Abstract
A multiple reaction monitoring liquid chromatography method with tandem mass spectrometric detection for quantitation of Staphylococcus aureus via phage amplification detection is described. This phage amplification detection method enables rapid and accurate quantitation of viable S. aureus by detecting an amplified capsid protein from a specific phage. A known amount of metabolically labeled 15N reference bacteriophage, utilized as the input phage and as the internal standard for quantitation, was spiked into S. aureus samples. Following a 2-h incubation, the sample was subjected to a 3-min rapid trypsin digest and analyzed by high-throughput liquid chromatography tandem mass spectrometric detection targeting peptides unique to both the 15N (input phage) and 14N (progeny phage) capsid proteins. Quantitation was achieved by comparing peak areas of target peptides from the metabolically labeled 15N bacteriophage peptide internal standard with that of the wild-type 14N peptides that were produced by phage amplification and subsequent digestion when the host bacteria was present. This approach is based on the fact that a labeled species differs from the unlabeled one in terms of its mass but exhibits almost identical chemical properties such as ion yields and retention times. A 6-point calibration curve for S. aureus concentration was constructed with standards ranging from 5.0 × 104 colony forming units (CFU) ml−1 to 2.0 × 106 CFU ml−1, with the 15N reference phage spiked at a concentration of 1.0 × 109 plaque forming units (PFU) ml−1. Amplification with 15N bacteriophage coupled with LC-MS/MS detection offers speed (3 h total analysis time), sensitivity (LOD: < 5.0 × 104 CFU ml−1), accuracy, and precision for quantitation of S. aureus.
Staphylococcus aureus (S. aureus) is a versatile pathogenic bacterium (1) responsible for a significant number of healthcare-associated and community-acquired infections (2). It causes a broad spectrum of infections ranging from acute to chronic disease (3, 4) and is a common etiological agent of opportunistic infections. An increasing prevalence of antibiotic resistant S. aureus strains are emerging, posing an even greater threat to the general public worldwide (5). For these reasons, the development and improvement of diagnostic methods that allow rapid identification and quantitation of this bacterium are highly critical (2).
Several methods are available for definitive identification of S. aureus. Traditional identification through plate-culture requires 2–3 days, can require subculturing or biochemical analysis (6), and necessitates blood sample volumes difficult to obtain in pediatric patients (7). Molecular methods, such as polymerase chain reaction, reverse transcription-polymerase chain reaction, and quantitative reverse transcription-polymerase chain reaction (PCR, RT-PCR, and qRT-PCR) are primarily based on particular genes specific to S. aureus (2) and offer the most sensitive measurements in the least amount of time, with the least amount of sample, but can be prone to ambiguous results that can only be resolved through sample cultivation (5). Additionally PCR is unable to distinguish dead bacteria from live cells, and the required reagents are relatively costly. Gene probe assays are promising in that they offer simple, rapid, and sensitive measurements and are lower in cost than PCR techniques (2, 6). Several rapid methods (culture-based and molecular-based screening methods) for S. aureus are also available, allowing diagnostics within hours of collection time; however these tests can be costly and the majority yield qualitative, not quantitative results (8–10).
Isotope labeling MS has long been used for quantitation of small molecules in a variety of matrices (11). More recently, MS-based stable isotope tagging of proteins and peptides followed by MS/MS experiments (12) has emerged as an important tool in quantitative proteomic experiments (13–19). Quantitation is achieved by adding a known amount of stable isotope-labeled protein or peptide to a sample as an internal standard and comparing instrument response to an unlabeled counterpart. Because species tagged with heavy isotopes differ from the unlabeled light ones in terms of their mass but exhibit almost identical chemical properties such as ion yields and retention times (20), ionic signals from tagged ion pairs can be accurately compared independently from instrument response. Labeling strategies include the use of chemical reactions to introduce an isotopic or isobaric tag at specific functional groups on polypeptides (21–24), metabolic isotope labeling using heavy amino acids (25–29), and methods that introduce stable isotope tags via enzymatic reactions (30, 31, 32). Each of these methods has specific strengths and weaknesses (32); however, metabolic incorporation of stable isotopes in whole organisms using cell culturing in heavy media is attractive in that it allows labels to be introduced at the earliest time point possible, during protein synthesis, and offers the most comprehensive way to cover proteomes completely (33, 34). Heavy isotopes, such as 15N and 13C, in nutrients fed to organisms during growth result in incorporation of heavy labels into all proteins over the course of doubling (35). Labeled controls and unlabeled samples are then combined prior to sample preparation, providing an internal control to reduce variability in the comparison of two proteomes (36).
The use of phages for bacterial detection is well documented (37). Bacteriophages are specific to their target host, self-replicate, have extensive shelf lives, are inexpensive (38), and infect only metabolically active cells (39). Bacteriophage amplification technology has emerged as a means of rapid bacterial detection, using modern protein analytical techniques to monitor changes in sample bacteriophage concentration (40–43). Standard phage amplification detection (PAD)1 techniques are highly specific, but the progeny phage produced through infection cannot be differentiated from the input parent phage initially added to the sample. For this reason, standard PAD techniques must use low-level concentrations of input phage, well below the detection limit of the detection device, thus prolonging incubation time prior to analysis, decreasing analysis frequency, increasing detection limits, and potentially increasing the probability of false positive results. To overcome these limitations, we have recently developed a rapid 15N PAD approach for analysis of intact phage proteins as a means of detecting S. aureus via top-down matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) (44). Although this 15N PAD MALDI-TOF MS method has proven successful for detecting viable S. aureus via amplified phage capsid proteins, it is unable to quantify S. aureus concentration because of the inherent problems in obtaining quantitative MALDI results.
Here, we present a “bottom-up” approach that combines PAD, stable isotope metabolic labeling, and liquid chromatography-multiple reaction monitoring tandem mass spectrometry (LC-MRM MS/MS) to quantify Staphylococcal bacteriophage 53 peptides from a phage capsid head protein, which when present, are indicative of the concentrations of viable host S. aureus bacteria (ATCC 27694). We show that our 15N PAD LC-MS/MS method offers good accuracy, precision, and sensitivity for high-throughput quantitation of viable S. aureus. To our knowledge, this is the first study employing 15N PAD combined with LC-MS/MS to quantify target bacteria.
EXPERIMENTAL PROCEDURES
S. aureus and Bacteriophage 53
Stock cultures of lyophilized S. aureus (ATCC 27694) and bacteriophage 53 (ATCC 27694-B1) were obtained from the American Type Culture Collection (ATCC, Manassas, VA). All microbiological procedures were performed in a U.S. biological safety level 2 (BSL2) facility following standard biosafety protocols. 14N bacteriophage stocks were generated by combining 500 μl of dense ATCC 27694 culture with 500 μl of wild-type bacteriophage 53 (1.0 × 106 PFU ml−1) in a test tube, pouring the contents onto a tryptic soy agar plate, and incubating the plate overnight at 37 °C. Following incubation, the plate's content was collected into a 50-ml conical tube by washing with tryptic soy broth (TSB) (Bacto™ TSB, BD Diagnostics, Franklin, NJ), scraping with a sterile plastic loop, and aspirating with a pipette. The conical tube was then centrifuged for 20 min at 3500 rpm to pellet out the debris, and the supernatant was filtered through an Autovial 0.2 μm polyvinylidene difluoride membrane syringeless filter device (Whatman, Inc, Clifton, NJ) to remove extraneous debris. The final phage filtrate was quantified using a traditional plaque assay (45).
