This article shows that unprotected telomeres in plant cells are recognized as DNA breaks and elicit a DNA damage signal that is transduced by the ATM and ATR kinases, with differing specificities for eroded and for uncapped telomeres. It finds that both kinases contribute to genomic stability by inducing programmed cell death and elimination of genetically unstable cells.
Abstract
The ends of linear eukaryotic chromosomes are hidden in nucleoprotein structures called telomeres, and loss of the telomere structure causes inappropriate repair, leading to severe karyotypic and genomic instability. Although it has been shown that DNA damaging agents activate a DNA damage response (DDR), little is known about the signaling of dysfunctional plant telomeres. We show that absence of telomerase in Arabidopsis thaliana elicits an ATAXIA-TELANGIECTASIA MUTATED (ATM) and ATM AND RAD3-RELATED (ATR)-dependent DDR at telomeres, principally through ATM. By contrast, telomere dysfunction induces an ATR-dependent response in telomeric Conserved telomere maintenance component1 (Ctc1)-Suppressor of cdc thirteen (Stn1)-Telomeric pathways in association with Stn1 (CST)–complex mutants. These results uncover a new role for the CST complex in repressing the ATR-dependent DDR pathway in plant cells and show that plant cells use two different DNA damage surveillance pathways to signal telomere dysfunction. The absence of either ATM or ATR in ctc1 and stn1 mutants significantly enhances developmental and genome instability while reducing stem cell death. These data thus give a clear illustration of the action of ATM/ATR-dependent programmed cell death in maintaining genomic integrity through elimination of genetically unstable cells.
INTRODUCTION
Telomeres are the nucleoprotein complex structures present at the ends of eukaryotic chromosomes. They play an essential role in the maintenance of genome integrity by protecting chromosome ends against degradation and, most important, prevent recognition of the ends of linear chromosomes as DNA double-strand breaks (DSBs). Telomeres also resolve the end-replication problem associated with the inability of the DNA polymerase machinery to replicate completely the lagging chromosome strand with the consequent progressive erosion of chromosomal ends at each cell division. To avoid this, most eukaryotes maintain telomere length by the action of a reverse transcriptase called telomerase. The basic telomere structure consists of long tracts of short tandem G-rich DNA repeats and ends with a single-stranded protrusion called the 3′ G-overhang (Palm and de Lange, 2008; Jain and Cooper, 2010).
Mammalian telomeres are bound by shelterin, a complex composed of six proteins that bind double-stranded and single-stranded DNA (ssDNA): Telomeric repeat binding factor1 (TRF1), TRF2, TRF1 and TRF2 Interacting Nuclear Factor2 (TIN2), Protection Of Telomeres1 (POT1), TPP1 (formerly known as TINT1, PTOP, or PIP1), and Repressor/Activator Protein1 (RAP1). TRF1 and TRF2 proteins bind the double-stranded telomeric DNA, whereas POT1 interacts specifically with the ssDNA that constitutes the 3′ G-overhang. TRF1 and TRF2 bind to DNA through a C-terminal SANT/Myb domain, and POT1 binds ssDNA through two oligonucleotide/oligosaccharide binding folds (OB-folds) in its N terminus. The shelterin protein TIN2 simultaneously binds TRF1 and TRF2, stabilizing their interaction at telomeres. TIN2 also binds to the TPP1-POT1 complex, establishing a bridge between the double- and single-strand region of the shelterin complex (Palm and de Lange, 2008; Jain and Cooper, 2010). The protein composition of budding yeast telomeres differs from that of mammals. Budding yeast double-stranded DNA is bound directly by RAP1, which contains two myb domains with little homology to those present in TRF1-TRF2. In Saccharomyces cerevisiae, the trimeric protein complex Cell division cycle13 (Cdc13), Suppressor of cdc13 (Stn1), and Ten1 (CST) associates with the G-overhang through Cdc13 protein, which binds the ssDNA via an OB-fold domain. However, Cdc13 is not an ortholog of POT1. Absence of any of the three CST complex proteins generates accumulation of long G-strands and activation of a DDR (Weinert and Hartwell, 1993; Garvik et al., 1995; Grandin et al., 1997; Grandin et al., 2001). Recently, it has been shown that the CST complex is structurally related to the heterotrimeric replication protein A (RPA) complex that binds ssDNA and plays an essential role in DNA replication, recombination, and repair (Gao et al., 2007; Gelinas et al., 2009; Paschini et al., 2010).
Relatively little is known of shelterin components in plants. Searches for TRF-like proteins have been based on the identification of proteins possessing a Myb-like DNA binding domain containing the telebox motif needed for specific telomeric sequence recognition. Arabidopsis thaliana has 12 proteins with a single C-terminal telebox Myb domain, and those presenting a Myb extension domain are able to bind double-stranded telomeric DNA (Karamysheva et al., 2004). None of these proteins seem to be essential for telomere protection, suggesting redundancy of double-stranded DNA binding telomeric proteins in plants. Two POT1 paralogs have been identified in Arabidopsis, POT1a and POT1b; however, these proteins do not bind single-stranded telomeric DNA in vitro and are not essential for telomere capping (Surovtseva et al., 2007; Shakirov et al., 2009). Recently, two components of the CST complex have been identified in Arabidopsis: STN1 and CTC1 (a putative Cdc13 homolog). Plants lacking STN1 and CTC1 show severe phenotypic defects and genomic instability. As is the case in yeast, plant cells mutated for STN1 and CTC1 display extended telomeric G-overhangs (Song et al., 2008; Surovtseva et al., 2009).
The primary signal transducers of DNA breakage are two phosphatidylinositol 3 kinase-like (PI3K) protein kinases: ATAXIA-TELANGIECTASIA MUTATED (ATM) and ATM AND RAD3-RELATED (ATR). In general, DSBs activate the ATM kinase in a manner dependent on the Mre11, Rad50, Nbs1 (MRN) complex, and ATR is activated by RPA-coated ssDNA (Jazayeri et al., 2006; Cimprich and Cortez, 2008). Both kinases initiate a phosphorylation-mediated signal transduction cascade that leads to cell-cycle arrest and repair of DSBs (Sancar et al., 2004; Su, 2006). They phosphorylate the histone variant H2AX in a large chromatin domain around the damage, inducing the accumulation of other damage-response factors and producing cytologically detectable foci.
Dysfunctional telomeres, resulting from replicative attrition of the telomeric DNA in cells lacking telomerase activity or by absence of the shelterin proteins, are recognized as DSBs and thus activate a DNA damage response (DDR). In mammalian cells with dysfunctional telomeres, this response can be detected by the presence of DNA damage proteins at telomeres, forming telomere dysfunction-induced foci (TIFs). TIFs are similar in protein content to the foci induced at DSB and they generate similar downstream effects, including cell-cycle arrest, senescence, or cell death. The earliest detectable response to dysfunctional telomeres (as for DSBs) is the phosphorylation of the histone variant H2AX by ATM/ATR, generating γ-H2AX foci that colocalize with telomeres (d’Adda di Fagagna et al., 2003; Hao et al., 2004; Herbig et al., 2004). That the shelterin complex is involved in the repression of both kinases is shown by the observation that suppression of TRF2 induces ATM activation (Takai et al., 2003; Celli and de Lange, 2005). Furthermore, ATM is essential for efficient nonhomologous end joining of chromosome ends observed after TRF2 depletion in mouse cells (Denchi and de Lange, 2007). By contrast, the ATR kinase is activated after removal of POT1 from telomeres independently of ATM, leading to the suggestion that the main function of POT1 is to prevent binding of RPA to the telomeres’ single-stranded overhangs and thus block ATR activation (Denchi and de Lange, 2007; Guo et al., 2007). TRF2 thus acts mainly to protect telomeres against ATM activation, and POT1 is principally involved in repression of the ATR pathway.
