV in normal zebrafish embryo development at the stage when LR asymmetry is initiated, and this flux requires an intact KV. Interesting, Ipk1 knockdown leads to the disappearance of this asymmetrical Ca2+ flux, suggesting Ipk1 may regulate LR positioning through generation of asymmetrical Ca2+ flux.
GFP-Ipk1 is enriched in centrosomes and basal bodies, and the Ipk1 knockdown also results in reduction of KC ciliary beating and length (Sarmah et al., 2007). These defects can be rescued by co-injection of wild type Ipk1 mRNA but not a kinase-dead version, demonstrating the Ipk1 kinase activity is critical for proper KV cilia functions. Ipk1 knockdown also leads to decrease of cilia length in the pronephric duct and spinal canal cilia, indicating Ipk1 is important for cilia length maintenance in multiple organs. These data suggest a new pathway by which IPs induce local change in Ca2+ concentrations by regulating biased cilia movement.
IP4 AND IP7 REGULATES CHEMOTAXIS IN DICTYOSTELIUM AND NEUTROPHILS BY ANTAGONIZING PI(3,4,5)P3 SIGNALING
Under starvation, the slime mold D. discoideum, release cAMP which acts as chemoattractant to induce chemotaxis and hence cell aggregation (Chung et al., 2001; Devreotes and Janetopoulos, 2003; Iijima et al., 2002). Upon binding to its surface receptor, cAMP elicits a localized, G-protein dependent activation of a phosphatidylinositol 3-kinase (PI3K) and inhibition of a phosphatidylinositol 3-phosphatase (PTEN), resulting in an increase in levels of phosphatidylinositol 3,4,5,-trisphosphate [PI(3,4,5)P3]. The local increase in PI(3,4,5)P3 then recruits a subset of PH domain-containing proteins to the lead edge of the chemotaxing cells. Snyder group cloned the Dictyostelium IP6K and discovered that IP6K and its products IP7 regulate chemotaxis in Dictyostelium (Luo et al., 2003). Disruption of IP6K results in drastic reductions in the IP7 and IP8 levels, while the concentrations of IP3, IP5 and IP6 remains unchanged, indicating IP6K is required for IP7 and IP8 synthesis in Dictyostelium. The chemoattractant cAMP can induce increase in IP7 and IP8 levels in WT cells, while the ip6k- cells have increased sensitivity to cAMP, suggesting IP7/IP8 negatively regulates cAMP-induced chemotaxis. Indeed, in vitro, IP7 can compete with PI(3,4,5)P3 for binding to PH domain containing proteins, and deletion of IP6K enhances the translocation of a GFP-PH marker to the leading edge of the chemotaxing cells. Moreover, GFP-PH protein isolated from amoeba cells was found associated with IP6, IP7 and IP8, indicating IPs and PP-IPs can bind to PH domain in vivo. This study shows that IP7 can negatively regulate PI(3,4,5)P3 signaling through competition with this inositol lipid for PH domain binding. In another study by Luo group, chemotaxis of neutrophils through activation of PI(3,4,5)P3 signaling is also negatively regulated by an IP, namely I(1,3,4,5)P4 (Jia et al., 2007). I(1,3,4,5)P4 also inhibits PI(3,4,5)P3 by competition for binding to PH domain proteins. These two studies highlights a novel role of IPs and PP-IPs in regulating PI(3,4,5)P3 signaling by competition for binding to downstream protein partners.
IP6 IS A CO-FACTOR OF ADAR2 AND IS REQUIRED FOR RNA EDITING
An IP6 molecule was found to be buried in the interior of the structure of hADAR2 by Bass group (Macbeth et al., 2005). ADAR2 encodes adenosine deaminases that act on RNA (Bass, 2002) and catalyze RNA editing by deaminating adenosine to inosine in double stranded RNA. IP6 binds in the core of the catalytic domain, and is required for protein stability and enzyme activity. IP6 is also required for activity of another class of RNA editing enzymes, ADAT1, which works on tRNA. These data uncovered an unexpected role for IP6 as a structural co-factor of RNA editing enzymes.
DROSOPHILA PLC HOMOLOGS REGULATE PHOTOTRANSDUCTION, AND EYE AND WING DEVELOPMENT
There are three PLC homologs in fruit fly: small wing (sl), no receptor potential A (norpA) and PLC21C. sl belongs to the PLC-γ sub-type. Genetically null mutant of sl exhibit reduction of adult wing size and extra R7 photoreceptors in the compound eye (Thackeray et al., 1998). Each ommatidium of a compound eye consists of a precise assembly of eight photoreceptors, R1-R8. In many fly mutants that have altered MAPK signaling, there is an increase in R7 cells (Basler and Hafen, 1989; Brunner et al., 1994; Buckles et al., 1992). In fact, attenuation of MAPK signaling in the sl mutant by reduction of the gene dosage of EGFR by half, can almost completely rescue the extra R7 cell phenotype. This suggests that sl is a negative regulator of MAPK signaling in Drosophila eye development.
norpA is a PLCβ isoform, and is required for phototransduction in Drosophila eye (Bloomquist et al., 1988). Upon light activation, rhodopsin activates heteromeric Gq protein, which subsequently turns on norpA. norpA hydrolyze PIP2 into IP3 and DAG (Hardie and Raghu, 2001; Minke, 2001; Montell, 1999). Evidence suggests that DAG and its downstream metabolite, rather than IP3 is responsible for opening of the light activated TRP channel to produce quantum pump (Hardie et al., 2002). On the other hand, norpA can inactivate its upstream G protein by acting as GTPase-activating protein (GAP) when associated with the PDZ scaffold protein INAD (Cook et al., 2000). The negative feedback mechanism was proposed to maintain high signal resolution of the response to light.