Biosynthetic Production of 15N S. aureus and 15N Bacteriophage 53
15N S. aureus cells were grown in Bioexpress Cell Growth Media U-15N, 98% (Cambridge Isotope Laboratories, Inc., Andover, MA) according to manufacturer's instructions with only minor modifications. A 1-ml sample of 15N cell growth media was added to 9 ml of 0.1 m phosphate buffered saline (PBS), inoculated with 500 μl of S. aureus ATCC 27694, and allowed to grow overnight at 37 °C. Following overnight culture, the same procedure was repeated with the exception of inoculating the fresh 15N cell growth media with 500 μl of the dense overnight bacterial growth. This double-round inoculation of ATCC 27694 with 15N cell growth media ensured maximum labeling efficiency. Following the second overnight incubation, the dense 15N labeled bacteria were combined with 500 μl of wild-type bacteriophage (1.0 × 106 PFU ml−1) and added to a 15N-labeled agar plate. The 15N-labeled agar plates were made by combining 10 ml of 15N cell growth media, 90 ml of de-ionized water, and 1.5 grams of agar. The solution was autoclaved for 60 min and poured into sterile Petri dishes. After allowing the 15N S. aureus and 15N bacteriophage to incubate overnight on the 15N-labeled agar plate, the plate's contents was collected into a 50-ml conical tube by washing with 5 ml TSB, scraping with a sterile plastic loop, and aspirating with a pipette. The conical tube was then centrifuged for 20 min at 3500 rpm to pellet out the debris, and the supernatant was filtered through an Autovial 0.2 μm polyvinylidene difluoride membrane syringeless filter device to remove extraneous debris. The 15N-labeled bacteriophages were quantified using a standard plaque assay (45). Phage stock suspensions (2.1 × 109 PFU ml−1) were stored at 4 °C for further analyses.
Nano-LC-MS/MS
Protein identification was performed as previously reported (44). Briefly, 5-μl tryptic digests of the 14N and 15N phage stock solutions were analyzed using a Waters NanoAquity liquid chromatography system (Waters Corporation, Milford, MA) utilizing a 100 μm × 100 mm BEH130 C18 analytical column (1.7 μm particle size) (Waters Corporation). The aqueous mobile phase (A) consisted of HPLC-grade water with 0.2% formic acid (FA) and 0.005% trifluoroacetic acid, whereas the organic phase (B) was acetonitrile with 0.2% FA and 0.005% trifluoroacetic acid. A gradient profile was used at a flow rate of 400 nL min−1. Initially, the mobile phase consisted of 5% B and 95% A. After 5 min, the gradient was ramped to 30% B over the next 100 min, continuing up to 90% B over the next 5 min and holding for 2 min. After 112 min total run time, the gradient was returned to 5% B and 95% A for the next 20 min to equilibrate the column to initial conditions. The total run time was 132 min.
The column eluent was introduced into a Thermo Scientific LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific, Waltham, MA) equipped with an electrospray interface. The LTQ-Orbitrap instrument performed data-dependent acquisition of precursor and product ion spectra. The nominal resolution of the Orbitrap analyzer was set at 60,000 with scan range from m/z 400 to 1600. The top nine most intense ions were selected for fragmentation; the selection window was set at m/z = 2. Conventional collision induced dissociation was used for ion activation purposes. Dynamic exclusion was enabled with a repeat count of 1 and an exclusion duration of 120 s. Charge state screening and monoisotopic precursor ion selection were enabled along with charge state rejection, whereby singly charged ions were not selected for MS/MS analysis.
Protein Identification and Target Peptide Determination
Protein identification was performed by matching acquired peptide tandem mass spectra to theoretical digests found in a protein database. Prior to database searching, MS/MS spectra were converted to Mascot generic format by using Mascot Distiller version 2.3.2.0 (Matrix Science, London, UK). Charge state deconvolution and de-isotoping were not performed. Database searching utilized the Mascot version 2.2.0 algorithm (Matrix Science, London, UK) with the following parameters: fragment ion mass tolerance of 0.50 Da, parent ion tolerance of 200 ppm, trypsin enzyme, two missed or nonspecific cleavage permitted, and deamidation of asparagine and oxidation of methionine specified as variable modifications. A database of 13,767,831 entries was generated by extracting sequences from the National Center for Biotechnology (NCBI) protein database (downloaded on 04/22/2011) that contained at least one of the following strings in the entry description; “staphylococcus phage,” “aureus,” “trypsin,” “keratin.” To validate MS/MS-based peptide and protein identification, Scaffold version 3.00.02 (Proteome Software Inc., Portland, OR) with PeptideProphet™ (46) and ProteinProphet™ (47) algorithms was used. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by PeptideProphet™. Protein identifications were accepted if they could be established at a 99.0% probability based on ProteinProphet™, and if they were based on at least 3 peptide matches. Proteins containing similar peptides and that could not be differentiated based on MS/MS analysis alone were grouped into a single match.
The peptide sequences SIAQSIEK, LGVILPVTK, and LIYGIPQLIEYK were selected for quantitation purposes. The metabolically labeled SIAQSIEK and LGVILPVTK were 10 Da heavier than the naturally occurring peptides, because of incorporation of serine, isoleucine, alanine, glutamic acid, leucine, glycine, valine, proline, and threonine each with a 15N nitrogen count of 1, and glutamine and lysine amino acids with a 15N nitrogen count of 2. Similarly, labeled LIYGIPQLIEYK was 14 Da heavier than the naturally-occurring peptide.
Preparation of S. aureus Working Stock and Standard Solutions
To determine the S. aureus concentration (CFU ml−1) and to prepare calibrated standards, serial dilutions of S. aureus concentrated overnight suspensions (working stocks) were made and absorbance spectra for each dilution were recorded at 650 nm using a SpectraMax 2 (Molecular Devices, Inc., Sunnyvale, CA) spectrophotometer. The bacterial dilutions were then plated for viable cell counting. After overnight growth at 37 °C, the relationship between absorbance and the CFU ml−1 was graphed and values in the linear range of this graph were used to calculate the concentration of S. aureus working stocks solutions. This calibration curve was then used in daily analyses to calculate the number of viable cells in the S. aureus working stock by optical density readings.
For quantitation, an initial absorbance reading was taken of the S. aureus working stock solution and was used as the starting point when making serial dilutions for calibration curves. From the working stock, six 1-ml serial dilutions in TSB were prepared ranging from 5.0 × 105 CFU ml−1 to 2.0 × 106 CFU ml−1. The six 0.5-ml calibration standards, ranging from 5.0 × 104 CFU ml−1 to 2.0 × 105 CFU ml−1, were prepared by adding 50 μl of the corresponding S. aureus dilution, 250 μl of stock 15N phage (2.1 × 109 PFU ml−1), 50 μl of 1 m magnesium chloride, and 150 μl TSB, to make the final volume, 0.5 ml. The 250 μl 500 ml−1 of stock 15N phage was used as the internal standard (ISTD).
15N Bacteriophage 53 Infection and Amplification Procedures
The infected sample was incubated at 37 °C with gentle rotation for 2 h. Following incubation, samples were centrifuged (5 min at 10,000 × g) to pellet bacteria out of solution. The supernatant was recovered and phage samples were centrifuged with washing (5 min at 14,000 × g), using 30-kDa molecular weight cutoff filters (Amicon Ultra; Millipore, Billerica, MA) and 500 μl of 50 mm ammonium bicarbonate, pH 9 (×2). The filtrate (20 μl) was recovered for subsequent enzymatic digestion.