As for mammals, budding yeast cells with critically short telomeres trigger a DDR. In S. cerevisiae, the mitosis entry checkpoint1 (Mec1) kinase, the ATR homolog, has a predominant role in response to DSBs. In agreement with this, S. cerevisiae cells deficient in telomerase or the ssDNA binding protein Cdc13 activate a Mec1-dependent checkpoint (Enomoto et al., 2002; IJpma and Greider, 2003; Hirano and Sugimoto, 2007).
The role of the ATM and ATR kinases in plants seems to be conserved. Arabidopsis atm mutants are sensitive to agents that induce DSBs (ionizing radiation or methyl methane sulphonate), whereas atr mutant plants are sensitive to replication stress (Garcia et al., 2003; Culligan et al., 2004; Culligan et al., 2006). A role of these proteins in signaling in plants comes from experiments showing that H2AX phosphorylation in response to irradiation-induced DSBs is dependent on ATM and ATR with a preponderant role for ATM (Friesner et al., 2005). Furthermore, we have recently shown that activation of both kinases in response to irradiation-induced DSBs is dependent on the MRN complex (Amiard et al., 2010).
Given the conservation in the activation of the DDR to DSBs in plants, we asked in this article whether dysfunctional telomeres in plants would activate a similar DDR. We show the presence of γ-H2AX foci in late generations of plants lacking telomerase activity or the CST complex. Fluorescence in situ hybridization (FISH) analysis using specific subtelomeric probes shows that these colocalize with telomeres. Thus, unprotected telomeres in plant cells are recognized as DSBs and elicit a DNA damage signal. Short telomeres in telomerase-deficient plants activate both the ATM and ATR kinases, whereas absence of the CST complex initiates an ATR-dependent DDR. We show that chromosomal instability and fusion promoted by dysfunctional telomeres do not require ATM/ATR activation. ctc1 mutant cells deficient for ATM/ATR kinases show increased chromosomal instability, and we observe ATM/ATR-dependent programmed cell death (PCD) of ctc1-mutated plants. Furthermore a correlation exists between the extent of dead cells in meristems and reduction of end-to-end chromosome fusions in pistil cells of mature plants as compared with dividing root tip cells of young plantlets.
RESULTS
Dysfunctional Telomeres in Plant Cells Activate a DNA Damage Response
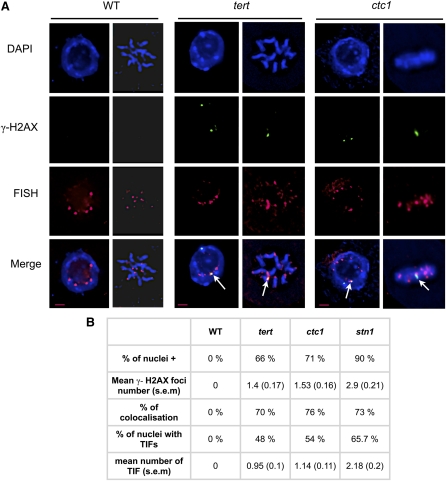
Plant cells lacking telomerase activity show progressive telomere loss, which causes cytogenetic defects, detected as the appearance of dicentric chromosome bridges at mitotic anaphase in later generations (from generation 5) of telomerase mutant plants. Depletion of either STN1 or CTC1 (components of the CST complex) causes rapid telomere erosion (first generation) and increased end-to-end fusions (Song et al., 2008; Surovtseva et al., 2009). Nonfunctional telomeres in Arabidopsis plants are thus recognized as DSBs and are repaired to generate end-to-end chromosome fusions. To test whether the DDR is activated in plant cells lacking functional telomeres, we monitored γ-H2AX foci formation in plants mutated for the telomerase (tert) or components of the CST complex. Given the variable intensity of the phenotype of sibling mutant plants, all analyses were performed on the most affected plants with root lengths from 0.5 to 1.5 cm, 5 d after germination. Examples of roots of the wild type, tert G7, and ctc1 G2 are shown in Supplemental Figure 1 online. In situ immunostaining of root tip nuclei using γ-H2AX antiserum showed no foci in root tip nuclei of the wild-type plants (Figure 1A). Foci were however detected in tert, ctc1, and stn1 mutants, with mean numbers of foci/nuclei of 1.4, 1.53, and 2.9, respectively (Figure 1B). Mutant plants with deprotected telomeric ends thus activate a DDR, detectable by the appearance of γ-H2AX foci.
Figure 1.
Dysfunctional Telomeres in Plant Cells Activate a DDR.
(A) Immunostaining and subtelomeric FISH labeling of root tip nuclei of the wild type (WT), tert, and ctc1 mutants. Nuclei were stained with DAPI (blue), γ-H2AX foci are colored in green, and FISH signals are colored in magenta. Images are collapsed Z-stack projections of a deconvolved three-dimensional image stack. Colocalized foci are indicated with white arrows.
(B) Percentage of nuclei containing at least one γ-H2AX focus (% of nuclei+), the mean number of foci per nucleus (with se), the percentage of colocalization of the foci with the subtelomeric probes, the percentage of nuclei containing at least one TIF, and the mean number of TIFs per nucleus. The number of telomeric or nontelomeric foci and the percentage of nuclei were counted in 75 nuclei (15 nuclei from five different root tips) in each case.
Bar in (A) = 2 μm.
To confirm that the DDR originated at telomeres, we performed FISH of root tip nuclei using nine subtelomeric-specific probes out of 10 ends of the five Arabidopsis chromosomes. The probe to the subtelomeric region of the short arm of chromosome 2 was excluded, because of the presence of rDNA repeats. As shown in Figure 1B, most of the γ-H2AX foci present in the tert, ctc1, and stn1 mutants do colocalize with the chromosome-end probes, confirming that they are TIFs. In absence of telomerase, 48% of the cells exhibit at least one TIF, with a mean number of 0.95 TIF per nucleus. In absence of CTC1 or STN1, 54% and 65.7% of the cells exhibit at least one TIF, and the mean numbers of TIFs per nucleus are 1.14 and 2.18, respectively. We conclude that uncapped telomeres do activate a DDR in Arabidopsis plants.
Short Telomeres in tert Mutant Plants Activate Both ATM and ATR
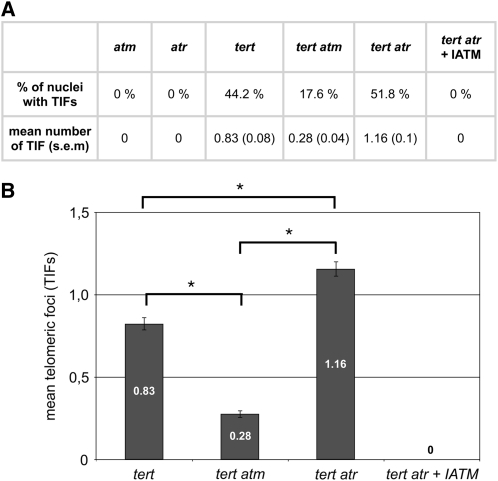
In plant cells, formation of γ-H2AX foci at irradiation-induced DSBs has been shown to be dependent on the activation of both kinases ATM and ATR (Friesner et al., 2005; Amiard et al., 2010). We next asked whether the DDR response activated by dysfunctional telomeres arising from telomere attrition in the absence of telomerase was ATM- or ATR-dependent. To determine which kinase was responsible for γ-H2AX phosphorylation, we generated double mutant tert atm and tert atr plants by crossing G4 tert mutant plants with atr/atr and atm/ATM. G2 (from these crosses) tert, tert atr, and tert atm plants show the expected phenotypes associated with late-generation tert plants. Root tip nuclei from these same-generation tert, tert atm, and tert atr mutant plants were immunostained using the γ-H2AX antiserum, and the numbers of foci that colocalize with the telomeres (TIFs) were counted. No γ-H2AX foci are detected in single mutant atr and atm plants (this article and Amiard et al., 2010), whereas 44% of cells exhibit at least one TIF in the absence of telomerase (Figure 2). In the absence of ATM (tert atm), this proportion is reduced to 17.6%, and more importantly, the mean number of TIFs per nucleus is decreased threefold in the double mutant (0.83 versus 0.28) (Figure 2). Short telomeres in plant cells thus activate an ATM-dependent DDR.