In addition to Plc, Ipk2 and Ipk1 homologs have been identified in Drosophila and the molecular basis of IP synthesis has been elucidated (Seeds et al., 2004). However, the developmental roles of fly IPKs remain elusive and are currently under active research in our lab.
SUMMARY
Inositol phosphates and inositol pyrophosphates are small molecule metabolites that play important roles in nuclear processes such as transcription control, mRNA export and DNA repair. On this wonderful occasion of the fiftieth anniversary of the Advances in Enzyme Regulation conference, it is a privilege to participate through presenting recent developments in the area of inositol phosphate and pyrophosphate regulatory biology. This article summarizes recent advances in understanding IPs and PP-IPs biology in development and nuclear cell signaling. Data obtained from various model organisms hint at the emerging modes of mechanisms of these versatile molecules.
Studies in model organisms revealed that IPs and PP-IPs are critical for development and signaling in plants, mice, zebrafish and slime mold. In plants, IP molecules are required for signaling in response to stress, hormone, and nutrient, by transcriptional regulation of the stimuli-responsive genes. In some cases, IPs act as regulatory molecules as the levels of IPKs/IPs are induced by those stimuli and a constitutively induction of downstream events can be accomplished by over-expressing the IPKs. Nutrient sensing is a recurring theme in IP biology as IPs and PP-IPs are also involved in amino acid and phosphate signaling in yeast and possible glucose sensing in pancreatic β cells, indicating it maybe acquired early during evolution. There are some newly discovered nuclear roles of IPs: 1) binding to and possibly regulation of the SCFTIR1 ubiquitin ligase complex, and 2) RNA editing by acting as structural co-factor of ADAR2 and ADAT1. On the other hand, IPs and PP-IPs are also implicated in some novel non-nuclear processes: 1) insulin secretion, 2) negative regulation of PIP3 signaling and 3) modulation of intracellular Ca2+ concentration. Following the discovery of new biological roles of IPs and PP-IPs, a few new receptors for IPs and PP-IPs were identified in the process. Three modes of receptor binding by IPs and PP-IPs can be summarized: 1) Direct binding to the receptor (e.g. TIR1), 2) displacement of a ligand (e.g. PIP3) already bound to the receptor (e.g. PH domain containing proteins, and 3) acting as a structural co-factor and possibly incorporated inside the receptor during protein folding (e.g. ADAR2). Hopefully, more experimentation will uncover more receptors for and biological roles of these versatile molecules.
Acknowledgments
We wish to thank members of the lab, past and present, and numerous colleagues for helpful discussions. This work is supported by funds from the Howard Hughes Medical Institute, and from the National Institutes of Health grants R01 HL055672.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcazar-Roman AR, Tran EJ, Guo S, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol. 2006;8:711–716. doi: 10.1038/ncb1427. [DOI] [PubMed] [Google Scholar]
- Alcazar-Roman AR, Wente SR. Inositol polyphosphates: a new frontier for regulating gene expression. Chromosoma. 2008;117:1–13. doi: 10.1007/s00412-007-0126-4. [DOI] [PubMed] [Google Scholar]
- Azevedo C, Burton A, Ruiz-Mateos E, Marsh M, Saiardi A. Inositol pyrophosphate mediated pyrophosphorylation of AP3B1 regulates HIV-1 Gag release. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0909176106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge K, Sorefan K, Ward S, Leyser O. Hormonally controlled expression of the Arabidopsis MAX4 shoot branching regulatory gene. Plant J. 2005;44:569–580. doi: 10.1111/j.1365-313X.2005.02548.x. [DOI] [PubMed] [Google Scholar]
- Basler K, Hafen E. Dynamics of Drosophila eye development and temporal requirements of sevenless expression. Development. 1989;107:723–731. doi: 10.1242/dev.107.4.723. [DOI] [PubMed] [Google Scholar]
- Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechet J, Greenson M, Wiame JM. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur J Biochem. 1970;12:31–39. doi: 10.1111/j.1432-1033.1970.tb00817.x. [DOI] [PubMed] [Google Scholar]
- Bercy J, Dubois E, Messenguy F. Regulation of arginine metabolism in Saccharomyces cerevisiae: expression of the three ARGR regulatory genes and cellular localization of their products. Gene. 1987;55:277–285. doi: 10.1016/0378-1119(87)90287-3. [DOI] [PubMed] [Google Scholar]
- Berridge MJ. Cell signalling. A tale of two messengers. Nature. 1993;365:388–389. doi: 10.1038/365388a0. [DOI] [PubMed] [Google Scholar]