Rapid Enzymatic Digestion Procedure
In-solution digestion was performed using 20 μl aliquots of the phage/bacteria filtered samples. A modification of an acid-labile surfactant digestion method described before (44) was used. The modification included incubation at a higher temperature (52 °C) for a short period of time (3 min) and the use of a thermocycler (Applied Biosystems) as described originally by Turapov et al. (48) and later by Moura et al. (49). Briefly, a volume of 10 μl of a 0.1% solution of sodium 3-[(2-methyl-2-undecyl-1,3-dioxolan-4-yl)methoxyl]-1-propanesulfonate (Rapigest, Waters Corporation, Milford, MA) in 50 mm ammonium bicarbonate digestion buffer was added to each 20 μl phage aliquot to solubilize proteins and facilitate protein digestion (50, 51). The solution was then incubated at 100 °C for 5 min and rapidly cooled to room temperature. A 10 μl aliquot (∼172 pmol) of sequence-grade Promega trypsin was added, and samples were incubated in a thermocycler at 52 °C for 3 min to achieve complete digestion. Following digestion, the samples were allowed to cool, 10 μl of 450 mm HCl were added, and the samples were incubated for 30 min at 37 °C to reduce the pH and cleave the acid-labile surfactant. The digested samples were the transferred to autosampler vials for LC-MS/MS analysis.
LC-MS/MS Instrumentation and Methodology
An Agilent 1200 series LC system (Agilent Technologies, Inc., Santa Clara, CA) was configured for alternating column regeneration to increase sample throughput. A dual column, dual pump system coupled to an Agilent 1200 Series 2 position/10 port valve allowed simultaneous analysis of one column eluent whereas a second identical column was flushed and equilibrated (supplemental Fig. S1). The analytical columns utilized were 150 mm × 1 mm i.d. Symmetry300 reverse phase C18 (3.5 μm particle size, Waters Corporation, Milford, MA). The injection volume was 8 μl, and a 2-μl full loop injection with three-time loop overfill was utilized for injections. The aqueous mobile phase (A) consisted of 0.1% formic acid in HPLC-grade water, whereas the organic mobile phase (B) was 0.1% formic acid in acetonitrile. A gradient profile for the analysis column was utilized at a flow rate of 75 μl min−1. Initially, the mobile phase consisted of 98% A and 2% B. At 3 min the gradient was stepped to 80% A and 20% B over the next 7 min. After 10 min the gradient was stepped to 75% A and 25% B over the next 5 min and then held constant for 2 min. After 17 min total the gradient was stepped to 2% A and 98% B for 7 min to clean the column, then stepped to 98% A and 2% B for the next 3 min to begin equilibrating the column to initial conditions. The isocratic gradient for the regeneration column utilized a 50 μl min−1 flow rate and consisted of a constant eluent composition of 98% A and 2% B. The total analysis run time was 28 min.
The column eluent was introduced into a Thermo Scientific Vantage TSQ triple quadrupole tandem mass spectrometer with an electrospray interface (Thermo Scientific, Waltham, MA). The instrument was operated in positive ion multiple reaction monitoring mode. The precursor→fragment ion transitions were m/z 438.2 → 675.4, m/z 470.3 → 670.5, and m/z 725.4 → 890.5 for the native peptides and m/z 443.2 → 683.4, m/z 475.4 → 677.4, and m/z 732.4 → 899.5 for the 15N corresponding labeled peptides. For each peptide, two additional transitions were monitored for confirmation purposes (Table I). Instrument parameters were as follows: spray voltage 4000 V, sheath gas 4, auxiliary gas 2, capillary tube temperature 300 °C, and collision gas pressure of 1.5 mTorr. Collision energies and tube lens settings were optimized for each peptide. Instrument control was performed via the Thermo Scientific Xcalibur software.
Table I. Target Staphylococcal bacteriophage 53 capsid protein peptide sequences and their corresponding 15N metabolically labeled counterparts. The unique peptide sequences were chosen to identify the 35 kDa (14N) and 35.5 kDa (15N) capsid protein from Staphylococcal bacteriophage 53. MS/MS = mass spectrometry/mass spectrometry.
| Target peptide | Actual Mass Da | Precursor Ion m/z | Fragment Ion (quantitation) | Fragment Ion (confirmation) | Fragment Ion (confirmation) |
|---|---|---|---|---|---|
| SIAQSIEK-14N | 874.5 | 438.2 (+2) | 675.4 (y6) | 476.3 (y4) | 604.3 (y5) |
| SIAQSIEK-15N | 884.5 | 443.2 (+2) | 683.4 (y6) | 481.3 (y4) | 611.3 (y5) |
| LGVILPVTK-14N | 938.6 | 470.3 (+2) | 670.5 (y6) | 557.4 (y5) | 444.3 (y4) |
| LGVILPVTK-15N | 948.8 | 475.4 (+2) | 677.4 (y6) | 563.3 (y5) | 449.3 (y4) |
| LIYGIPQLIEYK-14N | 1448.8 | 725.4 (+2) | 890.5 (y7) | 1223.7 (y10) | 1060.6 (y9) |
| LIYGIPQLIEYK-15N | 1462.8 | 732.4 (+2) | 899.5 (y7) | 1235.7 (y10) | 1071.6 (y9) |
Data Analysis
MRM data acquired on the triple quadrupole mass spectrometer were analyzed and processed by Thermo Scientific Xcalibur software. Typical ICIS peak integration settings were smoothing width of seven points, area noise factor of 10, peak noise factor of 5, and a tailing factor of 2. Peak integrations were reviewed manually, and transitions from analyte peptides were confirmed by having the same retention times of the heavy stable isotope-labeled peptides. Linear regression was performed without weighting in Xcalibur on the 14N/15N phage peak area ratio versus spiked S. aureus concentration to construct response curves. The most abundant transition for each peptide was selected as the quantitative transition to be used in quantitative and statistical analyses. The detection limit was based upon the lowest S. aureus concentration that repeatedly observed a signal-to-signal to noise (S/N) response of at least 500 for the SIAQSIEK quantitative transition. Percent coefficient of variation is expressed as percent relative standard deviation (standard deviation divided by the mean multiplied by 100).
RESULTS
A specific biomarker of bacteriophage 53, the 35 kDa bacteriophage 53 major capsid protein, was identified by MALDI-TOF MS of the intact protein and LC-MS/MS analysis of the tryptic digestion of the phage followed by database searching (44). Of the tryptic peptides identified, three candidates for MRM analysis were selected for further investigation: SIAQSIEK, LGVILPVTK, and LIYGIPQLIEYK. The amino acid sequence of the main capsid protein in Fig. 1 highlights peptide coverage (47%) and indicates candidate peptides in red, bolded font. These particular peptides were selected based on their signal intensities and absence of methionine, tryptophan, and cysteine residues. The presence of these residues is not desirable because they tend to be reactive and are prone to oxidation which would modify the peptide's molecular mass (52–54), leading to inconsistent MRM analyses.
Fig. 1.
Amino acid sequence of the major capsid head protein of interest present in Staphylococcal bacteriophage 53. The sequence coverage (47%) is highlighted and the three target peptides used for quantitation of S. aureus are indicated in bold red font. The sequence (gi | 66395381) was obtained from the National Center for Biotechnology Information database.