Figure 2.
Short Telomeres in tert Mutant Plants Activate Both ATM and ATR.
(A) The percentage of nuclei containing at least one TIF and the mean number of TIFs per nucleus (with se) after immunostaining and subtelomeric FISH labeling of root tip nuclei of atm, atr, tert, tert atm, and tert atr (± IATM) mutants. The number of telomeric foci and the percentage of nuclei were counted in 75 nuclei (15 nuclei of five different root tips) in each case.
(B) Mean numbers of telomeric foci. Error bars indicate se (n = 75), and an asterisk indicates significant differences (Student’s t test; P < 0.05) between the indicated mutants.
To check whether the remaining TIFs observed in the tert atm cells were associated with activation of the ATR kinase, we monitored TIFs in tert atr double mutant plants. The data shown in Figure 2 indicate that ATM-dependent signaling is responsible for ~0.55 TIF/nucleus in the tert mutant (0.83 − 0.28 = 0.55), and we should thus expect to find this number of TIFs per nucleus. Unexpectedly, we found more than twice the expected mean number of TIFs in the tert atr plants (1.16 per nucleus). Thus, absence of ATR in telomerase mutant cells contributes to telomere damage, and the DDR is ATM-dependent. To check the role of the ATM kinase in γ-H2AX foci generation in tert atr mutant plants, TIFs were monitored in root tips of tert atr mutant plants grown in the presence of the ATM-specific inhibitor (IATM) Ku55933. As expected, no γ-H2AX foci were detected in these cells (Figure 2). We conclude that telomere dysfunction in the absence of telomerase in plant cells activates a DNA damage signal, which is dependent upon both the ATM and the ATR kinases.
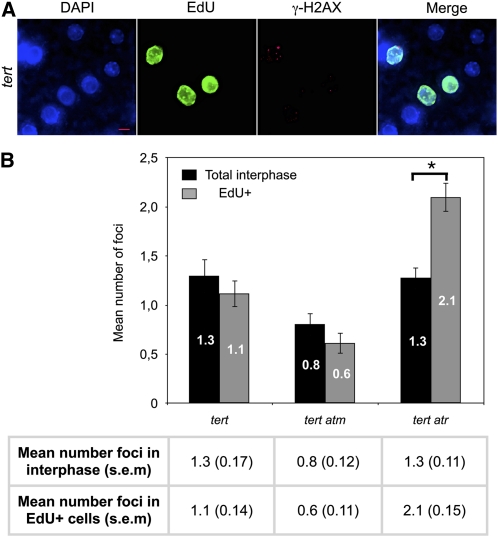
It has previously been shown that tert atr mutant plants show an increased rate of telomere loss compared with tert mutants (Vespa et al., 2005). These authors suggested a role for ATR in the maintenance of telomeric DNA, and this is in accordance with the increased number of TIF-positive cells that we detect in tert atr relative to tert mutant cells. A role for ATR in facilitating replication of telomeres could explain the dramatic loss of telomeric DNA as well as the increased number of TIF-positive cells. If this hypothesis is correct, we should expect an enrichment of γ-H2AX foci in replicating cells of the tert atr mutant plants. To identify cells in the S phase (or early G2 phase), root tips were incubated with the thymidine analog EdU for 60′, fixed, and immunostained for EdU and γ-H2AX detection. The results in Figure 3 show that the mean number of γ-H2AX foci is significantly increased in EdU-positive cells in the double tert atr mutants. Furthermore, no such increase was seen in the tert or the tert atm mutants. These results show the importance of ATR in S/G2 phase cells of the tert mutant, and concur with the suggested role for the ATR kinase in proper telomere replication.
Figure 3.
Increased H2AX Phosphorylation in S-Phase and Early G2-Phase Nuclei of tert atr Mutant Plants.
(A) Immunofluorescence of root tip nuclei labeled with EdU and γ-H2AX in the tert mutant. DNA is stained with DAPI (blue), EdU incorporation is detected in green, and γ-H2AX foci are colored in magenta.
(B) γ-H2AX foci in total interphase nuclei or in replicating (EdU+) nuclei (with se) in tert, tert atm, and tert atr mutants. Error bars indicate se (n = 75), and the asterisk indicates significant differences (Student’s t test; P < 0.05) between the tert atr mutant with or without EdU labeling. The number of foci was counted in 75 nuclei (15 nuclei of five different apices) in each case.
Bar in (A) = 5 μm.
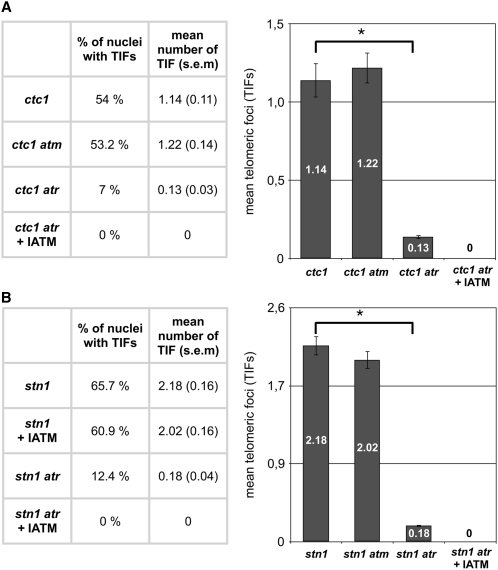
Arabidopsis CST Complex Mutants Activate an ATR-Dependent DNA Damage
Compared with telomerase mutants, Arabidopsis ctc1 and stn1 null mutants show a much more rapid reduction in telomere length associated with increased G-overhangs (Song et al., 2008; Surovtseva et al., 2009). To determine which kinase is activated by the uncapped telomeres in these mutants, we monitored the presence of TIF foci in nuclei of ctc1 atm and ctc1 atr mutant plants (no γ-H2AX foci are detected in single atm and atr mutants—see above). Root tip nuclei were immunostained for γ-H2AX, and foci that colocalized with the subtelomeric FISH signal were counted (TIF). As shown in Figure 4A, 54% of nuclei in ctc1 and 53.2% in ctc1 atm show at least one TIF, in contrast with only 7% of the ctc1 atr mutant nuclei. The role of ATR in signaling the presence deprotected telomeres generated in the absence of CTC1 is also illustrated by the very low mean number of TIFs per nucleus (0.13) in double ctc1 atr mutant plants compared with the mean number of TIFs (1.22) in the double ctc1 atm mutant. As expected, no γ-H2AX foci were observed in ctc1 atr root tip nuclei from plants grown in the presence of the IATM, Ku55933. Similar results were obtained with plants mutated for STN1 (Figure 4B). The mean numbers of TIFs in stn1 root tip nuclei from plants grown in the presence or absence of the IATM are equivalent (65.7% and 60.9% with at least one TIF, respectively). H2AX phosphorylation in cells lacking STN1 does not depend on ATM and is presumably ATR-dependent. This observation is in accordance with the dramatic decrease in mean number of TIFs in stn1 atr root tip nuclei (0.18; Figure 4B), with only 12.4% of the nuclei showing one TIF or more. Unprotected telomeres generated by the absence of the CST complex thus activate an ATR-dependent DDR.
Figure 4.
CST Complex Mutants Activate an ATR-Dependent DDR.
The percentage of nuclei containing at least one TIF and the mean number of TIFs per nucleus (with se) after immunostaining and subtelomeric FISH labeling of root tip nuclei of (A) ctc1, ctc1 atm, ctc1 atr, and ctc1 atr +IATM and (B) stn1, stn1 atm, stn1 atr, and stn1 atr +IATM mutant plants. The number of telomeric foci and the percentage of nuclei were counted in 75 nuclei (15 nuclei of five different apices) in each case. Error bars indicate se (n = 75), and an asterisk indicates significant differences (Student’s t test; P < 0.05).