- Bhandari R, Juluri KR, Resnick AC, Snyder SH. Gene deletion of inositol hexakisphosphate kinase 1 reveals inositol pyrophosphate regulation of insulin secretion, growth, and spermiogenesis. Proc Natl Acad Sci U S A. 2008;105:2349–2353. doi: 10.1073/pnas.0712227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari R, Saiardi A, Ahmadibeni Y, Snowman AM, Resnick AC, Kristiansen TZ, Molina H, Pandey A, Werner JK, Jr, Juluri KR, et al. Protein pyrophosphorylation by inositol pyrophosphates is a posttranslational event. Proc Natl Acad Sci U S A. 2007;104:15305–15310. doi: 10.1073/pnas.0707338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Maity IB, Chakrabarti S, Biswas BB. Purification and characterization of myo-inositol hexaphosphate-adenosine diphosphate phosphotransferase from Phaseolus aureus. Arch Biochem Biophys. 1978;185:557–566. doi: 10.1016/0003-9861(78)90201-1. [DOI] [PubMed] [Google Scholar]
- Bloomquist BT, Shortridge RD, Schneuwly S, Perdew M, Montell C, Steller H, Rubin G, Pak WL. Isolation of a putative phospholipase C gene of Drosophila, norpA, and its role in phototransduction. Cell. 1988;54:723–733. doi: 10.1016/s0092-8674(88)80017-5. [DOI] [PubMed] [Google Scholar]
- Bolger TA, Folkmann AW, Tran EJ, Wente SR. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell. 2008;134:624–633. doi: 10.1016/j.cell.2008.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brearley CA, Hanke DE. Metabolic evidence for the order of addition of individual phosphate esters in the myo-inositol moiety of inositol hexakisphosphate in the duckweed Spirodela polyrhiza L. Biochem J. 1996;314(Pt 1):227–233. doi: 10.1042/bj3140227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner D, Oellers N, Szabad J, Biggs WH, 3rd, Zipursky SL, Hafen E. A gain-of-function mutation in Drosophila MAP kinase activates multiple receptor tyrosine kinase signaling pathways. Cell. 1994;76:875–888. doi: 10.1016/0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- Buckles GR, Smith ZD, Katz FN. mip causes hyperinnervation of a retinotopic map in Drosophila by excessive recruitment of R7 photoreceptor cells. Neuron. 1992;8:1015–1029. doi: 10.1016/0896-6273(92)90124-v. [DOI] [PubMed] [Google Scholar]
- Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- Chamberlain PP, Qian X, Stiles AR, Cho J, Jones DH, Lesley SA, Grabau EA, Shears SB, Spraggon G. Integration of inositol phosphate signaling pathways via human ITPK1. J Biol Chem. 2007;282:28117–28125. doi: 10.1074/jbc.M703121200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SC, Miller AL, Feng Y, Wente SR, Majerus PW. The human homolog of the rat inositol phosphate multikinase is an inositol 1,3,4,6-tetrakisphosphate 5-kinase. J Biol Chem. 2002;277:43836–43843. doi: 10.1074/jbc.M206134200. [DOI] [PubMed] [Google Scholar]
- Chatfield SP, Stirnberg P, Forde BG, Leyser O. The hormonal regulation of axillary bud growth in Arabidopsis. Plant J. 2000;24:159–169. doi: 10.1046/j.1365-313x.2000.00862.x. [DOI] [PubMed] [Google Scholar]
- Chung CY, Funamoto S, Firtel RA. Signaling pathways controlling cell polarity and chemotaxis. Trends Biochem Sci. 2001;26:557–566. doi: 10.1016/s0968-0004(01)01934-x. [DOI] [PubMed] [Google Scholar]
- Cocco L, Capitani S, Maraldi NM, Mazzotti G, Barnabei O, Gilmour RS, Manzoli FA. Inositol lipid cycle and autonomous nuclear signalling. Adv Enzyme Regul. 1996;36:101–114. doi: 10.1016/0065-2571(95)00007-0. [DOI] [PubMed] [Google Scholar]
- Cook B, Bar-Yaacov M, Cohen Ben-Ami H, Goldstein RE, Paroush Z, Selinger Z, Minke B. Phospholipase C and termination of G-protein-mediated signalling in vivo. Nat Cell Biol. 2000;2:296–301. doi: 10.1038/35010571. [DOI] [PubMed] [Google Scholar]
- Desai P, Guha N, Galdieri L, Hadi S, Vancura A. Plc1p is required for proper chromatin structure and activity of the kinetochore in Saccharomyces cerevisiae by facilitating recruitment of the RSC complex. Mol Genet Genomics. 2009;281:511–523. doi: 10.1007/s00438-009-0427-9. [DOI] [PubMed] [Google Scholar]
- Devreotes P, Janetopoulos C. Eukaryotic chemotaxis: distinctions between directional sensing and polarization. J Biol Chem. 2003;278:20445–20448. doi: 10.1074/jbc.R300010200. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- Draskovic P, Saiardi A, Bhandari R, Burton A, Ilc G, Kovacevic M, Snyder SH, Podobnik M. Inositol hexakisphosphate kinase products contain diphosphate and triphosphate groups. Chem Biol. 2008;15:274–286. doi: 10.1016/j.chembiol.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Dubois E, Scherens B, Vierendeels F, Ho MM, Messenguy F, Shears SB. In Saccharomyces cerevisiae, the inositol polyphosphate kinase activity of Kcs1p is required for resistance to salt stress, cell wall integrity, and vacuolar morphogenesis. J Biol Chem. 2002;277:23755–23763. doi: 10.1074/jbc.M202206200. [DOI] [PubMed] [Google Scholar]
