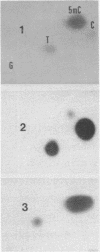
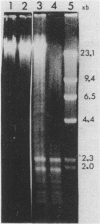
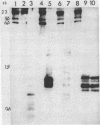
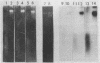
Abstract
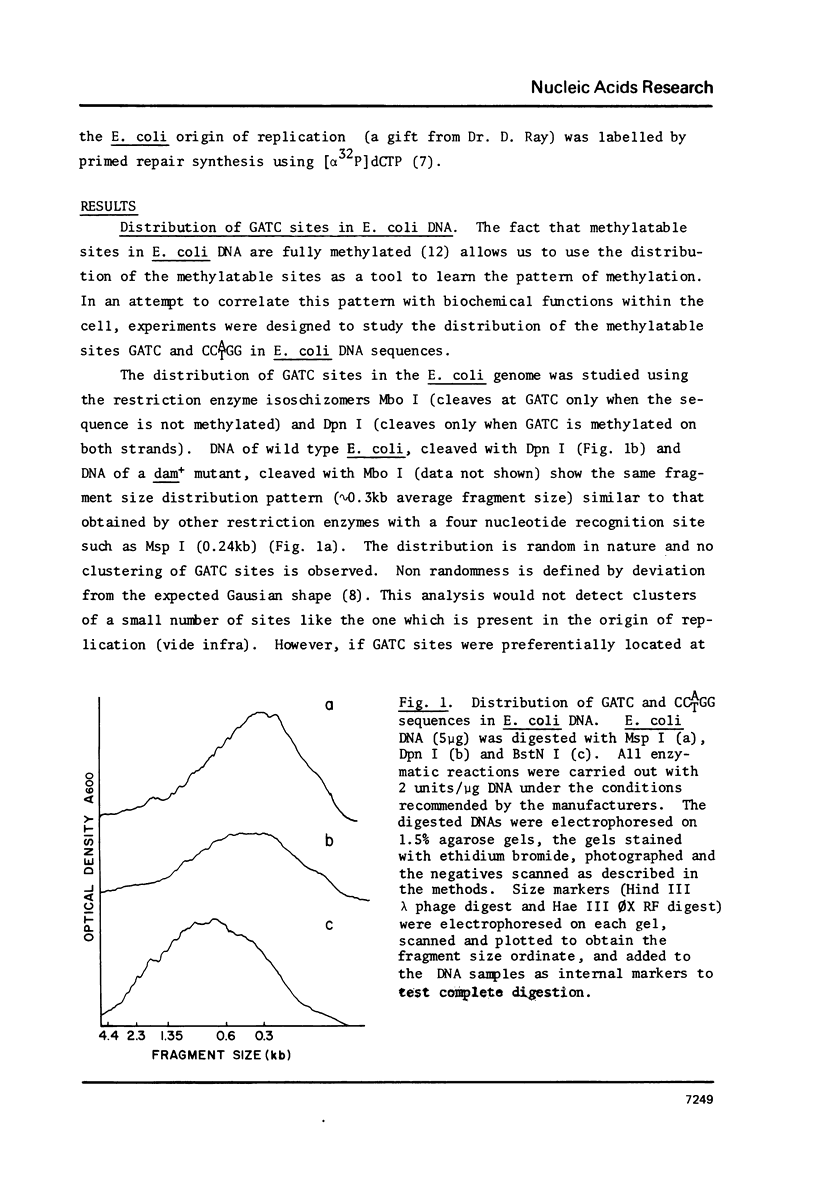
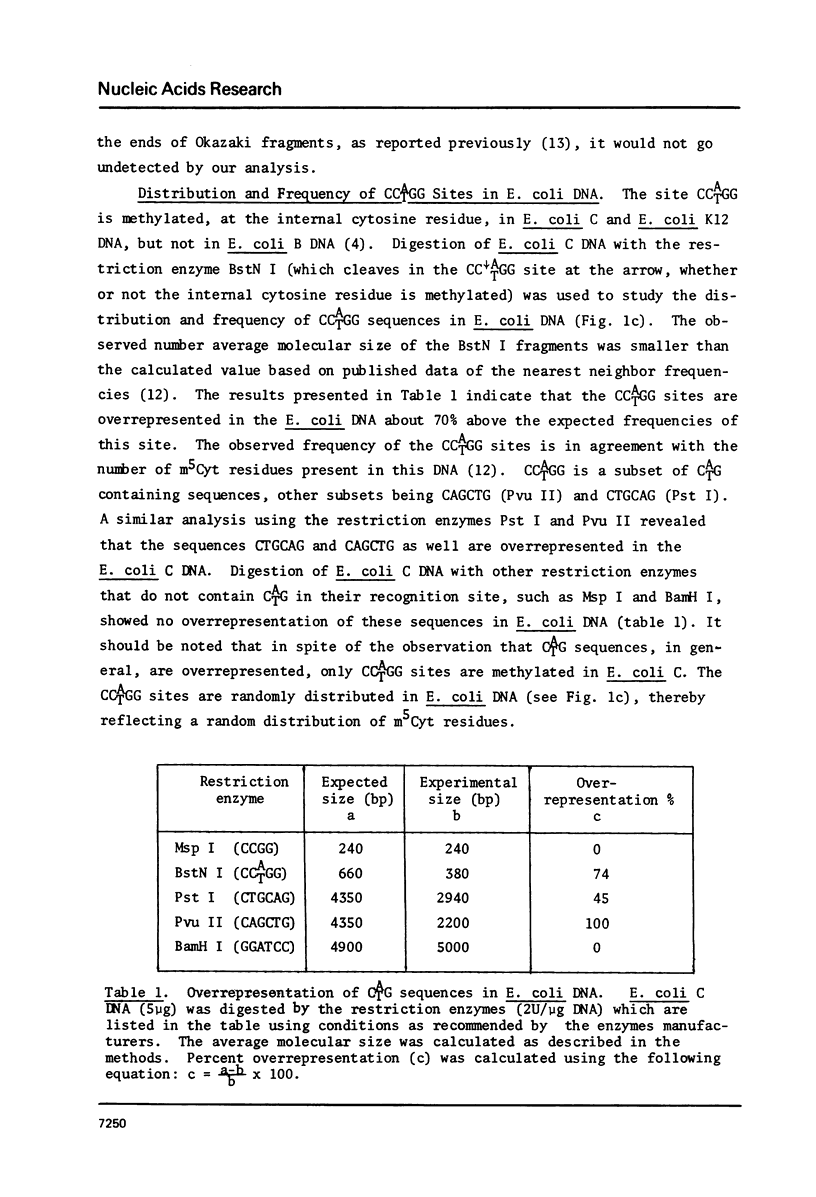
The distribution of the methylatable sites GATC and CCATGG was studied by analyzing the molecular average size of restriction fragments of E. coli DNA. Both sites were found to be randomly distributed, reflecting a random pattern of methylation. The methylation pattern of specific sequences such as the origin of replication and rRNA genes has been studied in wild type E. coli and a methylation deficient (dam- dcm-) mutant. These sequences were found to be methylated in wild type cells and unmethylated in the mutant indicating that there is no effect of the state of methylation of these sequences on their expression. Analysis of the state of methylation of GATC sites in newly replicating DNA using the restriction enzyme Dpn I (cleaves only when both strands are methylated) revealed no detectable hemimethylated DNA suggesting that methylation occurs at the replication fork. Taking together the results presented here and previously published data (5), we arrive at the conclusion that the most likely function of E. coli DNA methylations is probably in preventing nuclease activity.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amikam D., Razin S., Glaser G. Ribosomal RNA genes in Mycoplasma. Nucleic Acids Res. 1982 Jul 24;10(14):4215–4222. doi: 10.1093/nar/10.14.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros I., Kiss A., Venetianer P. Physical map of the seven ribosomal RNA genes of Escherichia coli. Nucleic Acids Res. 1979;6(5):1817–1830. doi: 10.1093/nar/6.5.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J. D., von Hippel P. H. Effects of methylation on the stability of nucleic acid conformations. Studies at the polymer level. J Biol Chem. 1978 Feb 10;253(3):927–934. [PubMed] [Google Scholar]
- Glaser G., Enquist L., Cashel M. ColE1 cloning of a ribosomal RNA promoter region from lambdarifd18 by selection for lambda integration and excision functions. Gene. 1977;2(3-4):159–172. doi: 10.1016/0378-1119(77)90015-4. [DOI] [PubMed] [Google Scholar]