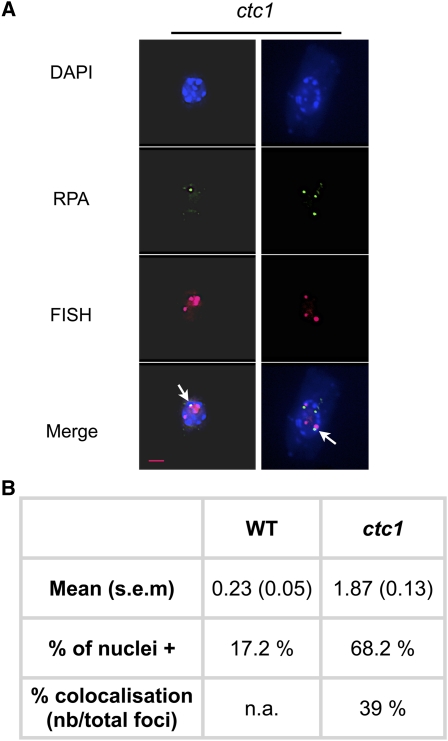
In mammals, ATR activation is dependent on the presence of a RPA-coated ssDNA (Cimprich and Cortez, 2008). Surovtseva et al. (2009) have shown that Arabidopsis ctc1 and stn1 mutants exhibit longer G-overhangs compared with the wild-type plants. Association of RPA with the ssDNA would activate the ATR kinase responsible for the H2AX phosphorylation that we detect in these mutant plants. We reasoned that if RPA associates with the telomere G-overhangs in the absence of the CST complex, we should be able to detect RPA foci. To investigate the presence of RPA foci in the nuclei from the ctc1 mutant, we used an antibody directed against a peptide comprising amino acid residues 116 to 135 of the Arabidopsis RPA1a protein (Osman et al., 2009; kindly provided by Chris Franklin). This analysis shows that 68% of the nuclei are positive for RPA foci, compared with 17% RPA-positive nuclei in wild-type cells (Figure 5). The absence of CTC1 thus induces accumulation of RPA-associated ssDNA. Furthermore, FISH with specific subtelomeric probes confirmed that a substantial fraction of these RPA foci are localized at telomeres (39%), showing the presence of RPA-bound ssDNA at telomeres in the ctc1 mutant. Activation of ATR by this telomeric ssDNA/RPA would provide a highly plausible mechanism for the activation of ATR by telomere dysfunction in the absence of CTC1.
Figure 5.
Increased Numbers of RPA Foci in the ctc1 Mutant.
(A) Images illustrating the presence of RPA foci in ctc1 mutants after immunostaining and subtelomeric BAC-FISH of root tip nuclei. Nuclei were stained with DAPI (blue), RPA foci are colored in green, and FISH signals are colored in magenta. Images are collapsed Z-stack projections of deconvolved three-dimensional image stacks. Colocalized foci are indicated with white arrows.
(B) Mean number of RPA foci per nucleus (with se), percentage of nuclei with at least one RPA focus (% of nuclei+), and the percentage of colocalization of RPA foci with the subtelomeric BAC FISH probes in the wild type (WT) and ctc1 mutants. The number of foci and the percentage of nuclei were counted in 100 nuclei (20 nuclei from five different slides) in each case. n.a., not analyzed; nb, the actual numbers.
Bar in (A) = 2 μm.
The CST complex thus protects Arabidopsis telomeres from initiating an ATR-dependent DDR. However, further studies will be needed to distinguish whether the action of the CST complex is to directly block RPA binding to ssDNA at telomeres or to assure proper telomere replication.
Increased Chromosomal Instability in ctc1 Mutant Plants in the Absence of ATR and/or ATM Kinases
Surovtseva et al. (2009) described the growth phenotypes of three ctc1 mutants (ctc1-1 has a point mutation resulting in a nonsense codon within exon 9; ctc1-2 and ctc1-3 have T-DNA insertions in the sixth exon and 10th intron, respectively). No full-length CTC1 mRNA was detected in these mutants, and all three show severe morphological defects in the first generation, with the few seeds they could recover being unable to germinate. Surprisingly, in our growth conditions, the ctc1-2/ctc1-2 mutants produced from self-pollination of heterozygous ctc1-2/CTC1 plants were phenotypically the wild type. Even the second mutant generation of ctc1-2 plants were mostly the wild type (class 1), with only 20% being sterile or semisterile (see Supplemental Figure 2 online). By striking contrast, ctc1-2 atm and ctc1-2 atr double mutants present severe morphological defects. A total of 65% of second-generation ctc1-2 atr plants were small and grossly deformed (class 3). Second-generation ctc1-2 atm plants were less severely affected, with 15% of the plants showing a wild-type phenotype. They are, however, completely sterile in contrast with the semisterility of atm mutant plants. Similarly, first-generation double stn1-2 atm mutants are completely sterile, and most display a fasciated phenotype (observed only in generation 2 for the single stn1-2 mutant). By contrast, double stn1-2 atr mutants are phenotypically wild type and are similar to the single stn1-2 mutant plants. Notwithstanding, only 35% (42 of 120) of the seeds from G1 stn1-2 atr mutant plants germinated, and of these, 69% (29 of 42) arrested at the cotyledon stage. The absence of either ATM or ATR thus accelerates the onset of developmental defects associated with the uncapped telomeres generated in ctc1-2 or stn1-2 mutant plants.
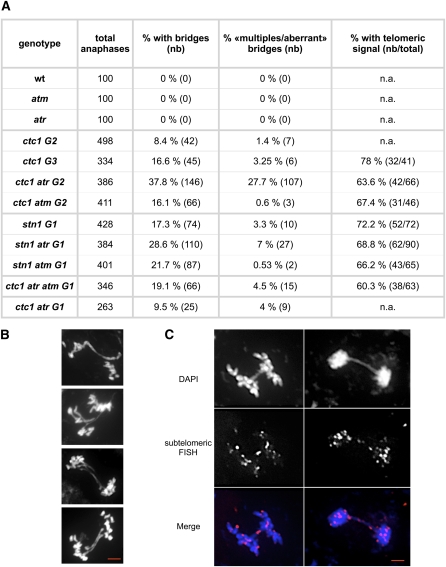
To investigate whether ATM and ATR are needed to prevent chromosomal instability in the absence of the CST complex, we quantified the presence of cytogenetic aberrations in double mutant plants. Recombination of uncapped telomeres leads to fused, dicentric chromosomes, detectable as chromosome bridging at mitotic anaphase. In accordance with previous work (Vespa et al., 2005), we detected no bridges in single atm and atr mutants. Mitotic figures were analyzed from pistils of G2 ctc1, ctc1 atr, and ctc1 atm mutant plants and of G1 stn1, stn1 atr, and stn1 atm mutants. As expected from their mild morphological phenotype, chromosome bridges were present in only 8% of anaphases in ctc1 single mutant plants (Figure 6). This increased to 16% in the double ctc1 atm mutant and to almost 38% in ctc1 atr double mutant plants. Furthermore 27% of anaphases show multiple/aberrant bridges, revealing a severe genomic instability in this double mutant. Similarly, stn1 mutant plants show increased numbers of anaphases, with bridges in the absence of either kinase ATM (21.7%) or ATR (28.6%) compared with the single mutant (17.3%). Both ATM and ATR thus contribute to the maintenance of chromosomal stability in the context of unprotected telomeres.
Figure 6.
Increased Chromosomal Instability in ctc1 and stn1 Mutant Plants in the Absence of ATR and/or ATM.
(A) The percentage of chromosomal bridges, the percentage of multiple bridges/aberrant, and the percentage of anaphases with subtelomeric signal in bridges observed after cytogenetic analysis of flower bud nuclei (from three different plants in each case) of atm, atr, ctc1, ctc1 atm, ctc1 atr, stn1, stn1 atm, stn1 atr, and ctc1 atm atr mutants. n.a., not analyzed; nb, actual numbers.
(B) Examples of multiple bridges/aberrant anaphases observed in ctc1 atr.