- El Alami M, Messenguy F, Scherens B, Dubois E. Arg82p is a bifunctional protein whose inositol polyphosphate kinase activity is essential for nitrogen and PHO gene expression but not for Mcm1p chaperoning in yeast. Mol Microbiol. 2003;49:457–468. doi: 10.1046/j.1365-2958.2003.03562.x. [DOI] [PubMed] [Google Scholar]
- Essner JJ, Amack JD, Nyholm MK, Harris EB, Yost HJ. Kupffer’s vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development. 2005;132:1247–1260. doi: 10.1242/dev.01663. [DOI] [PubMed] [Google Scholar]
- Essner JJ, Vogan KJ, Wagner MK, Tabin CJ, Yost HJ, Brueckner M. Conserved function for embryonic nodal cilia. Nature. 2002;418:37–38. doi: 10.1038/418037a. [DOI] [PubMed] [Google Scholar]
- Feoktistova A, McCollum D, Ohi R, Gould KL. Identification and characterization of Schizosaccharomyces pombe asp1(+), a gene that interacts with mutations in the Arp2/3 complex and actin. Genetics. 1999;152:895–908. doi: 10.1093/genetics/152.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick JP, Mattiske D, Wofford JA, Megosh LC, Drake LY, Chiou ST, Hogan BL, York JD. An essential role for an inositol polyphosphate multikinase, Ipk2, in mouse embryogenesis and second messenger production. Proc Natl Acad Sci U S A. 2005;102:8454–8459. doi: 10.1073/pnas.0503706102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridy PC, Otto JC, Dollins DE, York JD. Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol pentakisphosphate kinases. J Biol Chem. 2007;282:30754–30762. doi: 10.1074/jbc.M704656200. [DOI] [PubMed] [Google Scholar]
- Fujii M, York JD. A role for rat inositol polyphosphate kinases rIPK2 and rIPK1 in inositol pentakisphosphate and inositol hexakisphosphate production in rat-1 cells. J Biol Chem. 2005;280:1156–1164. doi: 10.1074/jbc.M412006200. [DOI] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol. 2002;49:373–385. [PubMed] [Google Scholar]
- Hardie RC, Martin F, Cochrane GW, Juusola M, Georgiev P, Raghu P. Molecular basis of amplification in Drosophila phototransduction: roles for G protein, phospholipase C, and diacylglycerol kinase. Neuron. 2002;36:689–701. doi: 10.1016/s0896-6273(02)01048-6. [DOI] [PubMed] [Google Scholar]
- Hardie RC, Raghu P. Visual transduction in Drosophila. Nature. 2001;413:186–193. doi: 10.1038/35093002. [DOI] [PubMed] [Google Scholar]
- Hermosura MC, Takeuchi H, Fleig A, Riley AM, Potter BV, Hirata M, Penner R. InsP4 facilitates store-operated calcium influx by inhibition of InsP3 5-phosphatase. Nature. 2000;408:735–740. doi: 10.1038/35047115. [DOI] [PubMed] [Google Scholar]
- Ho MW, Yang X, Carew MA, Zhang T, Hua L, Kwon YU, Chung SK, Adelt S, Vogel G, Riley AM, et al. Regulation of Ins(3,4,5,6)P(4) signaling by a reversible kinase/phosphatase. Curr Biol. 2002;12:477–482. doi: 10.1016/s0960-9822(02)00713-3. [DOI] [PubMed] [Google Scholar]
- Hokin LE. Receptors and phosphoinositide-generated second messengers. Annu Rev Biochem. 1985;54:205–235. doi: 10.1146/annurev.bi.54.070185.001225. [DOI] [PubMed] [Google Scholar]
- Hong Z, Lakkineni K, Zhang Z, Verma DP. Removal of feedback inhibition of delta(1)-pyrroline-5-carboxylate synthetase results in increased proline accumulation and protection of plants from osmotic stress. Plant Physiol. 2000;122:1129–1136. doi: 10.1104/pp.122.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KN, Symington LS. Suppressors of a Saccharomyces cerevisiae pkc1 mutation identify alleles of the phosphatase gene PTC1 and of a novel gene encoding a putative basic leucine zipper protein. Genetics. 1995;141:1275–1285. doi: 10.1093/genetics/141.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima M, Huang YE, Devreotes P. Temporal and spatial regulation of chemotaxis. Dev Cell. 2002;3:469–478. doi: 10.1016/s1534-5807(02)00292-7. [DOI] [PubMed] [Google Scholar]
- Illies C, Gromada J, Fiume R, Leibiger B, Yu J, Juhl K, Yang SN, Barma DK, Falck JR, Saiardi A, et al. Requirement of inositol pyrophosphates for full exocytotic capacity in pancreatic beta cells. Science. 2007;318:1299–1302. doi: 10.1126/science.1146824. [DOI] [PubMed] [Google Scholar]
- Irvine RF, Schell MJ. Back in the water: the return of the inositol phosphates. Nat Rev Mol Cell Biol. 2001;2:327–338. doi: 10.1038/35073015. [DOI] [PubMed] [Google Scholar]