Stable 15N isotope bacteriophage labeling was achieved by inoculating the wild-type bacteriophage with a 15N-enriched broth, producing a stable isotope-labeled bacteriophage 53 internal standard. Under the conditions employed, all nitrogen-containing amino acids biosynthesized were 15N-labeled. Following tryptic digest, the resulting heavy peptide mixture was analyzed by LC-MS/MS to determine the completion of 15N labeling. The SIAQSIEK and LGVILPVTK labeled peptides were shifted by 10 Da and the LIYGIPQLIEYK peptide was shifted by 14 Da from its unlabeled counterpart. These results were expected because SIAQSIEK and LGVILPVTK both have 10 nitrogen atoms whereas LIYGIPQLIEYK contains 14 nitrogen atoms. No evidence was seen for unlabeled or partially labeled peptides in the analysis. To evaluate mass spectrometric instrument response for the wild-type phage tryptic peptides of interest, and to establish the analytical limit of detection, twofold dilution series of the wild-type 14N bacteriophage-derived peptides were analyzed using the MRM transitions described in Table I. Typical chromatographic traces for quantitation and confirmation transitions are shown in supplemental Fig. S2. The resulting curves from the dilution series confirmed that the instrument response was proportional to the 14N bacteriophage concentration for the MRM quantitative transitions (supplemental Fig. S3) with a detection limit of ∼3.0 × 105 PFU ml−1, based upon the observed S/N of at least 500 for the SIAQSIEK quantitative transition (supplemental Fig. S2). This is 100 times more sensitive than the previously reported 15N PAD MALDI method (44).
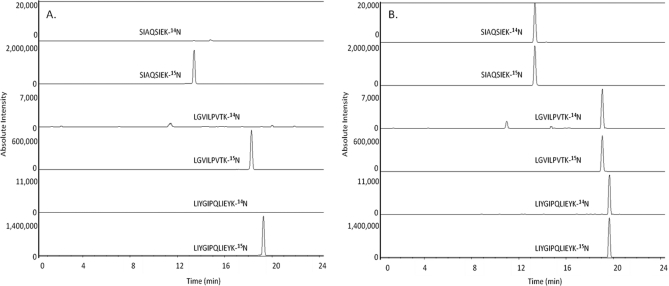
A 15N PAD experiment was conducted to confirm that the developed MRM instrumental method could detect an increase in progeny phage concentration, thereby implying the presence of S. aureus. In this experiment, duplicate samples containing 1.0 × 109 PFU ml−1 of 15N labeled bacteriophage and 2.0 × 105 CFU ml−1 of S. aureus were prepared. Following addition of the labeled bacteriophage, one replicate was immediately filtered, digested, and analyzed (t = 0 h), whereas the second replicate was subjected to a 2-h incubation period at 37 °C. Fig. 2 shows extracted ion chromatograms for this experiment. Fig. 2A (t = 0 h) shows high signal intensities for each 15N quantitative transition, whereas instrument response to the native bacteriophage (14N progeny) quantitative transition is negligible. Following the 2-h incubation, no significant differences were observed in 15N signal intensities, however, relative to the t = 0 h extracted ion chromatograms (Fig. 2A), a dramatic increase in signal intensity was observed for all three 14N quantitative transitions (Fig. 2B). Because the only means of generating the 14N phage peptides is through amplification of the progeny bacteriophage by a viable strain of S. aureus, these results confirm that the 15N PAD LC-MS/MS method can be used to positively detect the presence of S. aureus. Additionally, to look for background levels of the 14N phage peptides and to test for interferences, a t = 0 h S. aureus control sample was incorporated into each experimental sample set. This control sample was prepared identically to samples for 15N PAD, with the exception that it was not subjected to the t = 2 h phage amplification event. Following sample preparation, this control was immediately filtered, washed, digested, and analyzed by LC-MS/MS. Daily analyses showed no background levels (< 5.0 × 104 CFU ml−1) of 14N phage peptides present in any of the unamplified samples, and that there were not interferences to the analysis (Fig. 2A).
Fig. 2.
Liquid chromatography tandem mass spectrometry (LC-MS/MS) extracted ion chromatograms of the quantitative transitions monitored. The chromatograms show a standard solution (2.0 × 105 CFU ml−1) A, t = 0 h; prior to the phage amplification event and B, t = 2 h; the end point of the infection. The t = 0 h and t = 2 h chromatograms are displayed on identical scales based on ion counts.
The purpose of using 15N labeled bacteriophage in PAD experiments is twofold. First, to distinguish parent bacteriophage (15N labeled input) from progeny bacteriophage (14N wild type output) by their mass differences. The use of a heavy phage provides more confidence in the mass spectrometric analyses since the parent and progeny can be differentiated by mass. Second, by using the 15N labeled phage as an internal standard and a standard growth parameter, the number of bacteria can be quantified. To quantify the bacteria in culture, a high concentration of 15N bacteriophage must be used to ensure conditions where at least one infective 15N bacteriophage is attached to each S. aureus bacterium, preventing any further bacterial growth. For this condition to be met, a high multiplicity of infection (MOI); i.e. the ratio of infectious bacteriophage to bacteria, and a sufficiently dense concentration of bacteriophage must exist (55). The number of bacteriophages that infect a given bacterial cell can be calculated from the Poisson distribution, given as:
where, P(n) is the probability of bacterial cells being infected by n phage, and m is the MOI. Using overnight cultures, our experimentally determined mean bacterial density was found to be 3.6 × 108 CFU ml−1 ± 5% over the course of five different days. For the given 15N bacteriophage concentration of 1 × 109 PFU ml−1 used in this study, the lowest theoretical MOI that would be encountered in the experimental design is 2.77. supplemental Fig. S4 plots P(n) versus n for a MOI of 2.77. From the plot it can be seen that ≥95% of all bacteria will have at least one infectious bacteriophage attached for a sample of S. aureus at a concentration of the 3.6 × 108 CFU ml−1. Thus, theoretically if concentrations of bacteria are kept below 3.6 × 108 CFU ml−1, all bacteria can be assumed phage-infected, leading to accurate quantification. Empirically, as the concentration of viable S. aureus approaches 3.6 × 108 CFU ml−1, the number of live bacteria could be underestimated if high concentrations of dead S. aureus cells, in conjunction with viable S. aureus cells, exist in a given sample, and result in competitive 15N bacteriophage binding, shifting the MOI lower than 2.77. For the purposes of this quantitative study the dynamic range was 5.0 × 104 CFU ml−1 to 2.0 × 106 CFU ml−1, considerably lower than 3.6 × 108 CFU ml−1, to ensure that ≥ 95% of all (viable or otherwise) bacteria have at least one infectious bacteriophage attached even in the presence of a significantly high concentration of dead S. aureus cells.
Various modeling and experimental studies have been conducted that describe bacteriophage and bacteria proliferation concentration thresholds that must be met for a productive phage infection event (56). To ensure that at high phage concentrations effectively all bacteria are infected immediately after inoculation, various concentrations of bacteria (1 × 104 to 1 × 107 CFU ml−1) were inoculated at 1.0 × 109 PFU ml−1 bacteriophage, and allowed to incubate for 15 min. Following incubation, each sample was serially diluted and plated onto tryptic soy agar. After culturing the plates overnight, colonies on each plate were counted and compared against control plates that contained the same bacterial concentrations without bacteriophage infection. At each bacterial concentration tested, the cultures infected with 1.0 × 109 PFU ml−1 bacteriophage showed no bacterial growth, suggesting that all S. aureus were rapidly infected within the first minutes of phage infection.