(C) Examples of bridges containing a subtelomeric signal in ctc1 atr analyzed by FISH with the nine subtelomeric BAC fluorescent probes (magenta).
Bars in (B) and (C) = 2 μm.
To characterize further the anaphase bridges, FISH analysis was performed using a mixture of nine chromosome end-specific subtelomeric BAC probes (Figure 6A). In accordance with the results of Surovtseva et al. (2009), 78% of anaphase bridges contain a telomeric FISH signal in the ctc1 mutant, and we detect 67.4% and 63.6%, respectively, in ctc1 atm and ctc1 atr mutant plants. The chromosome bridges in these mutants thus result from end-to-end fusions. Similar proportions of bridges including chromosome ends were obtained for stn1 (72.2%), stn1 atr (68.8%), and stn1 atm (66.2%) double mutant plants. Furthermore and importantly, the proportion of chromosome end fusions observed in double ctc1 atr and ctc1 atm mutant cells show that the absence of ATM or ATR signaling does not block fusion of deprotected telomeres. To exclude the possibility that the absence of one kinase is compensated for by the other in the signaling of uncapped telomeres, we constructed the triple ctc1 atr atm mutant. Cytological analysis of the first-generation triple mutant shows that 19.1% of anaphases present bridges and that 60.3% of these contain a telomeric FISH signal. Activation of DNA damage signaling is thus not essential for the generation of end-to-end chromosome fusions in Arabidopsis plants.
Uncapped Telomeres Induce ATM/ATR-Dependent Cell Death of Root Initials
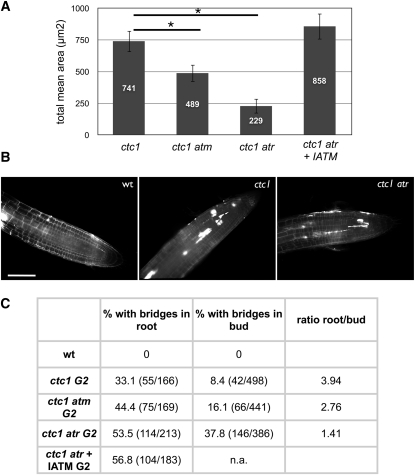
DNA damage checkpoints contribute to genome stability through delay or arrest of the cell cycle to allow DNA repair, or by inducing apoptosis or cell death in metazoans. We thus tested whether the increased genome instability associated with ATM and ATR deficiency in ctc1 and stn1 mutants results from a checkpoint defect. To check whether ATM/ATR-dependent PCD is activated in response to damaged telomeres, we stained plantlets with propidium iodide as a marker of cell death. Loss of membrane integrity in dead cells allows uptake of propidium and thus nuclear staining (Curtis and Hays, 2007). ctc1 mutant plants show an important fraction of dead cells around the quiescent center, and this cell death was strongly inhibited in the absence of the ATM or ATR kinases (Figures 7A and 7B). Uncapped telomeres thus activate ATM/ATR-dependent PCD in actively dividing cells in Arabidopsis. The absence of both kinases (Figure 7A; ctc1 atr +IATM) does not significantly alter levels of cell death in the ctc1 background, and this ATM- and ATR-independent cell death presumably derives from the accumulation of severe genetic damage (Figures 6A and 7C), leading to aneuploidy, mitotic catastrophe, etc.
Figure 7.
ATM/ATR-Dependent PCD of Stem Cells.
(A) ATM/ATR-dependent cell death in root tips. The values represent the mean of the cell death area calculated with eight different root tips. Error bars are ±se (n = 8). An asterisk indicates significant differences (Student’s t test; P < 0.05) between the ctc1 and the ctc1 atm or the ctc1 atr mutants.
(B) Representative images of root tips stained with propidium iodide, which stains dead cells (images are representative of eight root tips). No cell death is observed in wild-type (wt) plants, abundant cell death is observed in the region around the quiescent center in ctc1 or ctc1 atm mutants, and limited cell death is observed ctc1 atr mutants.
(C) Percentages of mitoses showing at least one chromosome bridge in root tips of young plantlets and floral tissue of mature plants and the ratios of values found in roots versus buds. Numbers in parentheses are the actual counts.
Bar in (B) = 50 μm.
The specific culling of genetically damaged stem cells would be expected to act to avoid accumulation of mutated cells that eventually could compromise continued development of the plant or germline. If this is so, in addition to the ATM/ATR-dependent cell death in the ctc1 mutant described above, we would expect to find lower levels of cytological abnormalities in the (surviving) ctc1 cells compared with the ctc1 atr and ctc1 atm cells. To test this prediction, we analyzed mitotic figures in meristematic root cells of ctc1, ctc1 atr, and ctc1 atm, as well as ctc1 atr mutant plants grown in the presence of the IATM. In all cases, anaphase bridges were observed in at least 33% of anaphases (Figure 7C). The striking, fourfold greater proportion of anaphases with bridges (33%) in root meristems of young ctc1 plantlets compared with cells from the mitotic floral pistil cells of mature plants (8%) strongly supports the suggestion that genetically unstable cells are preferentially eliminated in the meristems. That this is caused by an ATR-dependent response is shown by fewer stem cell deaths, and as a result of this, numbers of end-to-end chromosome fusions in pistil cells (38%) are reduced only 1.41-fold compared with the number of anaphase bridges in root tips in the absence of ATR (53%). Plant cells with uncapped telomeres thus activate an ATM/ATR-dependent response, and it is likely that this results in PCD to eliminate cells that would otherwise contribute to increased genomic instability.
DISCUSSION
We show here that cells of developing plants with uncapped telomeres activate a DDR as visualized by the presence of telomeric γ-H2AX foci. We detect the appearance of telomeric H2AX phosphorylation in late-generation telomerase mutant plants, as well as in plants deficient for the CST complex. The DDR in plant cells is thus conserved not only in response to DSBs, but also in the presence of nonfunctional telomeres. To characterize this response further, we have determined the roles of ATM and ATR in activation of DNA damage signaling in response to telomere dysfunction and examined whether these roles differ in uncapped telomeres resulting from the absence of telomerase or of the CST complex. We find that plant cells deficient in telomerase initiate an ATM- and ATR-dependent DDR response, with a predominant role for ATM. This result is in accordance with that found in diploid human fibroblasts undergoing replicative senescence, in which ATM plays a major role in transmitting the telomere DNA damage signal, and ATR is activated in cells lacking ATM (Herbig et al., 2004). The roles of both kinases in signaling the presence of uncapped telomeres caused by telomere erosion in the absence of telomerase thus seem to be conserved.
In contrast with the situation in telomerase mutants, signaling of telomere damage caused by the absence of either CTC1 or STN1, two components of the CST complex, is mediated through an ATR-dependent response. Previous work has shown that chromosome ends from ctc1 or stn1 mutant plants exhibit increased G-overhang ssDNA (Song et al., 2008; Surovtseva et al., 2009). The fact that activation of this ATR-dependent DNA damage signaling could result from association of RPA to this ssDNA is supported by our observation of RPA foci in ctc1 mutant plants, at least 39% of which are localized at telomeres. We thus suggest that a key function of the CST complex is to prevent RPA binding to telomeric 3′-ended ssDNA and thus inhibit activation of an ATR-dependent DDR. This is reminiscent of the role of POT1 protein in mammalian cells, in which deletion of either TPP1 or POT1 induces an ATR-dependent DNA damage signal (Denchi and de Lange, 2007; Guo et al., 2007). The suggestion that POT1 antagonizes RPA1 for interaction with the telomeric G-overhang is supported by the recent demonstration that TERRA and heterogeneous nuclear RNA binding proteins contribute to displace RPA from ssDNA after replication while promoting POT1 binding (Flynn et al., 2011). In Arabidopsis, no detectable interaction is seen between POT1 and single-stranded telomeric DNA, and no defects in telomere capping are found in the pot1 mutants (Surovtseva et al., 2007; Shakirov et al., 2009). Depletion of CTC1 in mammalian cells activates the DDR and phosphorylation of histone H2AX, detectable as formation of γ-H2AX foci. However, whether these γ-H2AX foci are at telomeres is uncertain, because they do not colocalize with TRF2 or TRF1—possibly explained by the complete loss of telomeric repeats (and thus TRF2 and TRF1 binding) from the subset of affected telomeres (Miyake et al., 2009; Surovtseva et al., 2009). Similarly, the kinase responsible for the observed H2AX phosphorylation in CTC1-depleted human cells has not been determined. The CST complex of the yeast S. cerevisiae binds to the single-stranded telomeric DNA through direct interaction with the Cdc13 protein (Nugent et al., 1996). That Cdc13, Stn1, and Ten1 all contribute to telomere end protection is confirmed by the activation of a DDR in the corresponding mutants (Garvik et al., 1995; Grandin et al., 2001). Structural similarities of the OB-fold binding domain of Cdc13 and the OB-folds in the POT1 proteins have led to the suggestion that yeast Cdc13 is the functional homolog of POT1 (Mitton-Fry et al., 2002).