- Glickman B., van den Elsen P., Radman M. Induced mutagenesis in dam- mutants of Escherichia coli: a role for 6-methyladenine residues in mutation avoidance. Mol Gen Genet. 1978 Jul 25;163(3):307–312. doi: 10.1007/BF00271960. [DOI] [PubMed] [Google Scholar]
- Gomez-Eichelmann M. C., Lark K. G. Endo R DpnI restriction of Escherichia coli DNA synthesized in vitro. Evidence that the ends of Okazaki pieces are determined by template deoxynucleotide sequence. J Mol Biol. 1977 Dec 15;117(3):621–635. doi: 10.1016/0022-2836(77)90061-4. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y., Cedar H., Razin A. Restriction enzyme digestion of hemimethylated DNA. Nucleic Acids Res. 1981 Jun 11;9(11):2509–2515. doi: 10.1093/nar/9.11.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberman A., Heywood J., Meselson M. DNA modification methylase activity of Escherichia coli restriction endonucleases K and P. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3138–3141. doi: 10.1073/pnas.69.11.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaguni J., LaVerne L. S., Ray D. S. Cloning and expression of the Escherichia coli replication origin in a single-stranded DNA phage. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6250–6254. doi: 10.1073/pnas.76.12.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korba B. E., Hays J. B. Novel mutations of Escherichia coli that produce recombinogenic lesions in DNA. V. Recombinogenic plasmids from arl mutants of Escherichia coli are unusually sensitive to nuclease S1 and partially deficient in cytosine methylation at C-C-(A/T)-G-G sequences. J Mol Biol. 1982 May 15;157(2):213–235. doi: 10.1016/0022-2836(82)90231-5. [DOI] [PubMed] [Google Scholar]
- Kühnlein U., Linn S., Arber W. Host specificity of DNA produced by Escherichia coli. XI. In vitro modification of phage fd replicative form. Proc Natl Acad Sci U S A. 1969 Jun;63(2):556–562. doi: 10.1073/pnas.63.2.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G. Adenine methylation of Okazaki fragments in Escherichia coli. J Bacteriol. 1976 Dec;128(3):853–854. doi: 10.1128/jb.128.3.853-854.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Biological function for 6-methyladenine residues in the DNA of Escherichia coli K12. J Mol Biol. 1974 May 15;85(2):309–322. doi: 10.1016/0022-2836(74)90366-0. [DOI] [PubMed] [Google Scholar]