Following these preliminary method characterization studies, 6-point calibration curves ranging from 5.0 × 104 CFU ml−1 to 2.0 × 106 CFU ml−1 were generated on five different days with three replicate LC-MS/MS injections for each standard. The metabolically 15N reference phage was spiked at a concentration of 1.0 × 109 PFU ml−1. The mean unlabeled and labeled MRM area ratios for each quantitative transition were plotted against expected S. aureus concentrations for each standard. Regression analysis showed good linearity (R2 = 0.99) over the 5.0 × 104 CFU ml−1 to 2.0 × 106 CFU ml−1 range for each quantitative peptide transition (supplemental Fig. S5) for all 5 days. As can be seen in supplemental Fig. S5, the calibration curves were highly reproducible from day to day, which allowed us to generate highly specific, sensitive, and reproducible data. All raw data were included in analyses. The concentrations of unknown samples were then determined using the slope and y-intercept calculated by linear regression analysis of the calibration curves constructed from each quantitative transition analyzed on that given day. To further evaluate precision and accuracy, samples with known amounts of S. aureus were spiked at low (1.0 × 105 CFU ml−1) and high levels (1.0 × 106 CFU ml−1), subjected to the 2-h phage amplification event, proteolytically digested, and analyzed by LC-ESI-MS/MS. Each sample preparation was analyzed in triplicate. All raw data were included in analyses. The intra- and interpeptide mean concentrations, standard deviations, and percent relative standard deviations (% RSD; (standard deviation/mean)*100) are reported in Table II. Mean S. aureus concentrations for five replicates spiked at 1.0 × 105 CFU ml−1 and 1.0 × 106 CFU ml−1 levels produced highly reproducible results with % RSDs of ≤ 15%, for all three quantitative transitions, demonstrating the effectiveness of the method. Similarly, interpeptide %RSDs of ≤ 2% and ≤ 9% for 1.0 × 105 CFU ml−1 and 1.0 × 106 CFU ml−1, respectively, show significant agreement among the three transitions used for quantification, indicating that the evaluated peptides did not differ with respect to precision. To ascertain the accuracy of the measurements the experimentally determined S. aureus concentrations were compared against the amounts spiked, as measured by optical density readings that were correlated to bacterial plate counts. Interpeptide accuracies were determined to be 31 and 1% for the 1.0 × 105 CFU ml−1 and 1.0 × 106 CFU ml−1 concentrations, respectively. Although the accuracy of the low-level spike appears to have a slight high bias, CFUs are only an estimate of the number of cells present (57), as the accuracy of the method is dependent on the reference curve obtained via direct plate count and optical density readings. Despite the limitations of plate counting and optical density readings we have rigorously standardized all steps in our 15N PAD method to control and minimize error in the analyses.
Table II. Precision and accuracy of S. aureus quantitative measurements. Three technical replicates were performed for n = 5 identical samples at two concentration levels. S.D. = standard deviation; % RSD = % relative standard deviation.
|
S. aureus Measurements (CFU ml−1) | ||||||
|---|---|---|---|---|---|---|
| Spike concentration (CFU ml−1) | Intra-peptide concentration (mean ± S.D.) (% RSD) | Inter-peptide concentration (mean ± S.D.) (% RSD) | Spike concentration (CFU ml−1) | Intra-peptide concentration (mean ± S.D.) (% RSD) | Inter-peptide concentration (mean ± S.D.) (% RSD) | |
| n = 5 | ||||||
| SIAQSIEK | 1.00 × 105 | 1.30 × 105 ± 1.16 × 104 (8.9) | 1.31 × 105 ± 1.73 × 103 (1.3) | 1.00 × 106 | 9.70 × 105 ± 1.04 × 105 (10.7) | 1.01 × 106 ± 1.16 × 104 (8.9) |
| LGVILPVTK | 1.00 × 105 | 1.33 × 105 ± 2.00 × 104 (15.0) | 1.00 × 106 | 1.02 × 106 ± 1.35 × 105 (13.2) | ||
| LIYGIPQLIEYK | 1.00 × 105 | 1.30 × 105 ± 1.70 × 104 (13.1) | 1.00 × 106 | 1.04 × 106 ± 1.53 × 105 (14.7) | ||
Finally, verification that complete digestion has been achieved is essential for accurately quantifying proteins using MS (58), and to ensure long term stability of the quantification method. Digestion parameters, including incubation time and temperature, amount of trypsin, and amount of acid-labile detergent, were varied and peptide recoveries were determined. Maximum peptide yields were assumed to have been achieved when no further increase in 14N or 15N peptide amounts could be observed. Three specific peptides from different regions of the protein were quantified to ensure complete digestion of the protein and accuracy of the measurements. Three digestion protocols were compared (1) overnight tryptic digestion at 37 °C; (2) 2-h tryptic digestion at 37 °C; and (3) 3-min tryptic digestion at 52 °C. For each protocol, triplicate S. aureus samples at 5.0 × 105 CFU ml−1, were subjected to the 2-h phage amplification event, proteolytically digested, and analyzed by LC-ESI-MS/MS. Each sample preparation was analyzed in triplicate. Table III shows the comparison of the different digestion conditions. The number of live S. aureus cells obtained from three independent analyses of a standard solution of 5.0 × 105 CFU ml−1 following phage infection were determined to be 5.0 × 105 CFU ml−1, 4.7 × 105 CFU ml−1, and 5.6 × 105 CFU ml−1 for the 3 min/52 °C, 2 h/37 °C and 18 h/37 °C digestion protocols, respectively, indicating that all digestion techniques yielded good precision and accuracy (Table III). Means for the three quantitative peptide transitions, at each digest condition, were reproducible with %RSDs of ≤ 13%. Interpeptide agreement at each digest condition resulted in %RSDs of ≤ 5%, indicating that each protocol was robust and suitable for the peptides evaluated. The inter-peptide accuracy of the 3-min 15N PAD MS method was calculated to be 100%, indicating that the rapid 3-min digest incubated at 52 °C was a good, rapid alternative to the longer traditional tryptic digestion methods that use 37 °C incubation temperature and 2–18 h digestion times. Furthermore, the accuracy of the method and the good agreement between values obtained independently on the three peptides suggests that all three digestion techniques yielded complete digestion of the major capsid protein of bacteriophage 53.
Table III. Verification of completeness of digestion for the SIAQSIEK, LGVILPVTK, and LIYGIPQLIEYK quantitative peptide transitions. For 15N PAD MS measurements, 5.0 × 105 CFU ml−1 of spiked S. aureus was subjected to phage amplification, tryptic digest, and LC-MS/MS analysis. Triplicate results for each preparation are shown and intra-peptide and inter-peptide means (CFU ml−1 tested), standard deviations, and percent relative standard deviations (% RSDs) for n = 3 sample preparations is reported. S.D. = standard deviation; % RSD = % relative standard deviation.