- Kamimura J, Wakui K, Kadowaki H, Watanabe Y, Miyake K, Harada N, Sakamoto M, Kinoshita A, Yoshiura K, Ohta T, et al. The IHPK1 gene is disrupted at the 3p21.31 breakpoint of t(3;9) in a family with type 2 diabetes mellitus. J Hum Genet. 2004;49:360–365. doi: 10.1007/s10038-004-0158-z. [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. Auxin-induced SCFTIR1-Aux/IAA interaction involves stable modification of the SCFTIR1 complex. Proc Natl Acad Sci U S A. 2004;101:12381–12386. doi: 10.1073/pnas.0402868101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- Larsson O, Barker CJ, Sjoholm A, Carlqvist H, Michell RH, Bertorello A, Nilsson T, Honkanen RE, Mayr GW, Zwiller J, Berggren PO. Inhibition of phosphatases and increased Ca2+ channel activity by inositol hexakisphosphate. Science. 1997;278:471–474. doi: 10.1126/science.278.5337.471. [DOI] [PubMed] [Google Scholar]
- Lee YS, Huang K, Quiocho FA, O’Shea EK. Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat Chem Biol. 2008;4:25–32. doi: 10.1038/nchembio.2007.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Mulugu S, York JD, O’Shea EK. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science. 2007;316:109–112. doi: 10.1126/science.1139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemtiri-Chlieh F, MacRobbie EA, Brearley CA. Inositol hexakisphosphate is a physiological signal regulating the K+-inward rectifying conductance in guard cells. Proc Natl Acad Sci U S A. 2000;97:8687–8692. doi: 10.1073/pnas.140217497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemtiri-Chlieh F, MacRobbie EA, Webb AA, Manison NF, Brownlee C, Skepper JN, Chen J, Prestwich GD, Brearley CA. Inositol hexakisphosphate mobilizes an endomembrane store of calcium in guard cells. Proc Natl Acad Sci U S A. 2003;100:10091–10095. doi: 10.1073/pnas.1133289100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyman A, Pouillon V, Bostan A, Schurmans S, Erneux C, Pesesse X. The absence of expression of the three isoenzymes of the inositol 1,4,5-trisphosphate 3-kinase does not prevent the formation of inositol pentakisphosphate and hexakisphosphate in mouse embryonic fibroblasts. Cell Signal. 2007;19:1497–1504. doi: 10.1016/j.cellsig.2007.01.024. [DOI] [PubMed] [Google Scholar]
- Lin H, Choi JH, Hasek J, DeLillo N, Lou W, Vancura A. Phospholipase C is involved in kinetochore function in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:3597–3607. doi: 10.1128/mcb.20.10.3597-3607.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Fridy PC, Ribeiro AA, Choi JH, Barma DK, Vogel G, Falck JR, Shears SB, York JD, Mayr GW. Structural analysis and detection of biological inositol pyrophosphates reveal that the family of VIP/diphosphoinositol pentakisphosphate kinases are 1/3-kinases. J Biol Chem. 2009;284:1863–1872. doi: 10.1074/jbc.M805686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Reed JW. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol. 2002;49:387–400. [PubMed] [Google Scholar]
- Luo HR, Huang YE, Chen JC, Saiardi A, Iijima M, Ye K, Huang Y, Nagata E, Devreotes P, Snyder SH. Inositol pyrophosphates mediate chemotaxis in Dictyostelium via pleckstrin homology domain-PtdIns(3,4,5)P3 interactions. Cell. 2003;114:559–572. doi: 10.1016/s0092-8674(03)00640-8. [DOI] [PubMed] [Google Scholar]
- Luo HR, Saiardi A, Yu H, Nagata E, Ye K, Snyder SH. Inositol pyrophosphates are required for DNA hyperrecombination in protein kinase c1 mutant yeast. Biochemistry. 2002;41:2509–2515. doi: 10.1021/bi0118153. [DOI] [PubMed] [Google Scholar]
- Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 2005;309:1534–1539. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerus PW. Inositol phosphate biochemistry. Annu Rev Biochem. 1992;61:225–250. doi: 10.1146/annurev.bi.61.070192.001301. [DOI] [PubMed] [Google Scholar]
- Manzoli L, Billi AM, Gilmour RS, Martelli AM, Matteucci A, Rubbini S, Weber G, Cocco L. Phosphoinositide signaling in nuclei of Friend cells: tiazofurin downregulates phospholipase C beta 1. Cancer Res. 1995;55:2978–2980. [PubMed] [Google Scholar]
- Maraldi NM, Zini N, Ognibene A, Martelli AM, Barbieri M, Mazzotti G, Manzoli FA. Immunocytochemical detection of the intranuclear variations of phosphatidylinositol 4,5-bisphosphate amount associated with changes of activity and amount of phospholipase C beta 1 in cells exposed to mitogenic or differentiating agonists. Biol Cell. 1995;83:201–210. doi: 10.1016/0248-4900(96)81309-8. [DOI] [PubMed] [Google Scholar]
- McGrath J, Somlo S, Makova S, Tian X, Brueckner M. Two populations of node monocilia initiate left-right asymmetry in the mouse. Cell. 2003;114:61–73. doi: 10.1016/s0092-8674(03)00511-7. [DOI] [PubMed] [Google Scholar]
- Menniti FS, Oliver KG, Putney JW, Jr, Shears SB. Inositol phosphates and cell signaling: new views of InsP5 and InsP6. Trends Biochem Sci. 1993;18:53–56. doi: 10.1016/0968-0004(93)90053-p. [DOI] [PubMed] [Google Scholar]