Whether it is direct interaction of the CST complex with the 3′ G-overhang or its action in regulating the length of the telomeric G-strand that prevents RPA binding is presently unknown. The Arabidopsis CTC1 protein has been shown to localize with telomeres in vivo (Surovtseva et al., 2009); however, whether the CST complex binds directly to the single-stranded telomeric G-overhang remains to be demonstrated. The CST complex is also known to play a role in coupling G- and C-strand replication in telomeres of yeast (Qi and Zakian, 2000; Grossi et al., 2004). Similarly, in Arabidopsis, coimmunoprecipitation experiments have recently shown interaction between CTC1 and the DNA polymerase alpha, suggesting a role for the Arabidopsis CST complex in telomeric DNA replication (Price et al., 2010).
Arabidopsis ctc1 and stn1 mutants display loss of telomeric and subtelomeric DNA and high levels of end-to-end chromosome fusions (Song et al., 2008; Surovtseva et al., 2009). As described above, ATR is required for signaling of the telomere damage in these mutants. This observation raises the question of whether the processing and recombination of dysfunctional telomeres generated by absence of the CST complex is dependent on ATR. We found that absence of either ATM or ATR increases the numbers of mitotic anaphase bridges and genomic instability in CST mutant plants. Thus, although unprotected telomeres are recognized as damaged DNA in Arabidopsis through activation of ATM and ATR, processing of these chromosome ends is clearly not dependent upon activation of the ATM and ATR signaling. Although it is possible that ATM compensates for the absence of ATR (and vice versa) to some extent in its role in chromosome end-to-end fusion, this is clearly not the case in G1 ctc1 atr atm triple mutant plants, which show even higher levels of anaphase chromosome bridges than do G1 ctc1 atr double mutants. These results from Arabidopsis thus contrast with the requirement for ATM or ATR signaling in fusion of dysfunctional telomeres in terf2−/− and Pot1−/− mouse cells (Denchi and de Lange, 2007). Our Arabidopsis results, however, concur with analyses of stably transformed p53−/− mouse cell lines, in which chromosomal instability (and fusions) resulting from telomere dysfunction induced by depletion of TPP1 is not dependent on ATM. In these ATM−/−, p53−/− mutant cells depleted for TPP1, 55% of mitoses contain aberrant chromosomes, including p-p fusions containing telomeric DNA at the site of fusion. These experiments show that mammalian cells stably depleted in Tpp1 can recognize and repair uncapped telomeres in the absence of ATM and p53 (Guo et al., 2007).
The DDR in mammalian cells activates p53, which in turn induces cellular senescence to eliminate genetically damaged cells. Telomere dysfunction leads to p53-dependent cell death and apoptosis in mice and thus acts as a p53-dependent tumor suppression mechanism (Perera et al., 2008; Sahin et al., 2011). PCD as a response to DNA damage induced by UV-B light, x-rays, or radiomimetic drugs has recently been described in Arabidopsis. This selectively affects dividing stem cells of roots and shoots and involves transduction of the DNA damage signal by the ATM and ATR kinases (Fulcher and Sablowski, 2009; Furukawa et al., 2010). We show here that plants with dysfunctional telomeres caused by absence of the CST complex also eliminate damaged cells from root meristems by eliciting ATM/ATR-dependent PCD.
ctc1 mutant cells have nonfunctional telomeres that are recognized as DSBs and are repaired to generate end-to-end chromosome fusions. Thus, dividing cells will enter into break and fusion cycles that activate the ATM/ATR kinases and induce cell death. Given this key role of ATM/ATR signaling in the elimination of genetically damaged cells, we were initially surprised to find similar proportions of root tip meristem cells showing chromosomal instability in ctc1, ctc1 atr, and ctc1 atm mutants (33 to 53.5% of mitoses with at least one visible chromosome bridge; Figure 7C). Similar analyses of mitotic pistil cells from ctc1 plants, however, show only 8% of the anaphases with detectable chromosome bridges (a fourfold decrease). The proportion of genetically unstable cells thus decreases dramatically through development, with very high levels found in the meristems of young plantlets and much lower levels in mitoses of the floral tissues of the adult plant. That this selective elimination of genetically unstable cells through development is caused by the action of ATR and ATM is confirmed by the lesser decrease observed in ctc1 atr and ctc1 atm mutants (ratios of proportions of anaphases with detectable chromosome bridges in root versus pistil of 1.41 and 2.76, respectively) (Figure 7C). These data thus provide a clear biological illustration of the action of ATM/ATR-dependent surveillance of genome integrity in maintaining genome stability through elimination of genetically unstable cells.
METHODS
Plant Material and Growth Conditions
All Arabidopsis thaliana plants used in this study are of Colombia ecotype. The T-DNA insertional Arabidopsis mutants and PCR-based genotyping of tert (Fitzgerald et al., 1999), ctc1-2 (Surovtseva et al., 2009), and stn1-2 (Song et al., 2008) have been described previously. Seeds of the Arabidopsis atm-2 line (Salk_006953-3-2) or of the atr-2 line (Salk _532841-8) were obtained from the European Arabidopsis Stock Centre (Nottingham, United Kingdom).
The double tert atr, ctc1-2 atr, and stn1-2 atr mutants were produced by crossing tert/tert, stn1-2/stn-2, or ctc1-2/ctc1-2 homozygotes with an atr/atr homozygote using standard techniques. The double tert atm, ctc1-2 atm, and stn1-2 atm mutants were produced by crossing tert/tert, stn1-2/stn1-2, or ctc1-2/ctc1-2 homozygotes with an atm/ATM heterozygote using standard techniques. The triple ctc1-2 atr-2 atm-2 was produced by crossing the double atr/atr ctc1-2/CTC1 with the double atr/atr atm/ATM plant.
Seeds were surface-sterilized by 7% calcium hypochlorite treatment for 15 min and rinsed four times with sterile water. Seeds were sown on Petri plates containing 1× Murashige and Skoog medium including vitamins and MES buffer (product no. M0255; Duchefa Biochemie), plus 1% Suc (Duchefa Biochemie), solidified with 0.8% agar (Bacto-Agar; DIFCO Laboratories). Petri dishes were placed at 4°C overnight (15 h) and transferred to a growth chamber (16 h light, 8 h in dark) at 23°C.
Plant Treatment
To inhibit ATM kinase, Arabidopsis plants were sown on Petri plates containing ku55933 (IATM; Calbiochem) at 10 μM (Amiard et al., 2010).
For EdU incorporation, Arabidopsis seedlings were germinated as usual and after 5 d were transferred 1 h in liquid medium containing 10 μM of EdU. Seedlings were then fixed in 3.7% formaldehyde in PBS and washed three times in PBS. After permeabilization in Triton X-100 0.5%, EdU detection was performed as indicated by the manufacturer (Invitrogen Click-iT EdU Alexa Fluor 594 Imaging kit) and as previously described (Amiard et al., 2010).
Slide Preparation and Immunostaining
γ-H2AX antiserum was raised and purified against a phospho-specific Arabidopsis H2AX peptide as previously described (Charbonnel et al., 2010).