| 3-min Digestion Protocol (5 × 105 CFU ml−1 spiked S. aureus) | |||||
|---|---|---|---|---|---|
| n = 3 | 3-min 15N PAD Digest 1 | 3-min 15N PAD Digest 2 | 3-min 15N PAD Digest 3 | 3-min 15N PAD Digest Intra-peptide (mean ± S.D.) (% RSD) | 3-min 15 N PAD Digest Inter-peptide (mean ± S.D.) (% RSD) |
| SIAQSIEK | 5.33 × 105 | 5.55 × 105 | 4.84 × 105 | 5.24 × 105 ± 3.64 × 104 (6.95) | 5.00 × 105 ± 2.09 × 104 (4.18) |
| LGVILPVTK | 4.83 × 105 | 5.03 × 105 | 4.86 × 105 | 4.91 × 105 ± 1.06 × 104 (2.18) | |
| LIYGIPQLIEYK | 4.72 × 105 | 5.20 × 105 | 4.64 × 105 | 4.85 × 105 5 ± 3.05 × 104 (6.29) | |
| 2-h Digestion Protocol (5 × 105 CFU ml−1 spiked S. aureus) | |||||
|---|---|---|---|---|---|
| n = 3 | 2-h 15N PAD Digest 1 | 2-h 15N PAD Digest 2 | 2-h 15N PAD Digest 3 | 2-h 15N PAD Digest Intra-peptide (mean ± S.D.) (% RSD) | 2-h 15N PAD Digest Inter-peptide (mean ± S.D.) (% RSD) |
| SIAQSIEK | 4.74 × 105 | 4.19 × 105 | 4.73 × 105 | 4.56 × 105 ± 3.13 × 104 (6.86) | 4.74 × 105 ± 1.82 × 104 (3.83) |
| LGVILPVTK | 4.96 × 105 | 4.66 × 105 | 5.14 × 105 | 4.92 × 105 ± 2.39 × 104 (4.85) | |
| LIYGIPQLIEYK | 4.80 × 105 | 4.57 × 105 | 4.90 × 105 | 4.76 × 105 ± 1.69 × 104 (3.56) | |
| Overnight Digestion Protocol (5 × 105 CFU ml−1 spiked S. aureus) | |||||
|---|---|---|---|---|---|
| n = 3 | Overnight 15N PAD Digest 1 | Overnight 15N PAD Digest 2 | Overnight 15N PAD Digest 3 | Overnight 15N PAD Digest Intra-peptide (mean ± S.D.) (% RSD) | Overnight 15N PAD Digest Inter-peptide (mean ± S.D.) (% RSD) |
| SIAQSIEK | 5.42 × 105 | 6.47 × 105 | 5.60 × 105 | 5.83 × 105 ± 5.64 × 104 (9.67) | 5.67 × 105 ± 1.48 × 104 (2.61) |
| LGVILPVTK | 5.06 × 105 | 6.38 × 105 | 5.53 × 105 | 5.66 × 105 ± 6.70 × 104 (11.84) | |
| LIYGIPQLIEYK | 5.04 × 105 | 6.30 × 105 | 5.26 × 105 | 5.54 × 105 ± 6.74 × 104 (12.17) | |
DISCUSSION
Rapid and accurate detection of pathogenic organisms is crucial to diagnosis, treatment, and prevention of disease. The use of 15N labeled bacteriophage in PAD experiments allows for greater accuracy in the detection of the phage progeny because the input and output phage are distinguishable by mass. The use of 15N labeled bacteriophage allows for greater amounts of input phage to be used, which decreases the time of analysis and the use of LC-MS/MS improves detection limits. Additionally, using 15N labeled bacteriophage is beneficial in that the 15N labeled phage proteins act as an internal standard allowing rapid, accurate, and sensitive quantitation of S. aureus. The rapid identification and accurate quantification of S. aureus required (1) developing a rapid 15N phage amplification step that could simultaneously infect all S. aureus cells; (2) developing a rapid and efficient proteolytic digestion method; and (3) employing high-throughput LC-ESI-MS/MS for rapid, sensitive, and specific quantitation. The ability of our method to inoculate with high 15N-labeled phage titers (higher than the LC-MS/MS detection limit) allows S. aureus cells to be infected simultaneously, permitting S. aureus quantitation and offering time-saving advantages over standard PAD methodologies. Traditional digestion protocols often include reduction and alkylation steps followed by lengthy trypsin incubation times that range from several hours to overnight. The rapid 3-min digest produced maximum peptide yields that showed no significant difference in peptide recoveries when compared with traditional digest preparations, thereby enhancing the rapidity of the method. Phage-amplified digest samples were quantified by LC-MS/MS configured for alternating column regeneration to further increase sample throughput. Using two columns, two pumps, and one 2-position 10-port valve allowed switching between columns for short cycle times from injection to injection. Extracted ion chromatograms of the labeled and unlabeled peptide isoforms showed co-elution of the peptide pairs with high retention time reproducibility and allowed the 15N signals to be used as retention time indicators for the native peptide signals, thus improving precision for quantifying peptide abundances. MRM MS allowed simultaneous quantitation of peptides from the phage capsid protein as a measure of S. aureus concentration and method specificity was enhanced by monitoring three ion transitional pairs for each peptide for a total of 12 independent ion transitions used for quantitation and confirmation of the phage protein of interest.
Although we have demonstrated the feasibility of this approach using cultivated S. aureus, Staphylococcal bacteriophage 53 and LC-MS/MS, this quantitative technique should be broadly applicable to other bacteria. The presented data encourages the continued development and use of isotopically labeled PAD MS-based methods for rapid bacterial quantification.
Although in recent years PAD has begun to emerge as an alternative to conventional assays for detection of bacterial pathogens, the technology is still in its infancy. The most prominent feature of PAD is the ability of the phage to amplify only in its bacterial target, allowing indirect detection with a high degree of specificity. The introduction of a combinatorial metabolic labeling PAD MS approach advances the field of PAD by providing quantitative results with short detection times, enhanced specificity, and improved sensitivity. Experimental design for quantitation of bacteria based on phage amplification used S. aureus strain 27694 and its corresponding bacteriophage. This strain was chosen solely as a representative test model and host range investigational studies would be necessary before diagnostic applications could be determined in a clinical context. There are many possible applications to the 15N PAD MS quantification method. The method uses the specificity of the bacteriophage to identify the bacteria of interest and should be able to accurately quantify target bacteria in co-infected samples and complex mixtures. Traditional techniques require an enrichment culture followed by plating and subsequent culturing of single colonies to obtain pure cultures. The 15N PAD MS method allows direct detection and accurate quantification of a target bacteria in the enrichment cultures. Further, the low limit of detection (<5.0 × 104 CFU ml−1) warrants investigation of analysis directly from some clinical and environmental samples. Because phage only amplify in viable bacterial cells, the use of bacteriophage as tools for drug, antimicrobial, and antibiotic susceptibility testing has been successfully demonstrated and testing the effectiveness of antibiotics on a S. aureus culture or clinical isolate is feasibly quantifiable with 15N PAD LC-MS/MS.
Bridging the gap between high complexity, low volume testing for research MS-based applications with less complex, rapid turnaround, and automated routine MS-based applications can be challenging. This method's current level of complexity makes it more compatible for complementing or confirming routine clinical diagnostics in higher end reference or specialized laboratories. However, isotopically labeled PAD coupled with MS possesses unique characteristics that are well-matched to be incorporated with ease into the workflow of microbiological laboratories and wider adoption of the technology is warranted. Mass spectrometers are becoming more common in clinical laboratories and all indications suggest that this trend will continue. Because of the simplicity of phage cultivation preparation and the low cost of required reagents, incorporating PAD into a user-friendly platform that offers cost-effective analysis is a practical outlook. Additionally, PAD's ability to quantify live, viable bacteria in a variety of sample matrices without the need for confirmatory analyses makes it an attractive alternative when compared with PCR and many rapid culture methods. LC-MS/MS instruments are becoming increasingly simple to operate, have improved sensitivity and selectivity than many traditional bioanalytical assays, and offer universal detection and multiplexing capabilities that support timely analyses and a cost-effective investment.