- Messenguy F, Dubois E. Genetic evidence for a role for MCM1 in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:2586–2592. doi: 10.1128/mcb.13.4.2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikoshiba K. The InsP3 receptor and intracellular Ca2+ signaling. Curr Opin Neurobiol. 1997;7:339–345. doi: 10.1016/s0959-4388(97)80061-x. [DOI] [PubMed] [Google Scholar]
- Miller GJ, Wilson MP, Majerus PW, Hurley JH. Specificity determinants in inositol polyphosphate synthesis: crystal structure of inositol 1,3,4-trisphosphate 5/6-kinase. Mol Cell. 2005;18:201–212. doi: 10.1016/j.molcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Minke B. The TRP channel and phospholipase C-mediated signaling. Cell Mol Neurobiol. 2001;21:629–643. doi: 10.1023/A:1015191702536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C. Visual transduction in Drosophila. Annu Rev Cell Dev Biol. 1999;15:231–268. doi: 10.1146/annurev.cellbio.15.1.231. [DOI] [PubMed] [Google Scholar]
- Morrison BH, Haney R, Lamarre E, Drazba J, Prestwich GD, Lindner DJ. Gene deletion of inositol hexakisphosphate kinase 2 predisposes to aerodigestive tract carcinoma. Oncogene. 2009;28:2383–2392. doi: 10.1038/onc.2009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulugu S, Bai W, Fridy PC, Bastidas RJ, Otto JC, Dollins DE, Haystead TA, Ribeiro AA, York JD. A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science. 2007;316:106–109. doi: 10.1126/science.1139099. [DOI] [PubMed] [Google Scholar]
- Murphy R, Wente SR. An RNA-export mediator with an essential nuclear export signal. Nature. 1996;383:357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- Nalaskowski MM, Deschermeier C, Fanick W, Mayr GW. The human homologue of yeast ArgRIII protein is an inositol phosphate multikinase with predominantly nuclear localization. Biochem J. 2002;366:549–556. doi: 10.1042/BJ20020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli CA, Beveridge CA, Snowden KC. Reevaluating concepts of apical dominance and the control of axillary bud outgrowth. Curr Top Dev Biol. 1999;44:127–169. doi: 10.1016/s0070-2153(08)60469-x. [DOI] [PubMed] [Google Scholar]
- Nishizuka Y. Studies and perspectives of protein kinase C. Science. 1986;233:305–312. doi: 10.1126/science.3014651. [DOI] [PubMed] [Google Scholar]
- Niu X, Chen Q, Wang X. OsITL1 gene encoding an inositol 1,3,4-trisphosphate 5/6-kinase is a negative regulator of osmotic stress signaling. Biotechnol Lett. 2008;30:1687–1692. doi: 10.1007/s10529-008-9730-5. [DOI] [PubMed] [Google Scholar]
- Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- Qin ZX, Chen QJ, Tong Z, Wang XC. The Arabidopsis inositol 1,3,4-trisphosphate 5/6 kinase, AtItpk-1, is involved in plant photomorphogenesis under red light conditions, possibly via interaction with COP9 signalosome. Plant Physiol Biochem. 2005;43:947–954. doi: 10.1016/j.plaphy.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Raya A, Izpisua Belmonte JC. Unveiling the establishment of left-right asymmetry in the chick embryo. Mech Dev. 2004;121:1043–1054. doi: 10.1016/j.mod.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Reed JW. Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 2001;6:420–425. doi: 10.1016/s1360-1385(01)02042-8. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Bhandari R, Resnick AC, Snowman AM, Snyder SH. Phosphorylation of proteins by inositol pyrophosphates. Science. 2004;306:2101–2105. doi: 10.1126/science.1103344. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Caffrey JJ, Snyder SH, Shears SB. Inositol polyphosphate multikinase (ArgRIII) determines nuclear mRNA export in Saccharomyces cerevisiae. FEBS Lett. 2000a;468:28–32. doi: 10.1016/s0014-5793(00)01194-7. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Caffrey JJ, Snyder SH, Shears SB. The inositol hexakisphosphate kinase family Catalytic flexibility and function in yeast vacuole biogenesis. J Biol Chem. 2000b;275:24686–24692. doi: 10.1074/jbc.M002750200. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Nagata E, Luo HR, Sawa A, Luo X, Snowman AM, Snyder SH. Mammalian inositol polyphosphate multikinase synthesizes inositol 1,4,5-trisphosphate and an inositol pyrophosphate. Proc Natl Acad Sci U S A. 2001;98:2306–2311. doi: 10.1073/pnas.041614598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiardi A, Resnick AC, Snowman AM, Wendland B, Snyder SH. Inositol pyrophosphates regulate cell death and telomere length through phosphoinositide 3-kinase-related protein kinases. Proc Natl Acad Sci U S A. 2005;102:1911–1914. doi: 10.1073/pnas.0409322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiardi A, Sciambi C, McCaffery JM, Wendland B, Snyder SH. Inositol pyrophosphates regulate endocytic trafficking. Proc Natl Acad Sci U S A. 2002;99:14206–14211. doi: 10.1073/pnas.212527899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmah B, Latimer AJ, Appel B, Wente SR. Inositol polyphosphates regulate zebrafish left-right asymmetry. Dev Cell. 2005;9:133–145. doi: 10.1016/j.devcel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Sarmah B, Winfrey VP, Olson GE, Appel B, Wente SR. A role for the inositol kinase Ipk1 in ciliary beating and length maintenance. Proc Natl Acad Sci U S A. 2007;104:19843–19848. doi: 10.1073/pnas.0706934104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeds AM, Sandquist JC, Spana EP, York JD. A molecular basis for inositol polyphosphate synthesis in Drosophila melanogaster. J Biol Chem. 2004;279:47222–47232. doi: 10.1074/jbc.M408295200. [DOI] [PubMed] [Google Scholar]