- Meijer M., Beck E., Hansen F. G., Bergmans H. E., Messer W., von Meyenburg K., Schaller H. Nucleotide sequence of the origin of replication of the Escherichia coli K-12 chromosome. Proc Natl Acad Sci U S A. 1979 Feb;76(2):580–584. doi: 10.1073/pnas.76.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveh-Many T., Cedar H. Topographical distribution of 5-methylcytosine in animal and plant DNA. Mol Cell Biol. 1982 Jul;2(7):758–762. doi: 10.1128/mcb.2.7.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki T., Okazaki R. Mechanism of DNA chain growth. IV. Direction of synthesis of T4 short DNA chains as revealed by exonucleolytic degradation. Proc Natl Acad Sci U S A. 1969 Dec;64(4):1242–1248. doi: 10.1073/pnas.64.4.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack Y., Stein R., Razin A., Cedar H. Methylation of foreign DNA sequences in eukaryotic cells. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6463–6467. doi: 10.1073/pnas.77.11.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Friedman J. DNA methylation and its possible biological roles. Prog Nucleic Acid Res Mol Biol. 1981;25:33–52. doi: 10.1016/s0079-6603(08)60482-1. [DOI] [PubMed] [Google Scholar]
- Razin A., Goren D., Friedman J. Studies on the biological role of DNA methylation: inhibition of methylation and maturation of the bacteriophage phichi174 by nicotinamide. Nucleic Acids Res. 1975 Oct;2(10):1967–1974. doi: 10.1093/nar/2.10.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Razin A., Urieli S., Pollack Y., Gruenbaum Y., Glaser G. Studies on the biological role of dna methylation; IV. Mode of methylation of DNA in E. coli cells. Nucleic Acids Res. 1980 Apr 25;8(8):1783–1792. doi: 10.1093/nar/8.8.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Oka A., Sugisaki H., Takanami M., Nishimura A., Yasuda Y., Hirota Y. Nucleotide sequence of Escherichia coli K-12 replication origin. Proc Natl Acad Sci U S A. 1979 Feb;76(2):575–579. doi: 10.1073/pnas.76.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyskind J. W., Smith D. W. Nucleotide sequence of the Salmonella typhimurium origin of DNA replication. Proc Natl Acad Sci U S A. 1980 May;77(5):2460–2464. doi: 10.1073/pnas.77.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]