As PAD matures and MS continues its emergence in clinical settings, this technique for detection and quantification of living pathogens should prove to be a powerful diagnostic tool for existing and emerging infectious diseases.
Acknowledgments
Reference in this article to any specific commercial products, process service, manufacturer, or company does not constitute an endorsement or a recommendation by the U.S. government or the Centers for Disease Control and Prevention. The findings and conclusions reported in this article are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
* FMF gratefully acknowledges funding by 3M through a Nontenured Faculty Award and the GT/CDC seed funding program.
 This article contains supplemental Figs. S1 to S5.
This article contains supplemental Figs. S1 to S5.
1 The abbreviations used are:
- PAD
- phage amplification detection
- CFU
- colony forming units
- FA
- formic acid
- MOI
- multiplicity of infection
- MRM
- multiple reaction monitoring
- PFU
- plaque forming units
- RSD
- relative standard deviation
- S/N
- signal-to-noise
- TSB
- tryptic soy broth.
REFERENCES
- 1. Schmidt F., Scharf S. S., Hildebrandt P., Burian M., Bernhardt J., Dhople V., Kalinka J., Gutjahr M., Hammer E., Völker U. (2010) Time-resolved quantitative proteome profiling of host-pathogen interactions: the response of Staphylococcus aureus RN1HG to internalisation by human airway epithelial cells. Proteomics 10, 2801–2811 [DOI] [PubMed] [Google Scholar]
- 2. Kurlenda J., Grinholc M. (2010) Current diagnostic tools for methicillin-resistant Staphylococcus aureus infections. Mol. Diagn. Ther. 14, 73–80 [DOI] [PubMed] [Google Scholar]
- 3. van den Berg S., Bowden M. G., Bosma T., Buist G., van Dijl J. M., van Wamel W. J., de Vogel C. P., van Belkum A., Bakker-Woudenberg I. A. (2011) A multiplex assay for the quantification of antibody responses in Staphylococcus aureus infections in mice. J. Immunol. Methods 365, 142–148 [DOI] [PubMed] [Google Scholar]
- 4. François P., Scherl A., Hochstrasser D., Schrenzel J. (2010) Proteomic approaches to study Staphylococcus aureus pathogenesis. J. Proteomics 73, 701–708 [DOI] [PubMed] [Google Scholar]
- 5. Durai R., Ng P. C., Hoque H. (2010) Methicillin-resistant Staphylococcus aureus: an update. Aorn. J. 91, 599–606; quiz 607–609 [DOI] [PubMed] [Google Scholar]
- 6. Oliveira K., Procop G. W., Wilson D., Coull J., Stender H. (2002) Rapid identification of Staphylococcus aureus directly from blood cultures by fluorescence in situ hybridization with peptide nucleic cid probes. J. Clin. Microbiol. 40, 247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wada M., Lkhagvadorj E., Bian L., Wang C., Chiba Y., Nagata S., Shimizu T., Yamashiro Y., Asahara T., Nomoto K. (2010) Quantitative reverse transcription-PCR assay for the rapid detection of methicillin-resistant Staphylococcus aureus. J. Appl. Microbiol. 108, 779–788 [DOI] [PubMed] [Google Scholar]
- 8. Fuchs B., Süss R., Schiller J. (2010) An update of MALDI-TOF mass spectrometry in lipid research. Prog. Lipid Res. 49, 450–475 [DOI] [PubMed] [Google Scholar]
- 9. Monagas M., Quintanilla-López J. E., Gomez-Cordovés C., Bartolomé B., Lebrón, Aguilar R. (2010) MALDI-TOF MS analysis of plant proanthocyanidins. J. Pharm. Biomed. Anal. 51, 358–372 [DOI] [PubMed] [Google Scholar]
- 10. Giebel R., Worden C., Rust S. M., Kleinheinz G. T., Robbins M., Sandrin T. R. (2010) Microbial fingerprinting using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) applications and challenges. Adv Appl Microbiol. 71, 149–184 [DOI] [PubMed] [Google Scholar]
- 11. Browne T. R., Van Langenhove A., Costello C. E., Biemann K., Greenblatt D. J. (1981) Kinetic equivalence of stable-isotope-labeled and unlabeled phenytoin. Clin. Pharmacol. Ther. 29, 511–515 [DOI] [PubMed] [Google Scholar]
- 12. Zhang H., Li X. J., Martin D. B., Aebersold R. (2003) Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 21, 660–666 [DOI] [PubMed] [Google Scholar]
- 13. Cravatt B. F., Simon G. M., Yates J. R., 3rd (2007) The biological impact of mass-spectrometry-based proteomics. Nature 450, 991–1000 [DOI] [PubMed] [Google Scholar]
- 14. Kristjansdottir K., Kron S. J. (2010) Stable-isotope labeling for protein quantitation by mass spectrometry. Current Proteomics 7, 144–155 [Google Scholar]
- 15. Jensen O. N. (2006) Interpreting the protein language using proteomics. Nat. Rev. Mol. Cell Biol. 7, 391–403 [DOI] [PubMed] [Google Scholar]
- 16. Yates J. R., 3rd, Gilchrist A., Howell K. E., Bergeron J. J. (2005) Proteomics of organelles and large cellular structures. Nat. Rev. Mol. Cell Biol. 6, 702–714 [DOI] [PubMed] [Google Scholar]
- 17. Domon B., Aebersold R. (2006) Mass spectrometry and protein analysis. Science 312, 212–217 [DOI] [PubMed] [Google Scholar]
- 18. Gingras A. C., Gstaiger M., Raught B., Aebersold R. (2007) Mass spectrometry and protein analysis. Nat. Rev. Mol. Cell Biol. 8, 645–654 [DOI] [PubMed] [Google Scholar]
- 19. Trinkle-Mulcahy L., Boulon S., Lam Y. W., Urcia R., Boisvert F. M., Vandermoere F., Morrice N. A., Swift S., Rothbauer U., Leonhardt H., Lamond A. (2008) Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 183, 223–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schulze W. X., Usadel B. (2010) Quantitation in mass-spectrometry-based proteomics. Annul. Rev. Plant Biol. 61, 491–516 [DOI] [PubMed] [Google Scholar]
- 21. Gygi S. P., Rist B., Gerber S. A., Turecek F., Gelb M. H., Aebersold R. (1999) Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17, 994–999 [DOI] [PubMed] [Google Scholar]
- 22. Zhou H., Ranish J. A., Watts J. D., Aebersold R. (2002) Quantitative proteome analysis by solid-phase isotope tagging and mass spectrometry. Nat. Biotechnol. 20, 512–515 [DOI] [PubMed] [Google Scholar]
- 23. Cagney G., Emili A. (2002) De novo peptide sequencing and quantitative profiling of complex protein mixtures using mass-coded abundance tagging. Nat. Biotechnol. 20, 163–170 [DOI] [PubMed] [Google Scholar]
- 24. Chakraborty A., Regnier F. E. (2002) Global internal standard technology for comparative proteomics. J. Chromatogr. A 949, 173–184 [DOI] [PubMed] [Google Scholar]
- 25. Veenstra T. D., Martinovic S., Anderson G. A., Pasa-Tolić L., Smith R. D. (2000) Proteome analysis using selective incorporation of isotopically labeled amino acids. J Am Soc. Mass Spectrom. 11, 78–82 [DOI] [PubMed] [Google Scholar]