- Shears SB. The versatility of inositol phosphates as cellular signals. Biochim Biophys Acta. 1998;1436:49–67. doi: 10.1016/s0005-2760(98)00131-3. [DOI] [PubMed] [Google Scholar]
- Shen X, Xiao H, Ranallo R, Wu WH, Wu C. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science. 2003;299:112–114. doi: 10.1126/science.1078068. [DOI] [PubMed] [Google Scholar]
- Shi J, Wang H, Wu Y, Hazebroek J, Meeley RB, Ertl DS. The maize low-phytic acid mutant lpa2 is caused by mutation in an inositol phosphate kinase gene. Plant Physiol. 2003;131:507–515. doi: 10.1104/pp.014258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart LB, Cameron KD, Bennett AB. Isolation of genes predominantly expressed in guard cells and epidermal cells of Nicotiana glauca. Plant Mol Biol. 2000;42:857–869. doi: 10.1023/a:1006480107407. [DOI] [PubMed] [Google Scholar]
- Snay-Hodge CA, Colot HV, Goldstein AL, Cole CN. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 1998;17:2663–2676. doi: 10.1093/emboj/17.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Haswell ES, Miller AL, Wente SR, O’Shea EK. Regulation of chromatin remodeling by inositol polyphosphates. Science. 2003;299:114–116. doi: 10.1126/science.1078062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens LR, Irvine RF. Stepwise phosphorylation of myo-inositol leading to myoinositol hexakisphosphate in Dictyostelium. Nature. 1990;346:580–583. doi: 10.1038/346580a0. [DOI] [PubMed] [Google Scholar]
- Stevenson-Paulik J, Bastidas RJ, Chiou ST, Frye RA, York JD. Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc Natl Acad Sci U S A. 2005;102:12612–12617. doi: 10.1073/pnas.0504172102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson-Paulik J, Odom AR, York JD. Molecular and biochemical characterization of two plant inositol polyphosphate 6-/3-/5-kinases. J Biol Chem. 2002;277:42711–42718. doi: 10.1074/jbc.M209112200. [DOI] [PubMed] [Google Scholar]
- Sun Y, Wilson MP, Majerus PW. Inositol 1,3,4-trisphosphate 5/6-kinase associates with the COP9 signalosome by binding to CSN1. J Biol Chem. 2002;277:45759–45764. doi: 10.1074/jbc.M208709200. [DOI] [PubMed] [Google Scholar]
- Sweetman D, Johnson S, Caddick SE, Hanke DE, Brearley CA. Characterization of an Arabidopsis inositol 1,3,4,5,6-pentakisphosphate 2-kinase (AtIPK1) Biochem J. 2006;394:95–103. doi: 10.1042/BJ20051331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetman D, Stavridou I, Johnson S, Green P, Caddick SE, Brearley CA. Arabidopsis thaliana inositol 1,3,4-trisphosphate 5/6-kinase 4 (AtITPK4) is an outlier to a family of ATP-grasp fold proteins from Arabidopsis. FEBS Lett. 2007;581:4165–4171. doi: 10.1016/j.febslet.2007.07.046. [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- Tantikanjana T, Yong JW, Letham DS, Griffith M, Hussain M, Ljung K, Sandberg G, Sundaresan V. Control of axillary bud initiation and shoot architecture in Arabidopsis through the SUPERSHOOT gene. Genes Dev. 2001;15:1577–1588. doi: 10.1101/gad.887301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao LZ, Cheung AY, Nibau C, Wu HM. RAC GTPases in tobacco and Arabidopsis mediate auxin-induced formation of proteolytically active nuclear protein bodies that contain AUX/IAA proteins. Plant Cell. 2005;17:2369–2383. doi: 10.1105/tpc.105.032987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teale WD, Paponov IA, Palme K. Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol. 2006;7:847–859. doi: 10.1038/nrm2020. [DOI] [PubMed] [Google Scholar]
- Thackeray JR, Gaines PC, Ebert P, Carlson JR. small wing encodes a phospholipase C-(gamma) that acts as a negative regulator of R7 development in Drosophila. Development. 1998;125:5033–5042. doi: 10.1242/dev.125.24.5033. [DOI] [PubMed] [Google Scholar]