Anti-RPA antiserum was kindly provided by Dr. Chris Franklin. The peptide antiserum was raised in rabbit against the 20 amino acid sequence ETDTEAQKTFSGTGNIPPPN (residues 116 to 135) conjugated to KLH (Sigma-Genosys) (Osman et al., 2009).
For γ-H2AX immunostaining, 5 d after germination, root tips were prepared as described previously (Liu et al., 1993; Friesner et al., 2005) with minor modifications: Root tips were fixed for 45 min in 4% paraformaldehyde in a solution of 1 X PME (50 mM Pipes, pH 6.9, 5 mM MgSO4, 1 mM EGTA) and then washed three times for 5 min in 1× PME. Tips were digested for 1 h in a 1% (w/v) cellulase, 0.5% (w/v) cytohelicase, 1% (w/v) pectolyase (Sigma-Aldrich; Refs. C1794, C8274, P5936) solution prepared in PME and then were washed three times for 5 min each time in PME. These tips were squashed gently onto slides as described previously (Liu et al., 1993), air dried, and stored at −80°C. The phenotype of the tert, tert atm, and tert atr mutant plants is heterogeneous between siblings; therefore, we decided to examine the most affected plants with root lengths between 0.5 and 1.5 cm 5 d after germination.
For RPA immunostaining, 5-d-old root tips were fixed for 45 min in 4% paraformaldehyde in solution of 1× PME and then were washed three times for 5 min in 1× PME. Tips were digested for 1 h in a 1% (w/v) cellulase, 0.5% (w/v) cytohelicase, 1% (w/v) pectolyase solution prepared in PME and then washed three times for 5 min in PME. Tips were then crushed in 10 μL of Lipsol 1%, pH 9, and incubated for 2 min with vigorous agitation. The suspension was gently dispersed over the slide using a glass spreader and was finally fixed by adding 4% paraformaldehyde, pH 8.0. The slides were then air dried, rinsed with water and 1× PBS, and stored at −80°C.
Each slide was incubated overnight at 4°C with 50 to 100 μL rabbit, anti-plant γ-H2AX antiserum diluted 1:600 (or 1:500 for anti-RPA) in fresh blocking buffer (3% BSA, 0.05% Tween-20 [Sigma-Aldrich] in 1× PBS). Slides were washed three times for 5 min in 1× PBS solution and then incubated for 2 to 3 h at room temperature in 50 to 100 μL blocking buffer consisting of Alexa 488-conjugated goat anti-rabbit (Molecular Probes; 1:1000) secondary antibodies. Finally, slides were washed three times for 5 min in 1× PBS and mounted in Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) (2 μg/mL) (Vector Laboratories).
DAPI Staining of Mitoses and FISH
Whole inflorescences were collected and fixed, and mitotic chromosomes of fixed flower pistils were squashed on a slide (Caryl et al., 2000). Slides were mounted using Vectashield (Vector Laboratories) mounting medium with 1.5 mg/mL DAPI.
FISH was performed according to Mokros et al. (2006) as previously described (Vannier et al., 2009) using BACs from subtelomeric regions of Arabidopsis chromosomes (F6F3, F23A5, F17A22, F4P13, T20O10, F6N15, T19P19, F7J8, K9I9), labeled with biotin (Amersham) by standard nick translation reactions (Roche). For the detection of biotin-labeled probes, avidin conjugated with Texas Red (1:500; Vector Laboratories) followed by goat anti-avidin conjugated with biotin (1:100; Vector Laboratories) and avidin-Texas Red (1:500) were used. FISH after immunostaining require a postfixation step of 30′ in 4% formaldehyde. Slides were observed by fluorescence microscopy, and images were further processed using Adobe Photoshop.
Microscopy and Analysis of γ-H2AX and RPA1 Foci
Images were acquired on a Zeiss AxioImager Z1 epifluorescence microscope using Axiovision software. Measurements were performed using the same software, and images were processed using Adobe Photoshop. Image stacks were captured in three dimensions (x, y, z) and deconvoluted with the deconvolution module (theoretical PSF, iterative algorithm) of AxioVision 4.6.2 (Zeiss) software to affine γ-H2AX foci, which were then counted by eye. For presentation, the pictures are collapsed Z-stack projections obtained using the Extended-focus module (projection method) of the AxioVision 4.6.2.
Cell Death Assay
A total of 5 d after germination, seedlings were immersed in propidium iodide solution (5 μg/mL in water) for 1 min and rinsed three times with water. Root tips were then transferred to slides in a drop of water and covered with a cover slip for observation under the fluorescence microscope with a Zeiss filter set 43HE (adapted from Curtis and Hays, 2007). To estimate the abundance of the cell death in the root meristem, we calculated the mean death areas of each root from collapsed Z-stack projections obtained using the Extended-focus module (projection method).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ATR (locus AT5G40820; GenBank NM_123447); ATM (locus AT3G48190; GenBank NM_ 114689.6); CTC1 (locus AT4G09680; GenBank NM_117036.5); STN1 (locus AT1G07130; GenBank NM_100586.2); TERT (locus AT5G16850; GenBank NM_121691.3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Examples of Root Material Used for Analyses of the Wild Type, tert, and ctc1 Mutants.
Supplemental Figure 2. Morphological Analysis of ctc1, ctc1 atm, and ctc1 atr Mutants.
Acknowledgments
We thank the members of the recombination group for their help and discussions. We thank Chris Franklin for the anti-RPA antibodies. This work was partly financed by a European Union research grant (LSHG-CT-2005-018785), a French Government ANR grant (ANR-07-BLAN-0068), the Centre National de la Recherche Scientifique, the Université Blaise Pascal, the Université d’Auvergne, and the Institut National de la Santé et la Recherche Medicale.
AUTHOR CONTRIBUTIONS
All authors contributed to this work. S.A., E.A., and A.D. carried out the experimental work, and S.A., C.I.W., and M.E.G. designed the experiments and wrote the text. All authors have seen and approved this article.