- 26. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 27. Foster L. J., De Hoog C. L., Mann M. (2003) Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc. Natl. Acad. Sci. U.S.A. 100, 5813–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Blagoev B., Kratchmarova I., Ong S. E., Nielsen M., Foster L. J., Mann M. (2003) A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat. Biotechnol. 21, 315–318 [DOI] [PubMed] [Google Scholar]
- 29. Ibarrola N., Kalume D. E., Gronborg M., Iwahori A., Pandey A. (2003) A proteomic approach for quantitation of phosphorylation using stable isotope labeling in cell culture. Anal. Chem. 75, 6043–6049 [DOI] [PubMed] [Google Scholar]
- 30. Mirgorodskaya O. A., Kozmin Y. P., Titov M. I., Körner R., Sönksen C. P., Roepstorff P. (2000) Quantitation of peptides and proteins by matrix-assisted laser desorption/ionization mass spectrometry using (18)O-labeled internal standards. Rapid Commun. Mass Spectrom. 14, 1226–1232 [DOI] [PubMed] [Google Scholar]
- 31. Yao X., Freas A., Ramirez J., Demirev P. A., Fenselau C. (2001) Proteolytic 18O labeling for comparative proteomics: model studies with two serotypes of adenovirus. Anal. Chem. 73, 2836–2842 [DOI] [PubMed] [Google Scholar]
- 32. Pan S., Aebersold R. (2007) Quantitative Proteomics by Stable Isotope Labeling and Mass Spectrometry In Methods in Molecular Biology, Vol. 367, pp. 209–218, Matthiesen, R., Totowa, NJ [DOI] [PubMed] [Google Scholar]
- 33. Krijgsveld J., Heck A. J. R. (2004) Quantitative proteomics by metabolic labeling with stable isotopes. Drug Discovery Today: TARGETS 3, 11–15 [Google Scholar]
- 34. Gouw J. W., Krijgsveld J., Heck A. J. (2010) Quantitative proteomics by metabolic labeling of model organisms. Mol. Cell Proteomics 9, 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Walther T. C., Mann M. (2010) Mass spectrometry-based proteomics in cell biology. J. Cell Biol. 190, 491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nelson C. J., Huttlin E. L., Hegeman A. D., Harms A. C., Sussman M. R. (2007) Implications of 15N-metabolic labeling for automated peptide identification in Arabidopsis thaliana. Proteomics 7, 1279–1292 [DOI] [PubMed] [Google Scholar]
- 37. Schofield D. A., Molineux I. J., Westwater C. (2009) Diagnostic bioluminescent phage for detection of Yersinia pestis. J. Clin. Microbiol. 47, 3887–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shabani A., Zourob M., Allain B., Marquette C. A., Lawrence M. F., Mandeville R. (2008) Bacteriophage-modified microarrays for the direct impedimetric detection of bacteria. Anal. Chem. 80, 9475–9482 [DOI] [PubMed] [Google Scholar]
- 39. Ripp S., Jegier P., Johnson C. M., Brigati J. R., Sayler G. S. (2008) Bacteriophage-amplified bioluminescent sensing of Escherichia coli O157:H7. Anal. Bioanal. Chem. 391, 507–514 [DOI] [PubMed] [Google Scholar]
- 40. Madonna A. J., Van Cuyk S., Voorhees K. (2003) Detection of Escherichia coli using immunomagnetic separation and bacteriophage amplification coupled with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 17, 257–263 [DOI] [PubMed] [Google Scholar]
- 41. Rees J. C., Voorhees K. J. (2005) Simultaneous detection of two bacterial pathogens using bacteriophage amplification coupled with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 19, 2757–2761 [DOI] [PubMed] [Google Scholar]
- 42. Reiman R. W., Atchley D. H., Voorhees K. J. (2007) Indirect detection of Bacillus anthracis using real-time PCR to detect amplified gamma phage DNA. J. Microbiol. Methods 68, 651–653 [DOI] [PubMed] [Google Scholar]
- 43. Goodridge L., Chen J., Griffiths M. (1999) Development and characterization of a fluorescent-bacteriophage assay for detection of Escherichia coli O157:H7. Appl. Environ. Microbiol. 65, 1397–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pierce C. L., Rees J. C., Fernández F. M., Barr J. R. (2011) Detection of Staphylococcus aureus Using 15N-labeled bacteriophage amplification coupled with matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry. Anal. Chem. 83, 2286–2293 [DOI] [PubMed] [Google Scholar]
- 45. Adams M. (1959) Bacteriophages, Interscience Publishers, NY [Google Scholar]
- 46. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 47. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 48. Turapov O. A., Mukamolova G. V., Bottrill A. R., Pangburn M. K. (2008) Digestion of native proteins for proteomics using a thermocycler. Anal. Chem. 80, 6093–6099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Moura H., Terilli R., Woolfitt A., Gallegos-Candela M., McWilliams L., Solano M., Pirkle J., Barr J. R. (2011) Studies on botulinum neurotoxins Type/C1 and mosaic/DC using Endopep-MS and proteomics. FEMS Immunol. & Medical Microbiol. 61, 288–300 [DOI] [PubMed] [Google Scholar]
- 50. Yu Y. Q., Gilar M., Lee P. J., Bouvier E. S., Gebler J. C. (2003) Enzyme-friendly, mass spectrometry-compatible surfactant for in-solution enzymatic digestion of proteins. Anal. Chem. 75, 6023–6028 [DOI] [PubMed] [Google Scholar]
- 51. Suder P., Bierczynska A., König S., Silberring J. (2004) Acid-labile surfactant assists in-solution digestion of proteins resistant to enzymatic attack. Rapid Commun. Mass Spectrom. 18, 822–824 [DOI] [PubMed] [Google Scholar]
- 52. Mo W., Ma Y., Takao T., Neubert T. A. (2000) Sequencing of oxidized methionine-containing peptides for protein identification. Rapid Commun. Mass Spectrom. 14, 2080–2081 [DOI] [PubMed] [Google Scholar]
- 53. Taylor S. W., Fahy E., Murray J., Capaldi R. A., Ghosh S. S. (2003) Oxidative post-translational modification of tryptophan residues in cardiac mitochondrial proteins. J. Biol. Chem. 278, 19587–19590 [DOI] [PubMed] [Google Scholar]
- 54. Atrih A., Turnock D., Sellar G., Thompson A., Feuerstein G., Ferguson M. A., Huang J. T. (2010) Stoichiometric quantification of Akt phosphorylation using LC-MS/MS. J. Proteome Res. 9, 743–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Payne R. J., Jansen V. A. (2001) Understanding bacteriophage therapy as a density-dependent kinetic process. J. Theor. Biol. 208, 37–48 [DOI] [PubMed] [Google Scholar]
- 56. Kasman L. M., Kasman A., Westwater C., Dolan J., Schmidt M. G., Norris J. S. (2002) Overcoming the phage replication threshold: a mathematical model with implications for phage therapy. J. Virol. 76, 5557–5564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sutton S. (2011) Counting Colonies, In The Microbiology Network, Accessed online February 17, 2011 http://www.microbiol.org/resources/monographswhite-papers/counting-colonies
- 58. Norrgran J., Williams T. L., Woolfitt A. R., Solano M. I., Pirkle J. L., Barr J. R. (2009) Optimization of digestion parameters for protein quantification. Anal. Biochem. 393, 48–55 [DOI] [PubMed] [Google Scholar]