- Thomashow MF. PLANT COLD ACCLIMATION: Freezing Tolerance Genes and Regulatory Mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell. 2001;13:2809–2822. doi: 10.1105/tpc.010289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbsky J, Lavine K, Majerus PW. Disruption of the mouse inositol 1,3,4,5,6-pentakisphosphate 2-kinase gene, associated lethality, and tissue distribution of 2-kinase expression. Proc Natl Acad Sci U S A. 2005a;102:8448–8453. doi: 10.1073/pnas.0503656102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbsky J, Majerus PW. Increased levels of inositol hexakisphosphate (InsP6) protect HEK293 cells from tumor necrosis factor (alpha)- and Fas-induced apoptosis. J Biol Chem. 2005;280:29263–29268. doi: 10.1074/jbc.M503366200. [DOI] [PubMed] [Google Scholar]
- Verbsky JW, Chang SC, Wilson MP, Mochizuki Y, Majerus PW. The pathway for the production of inositol hexakisphosphate in human cells. J Biol Chem. 2005b;280:1911–1920. doi: 10.1074/jbc.M411528200. [DOI] [PubMed] [Google Scholar]
- Verbsky JW, Wilson MP, Kisseleva MV, Majerus PW, Wente SR. The synthesis of inositol hexakisphosphate Characterization of human inositol 1,3,4,5,6-pentakisphosphate 2-kinase. J Biol Chem. 2002;277:31857–31862. doi: 10.1074/jbc.M205682200. [DOI] [PubMed] [Google Scholar]
- Webb SE, Miller AL. Calcium signalling during embryonic development. Nat Rev Mol Cell Biol. 2003;4:539–551. doi: 10.1038/nrm1149. [DOI] [PubMed] [Google Scholar]
- Wilson MP, Majerus PW. Characterization of a cDNA encoding Arabidopsis thaliana inositol 1,3,4-trisphosphate 5/6-kinase. Biochem Biophys Res Commun. 1997;232:678–681. doi: 10.1006/bbrc.1997.6355. [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. Auxin: regulation, action, and interaction. Ann Bot (Lond) 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia HJ, Brearley C, Elge S, Kaplan B, Fromm H, Mueller-Roeber B. Arabidopsis inositol polyphosphate 6-/3-kinase is a nuclear protein that complements a yeast mutant lacking a functional ArgR-Mcm1 transcription complex. Plant Cell. 2003;15:449–463. doi: 10.1105/tpc.006676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Brearley CA, Lin WH, Wang Y, Ye R, Mueller-Roeber B, Xu ZH, Xue HW. A role of Arabidopsis inositol polyphosphate kinase, AtIPK2alpha, in pollen germination and root growth. Plant Physiol. 2005;137:94–103. doi: 10.1104/pp.104.045427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Komori T, Hashimoto A, Kuwata S, Imaseki H, Kubo T. Differential expression of plastidic aldolase genes in Nicotiana plants under salt stress. Plant Sci. 2000;154:61–69. doi: 10.1016/s0168-9452(00)00188-6. [DOI] [PubMed] [Google Scholar]
- Yancey PH. Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J Exp Biol. 2005;208:2819–2830. doi: 10.1242/jeb.01730. [DOI] [PubMed] [Google Scholar]
- Yang L, Tang R, Zhu J, Liu H, Mueller-Roeber B, Xia H, Zhang H. Enhancement of stress tolerance in transgenic tobacco plants constitutively expressing AtIpk2beta, an inositol polyphosphate 6-/3-kinase from Arabidopsis thaliana. Plant Mol Biol. 2008;66:329–343. doi: 10.1007/s11103-007-9267-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SN, Yu J, Mayr GW, Hofmann F, Larsson O, Berggren PO. Inositol hexakisphosphate increases L-type Ca2+ channel activity by stimulation of adenylyl cyclase. FASEB J. 2001;15:1753–1763. doi: 10.1096/fj.00-0799com. [DOI] [PubMed] [Google Scholar]
- Yang X, Shears SB. Multitasking in signal transduction by a promiscuous human Ins(3,4,5,6)P(4) 1-kinase/Ins(1,3,4)P(3) 5/6-kinase. Biochem J. 2000;351(Pt 3):551–555. [PMC free article] [PubMed] [Google Scholar]
- York JD. Regulation of nuclear processes by inositol polyphosphates. Biochim Biophys Acta. 2006;1761:552–559. doi: 10.1016/j.bbalip.2006.04.014. [DOI] [PubMed] [Google Scholar]
- York JD, Guo S, Odom AR, Spiegelberg BD, Stolz LE. An expanded view of inositol signaling. Adv Enzyme Regul. 2001;41:57–71. doi: 10.1016/s0065-2571(00)00025-x. [DOI] [PubMed] [Google Scholar]
- York JD, Lew DJ. IP7 guards the CDK gate. Nat Chem Biol. 2008;4:16–17. doi: 10.1038/nchembio0108-16. [DOI] [PubMed] [Google Scholar]
- York JD, Odom AR, Murphy R, Ives EB, Wente SR. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science. 1999;285:96–100. doi: 10.1126/science.285.5424.96. [DOI] [PubMed] [Google Scholar]
- York SJ, Armbruster BN, Greenwell P, Petes TD, York JD. Inositol diphosphate signaling regulates telomere length. J Biol Chem. 2005;280:4264–4269. doi: 10.1074/jbc.M412070200. [DOI] [PubMed] [Google Scholar]
- Zenser N, Ellsmore A, Leasure C, Callis J. Auxin modulates the degradation rate of Aux/IAA proteins. Proc Natl Acad Sci U S A. 2001;98:11795–11800. doi: 10.1073/pnas.211312798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZB, Yang G, Arana F, Chen Z, Li Y, Xia HJ. Arabidopsis inositol polyphosphate 6-/3-kinase (AtIpk2beta) is involved in axillary shoot branching via auxin signaling. Plant Physiol. 2007;144:942–951. doi: 10.1104/pp.106.092163. [DOI] [PMC free article] [PubMed] [Google Scholar]