References
- Amiard S., Charbonnel C., Allain E., Depeiges A., White C.I., Gallego M.E. (2010). Distinct roles of the ATR kinase and the Mre11-Rad50-Nbs1 complex in the maintenance of chromosomal stability in Arabidopsis. Plant Cell 22: 3020–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caryl A.P., Armstrong S.J., Jones G.H., Franklin F.C. (2000). A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma 109: 62–71 [DOI] [PubMed] [Google Scholar]
- Celli G.B., de Lange T. (2005). DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 7: 712–718 [DOI] [PubMed] [Google Scholar]
- Charbonnel C., Gallego M.E., White C.I. (2010). Xrcc1-dependent and Ku-dependent DNA double-strand break repair kinetics in Arabidopsis plants. Plant J. 64: 280–290 [DOI] [PubMed] [Google Scholar]
- Cimprich K.A., Cortez D. (2008). ATR: An essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan K., Tissier A., Britt A. (2004). ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16: 1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan K.M., Robertson C.E., Foreman J., Doerner P., Britt A.B. (2006). ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 48: 947–961 [DOI] [PubMed] [Google Scholar]
- Curtis M.J., Hays J.B. (2007). Tolerance of dividing cells to replication stress in UVB-irradiated Arabidopsis roots: Requirements for DNA translesion polymerases eta and zeta. DNA Repair (Amst.) 6: 1341–1358 [DOI] [PubMed] [Google Scholar]
- d’Adda di Fagagna F., Reaper P.M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P. (2003). A DNA damage checkpoint response in telomere-initiated senescence. Nature 426: 194–198 [DOI] [PubMed] [Google Scholar]
- Denchi E.L., de Lange T. (2007). Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448: 1068–1071 [DOI] [PubMed] [Google Scholar]
- Enomoto S., Glowczewski L., Berman J. (2002). MEC3, MEC1, and DDC2 are essential components of a telomere checkpoint pathway required for cell cycle arrest during senescence in Saccharomyces cerevisiae. Mol. Biol. Cell 13: 2626–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald M.S., Riha K., Gao F., Ren S., McKnight T.D., Shippen D.E. (1999). Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl. Acad. Sci. USA 96: 14813–14818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn R.L., Centore R.C., O’Sullivan R.J., Rai R., Tse A., Songyang Z., Chang S., Karlseder J., Zou L. (2011). TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 471: 532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner J.D., Liu B., Culligan K., Britt A.B. (2005). Ionizing radiation-dependent gamma-H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3-related. Mol. Biol. Cell 16: 2566–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher N., Sablowski R. (2009). Hypersensitivity to DNA damage in plant stem cell niches. Proc. Natl. Acad. Sci. USA 106: 20984–20988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T., Curtis M.J., Tominey C.M., Duong Y.H., Wilcox B.W., Aggoune D., Hays J.B., Britt A.B. (2010). A shared DNA-damage-response pathway for induction of stem-cell death by UVB and by gamma irradiation. DNA Repair (Amst.) 9: 940–948 [DOI] [PubMed] [Google Scholar]
- Gao H., Cervantes R.B., Mandell E.K., Otero J.H., Lundblad V. (2007). RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 14: 208–214 [DOI] [PubMed] [Google Scholar]
- Garcia V., Bruchet H., Camescasse D., Granier F., Bouchez D., Tissier A. (2003). AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 15: 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvik B., Carson M., Hartwell L. (1995). Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15: 6128–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelinas A.D., Paschini M., Reyes F.E., Héroux A., Batey R.T., Lundblad V., Wuttke D.S. (2009). Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain. Proc. Natl. Acad. Sci. USA 106: 19298–19303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N., Damon C., Charbonneau M. (2001). Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 20: 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N., Reed S.I., Charbonneau M. (1997). Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 11: 512–527 [DOI] [PubMed] [Google Scholar]
- Grossi S., Puglisi A., Dmitriev P.V., Lopes M., Shore D. (2004). Pol12, the B subunit of DNA polymerase alpha, functions in both telomere capping and length regulation. Genes Dev. 18: 992–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Deng Y., Lin Y., Cosme-Blanco W., Chan S., He H., Yuan G., Brown E.J., Chang S. (2007). Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. EMBO J. 26: 4709–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L.Y., Strong M.A., Greider C.W. (2004). Phosphorylation of H2AX at short telomeres in T cells and fibroblasts. J. Biol. Chem. 279: 45148–45154 [DOI] [PubMed] [Google Scholar]
- Herbig U., Jobling W.A., Chen B.P., Chen D.J., Sedivy J.M. (2004). Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a). Mol. Cell 14: 501–513 [DOI] [PubMed] [Google Scholar]
- Hirano Y., Sugimoto K. (2007). Cdc13 telomere capping decreases Mec1 association but does not affect Tel1 association with DNA ends. Mol. Biol. Cell 18: 2026–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IJpma A.S., Greider C.W. (2003). Short telomeres induce a DNA damage response in Saccharomyces cerevisiae. Mol. Biol. Cell 14: 987–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D., Cooper J.P. (2010). Telomeric strategies: Means to an end. Annu. Rev. Genet. 44: 243–269 [DOI] [PubMed] [Google Scholar]
- Jazayeri A., Falck J., Lukas C., Bartek J., Smith G.C.M., Lukas J., Jackson S.P. (2006). ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 8: 37–45 [DOI] [PubMed] [Google Scholar]
- Karamysheva Z.N., Surovtseva Y.V., Vespa L., Shakirov E.V., Shippen D.E. (2004). A C-terminal Myb extension domain defines a novel family of double-strand telomeric DNA-binding proteins in Arabidopsis. J. Biol. Chem. 279: 47799–47807 [DOI] [PubMed] [Google Scholar]
- Liu B., Marc J., Joshi H.C., Palevitz B.A. (1993). A gamma-tubulin-related protein associated with the microtubule arrays of higher plants in a cell cycle-dependent manner. J. Cell Sci. 104: 1217–1228 [DOI] [PubMed] [Google Scholar]
- Mitton-Fry R.M., Anderson E.M., Hughes T.R., Lundblad V., Wuttke D.S. (2002). Conserved structure for single-stranded telomeric DNA recognition. Science 296: 145–147 [DOI] [PubMed] [Google Scholar]
- Miyake Y., Nakamura M., Nabetani A., Shimamura S., Tamura M., Yonehara S., Saito M., Ishikawa F. (2009). RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell 36: 193–206 [DOI] [PubMed] [Google Scholar]
- Mokros P., Vrbsky J., Siroky J. (2006). Identification of chromosomal fusion sites in Arabidopsis mutants using sequential bicolour BAC-FISH. Genome 49: 1036–1042 [DOI] [PubMed] [Google Scholar]
- Nugent C.I., Hughes T.R., Lue N.F., Lundblad V. (1996). Cdc13p: A single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274: 249–252 [DOI] [PubMed] [Google Scholar]
- Osman K., Sanchez-Moran E., Mann S.C., Jones G.H., Franklin F.C. (2009). Replication protein A (AtRPA1a) is required for class I crossover formation but is dispensable for meiotic DNA break repair. EMBO J. 28: 394–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W., de Lange T. (2008). How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42: 301–334 [DOI] [PubMed] [Google Scholar]
- Paschini M., Mandell E.K., Lundblad V. (2010). Structure prediction-driven genetics in Saccharomyces cerevisiae identifies an interface between the t-RPA proteins Stn1 and Ten1. Genetics 185: 11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera S.A., et al. (2008). Telomere dysfunction promotes genome instability and metastatic potential in a K-ras p53 mouse model of lung cancer. Carcinogenesis 29: 747–753 [DOI] [PubMed] [Google Scholar]
- Price C.M., Boltz K.A., Chaiken M.F., Stewart J.A., Beilstein M.A., Shippen D.E. (2010). Evolution of CST function in telomere maintenance. Cell Cycle 9: 3157–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H., Zakian V.A. (2000). The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 14: 1777–1788 [PMC free article] [PubMed] [Google Scholar]
- Sahin E., et al. (2011). Telomere dysfunction induces metabolic and mitochondrial compromise. Nature 470: 359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Lindsey-Boltz L.A., Unsal-Kaçmaz K., Linn S. (2004). Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73: 39–85 [DOI] [PubMed] [Google Scholar]
- Shakirov E.V., McKnight T.D., Shippen D.E. (2009). POT1-independent single-strand telomeric DNA binding activities in Brassicaceae. Plant J. 58: 1004–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Leehy K., Warrington R.T., Lamb J.C., Surovtseva Y.V., Shippen D.E. (2008). STN1 protects chromosome ends in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 19815–19820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T.T. (2006). Cellular responses to DNA damage: One signal, multiple choices. Annu. Rev. Genet. 40: 187–208 [DOI] [PubMed] [Google Scholar]
- Surovtseva Y.V., Shakirov E.V., Vespa L., Osbun N., Song X., Shippen D.E. (2007). Arabidopsis POT1 associates with the telomerase RNP and is required for telomere maintenance. EMBO J. 26: 3653–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva Y.V., Churikov D., Boltz K.A., Song X., Lamb J.C., Warrington R., Leehy K., Heacock M., Price C.M., Shippen D.E. (2009). Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol. Cell 36: 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H., Smogorzewska A., de Lange T. (2003). DNA damage foci at dysfunctional telomeres. Curr. Biol. 13: 1549–1556 [DOI] [PubMed] [Google Scholar]
- Vannier J.B., Depeiges A., White C., Gallego M.E. (2009). ERCC1/XPF protects short telomeres from homologous recombination in Arabidopsis thaliana. PLoS Genet. 5: e1000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vespa L., Couvillion M., Spangler E., Shippen D.E. (2005). ATM and ATR make distinct contributions to chromosome end protection and the maintenance of telomeric DNA in Arabidopsis. Genes Dev. 19: 2111–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T.A., Hartwell L.H. (1993). Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics 134: 63–80 [DOI] [PMC free article] [PubMed] [Google Scholar]