Abstract
Altered activity of the human dopamine transporter gene (hDAT) is asssociated with several common and severe brain disorders including cocaine abuse. However, there is little a priori information on whether such alteration was due to nature (genetic variantion) or nurture (human behaviors such as cocaine abuse). This study investigated the correlation between seven markers throughout hDAT and its mRNA levels in postmortem ventral midbrain tissues from 18 cocaine abusers and 18 strictly matched drug-free controls in the African American population. Here we show that one major haplotype with same frequency in cocaine abusers versus drug-free controls displays a 37.1%-reduction of expression levels in cocaine abusers, compared to matched controls (P = 0.0057). The most studied genetic marker, variable number tandem repeats (VNTR) located in Exon 15 (3′VNTR), is not correlated with hDAT mRNA levels. A 5′ upstream VNTR (rs70957367) has repeat numbers positively correlated with expression levels in controls (r2 0.9536, P = 0.0235) but this positive correlation disappears in cocaine abusers. The findings suggest that varying hDAT activity is attributed to both genetics and cocaine abuse.
Keywords: Addiction, DAT, expressional variation, pharmacogenomics, postmortem, epigenetics
INTRODUCTION
Dopamine (DA) neurotransmission regulates physiological states including motivation, cognition, and attention, and contributes to pathological processes including substance abuse. The human dopamine transporter (hDAT) is an important regulator of DA neurotransmission and a target of psychostimulant drugs of abuse such as cocaine. Conceivably, varying expression levels may be atributable to both genetic variation and substance abuse; and abnormal expression may confer risks for the related brain disorders.
In fact, reduced hDAT expression is correlated with both aging and diseases. In humans, decreased hDAT mRNA levels, in contrast to increased tyrosine hydroxylase (TH) mRNA levels, are found in residual DA neurons of Alzheimer’s disease patients (Joyce et al., 1997). Animal models show that different DAT dosages confer variations in locomotion activity (Giros et al., 1996), cocaine reward (Sora et al., 2001) and susceptibility to neurodegeneration in the neurotoxin MPTP models (Gainetdinov et al., 1997; Bezard et al., 1999), consistent with the view that reduced hDAT expression may be associated with various diseases.
In order to identify the underlying genetic risks, more than 150 association studies have investigated association of the hDAT-encoding gene hDAT (or SLC6A3) with various diseases. Most of the studies used markers located in the 3′ end of the gene, including the 40-bp variable number of tandem repeats located in the 3′ untranslated region (3′VNTR in 3′UTR) and more recently the 30-bp VNTR located in Intron 8 (Int8VNTR). Findings with 3′VNTR showed small effect size or are elusive for attention deficit/hyperactivity disorder (ADHD) (Faraone and Mick, 2010) and drug abuse (van der Zwaluw et al., 2009; Lohoff et al., 2010) among other disorders. More recently, the 6-repeat of 30-bp allele in Int8VNTR was associated with cocaine addiction in Brazilians, smoking in a British population and ADHD in both British and Taiwanese populations (Brookes et al., 2006; Guindalini et al., 2006; O’Gara et al., 2007). Despite the extensive investigations and variable associations of numerous CNS disorders with various hDAT markers, little is known about whether different alleles or haplotypes in hDAT are correlated with differences in gene expression in human brain tissues.
To better understand altered hDAT activity in related diseases, it is necessary to delineate the relationship between genetic variation (nature) or environmental experience (nurture) and gene expression in the brain. In this study, we used exonic polymorphisms to quantify haplotypic expression of hDAT in postmortem brain tissues of cocaine abusers and the matched drug-free control subjects.
SUBJECTS AND METHODS
Postmortem brain tissue acquisition and subject characterization
Postmortem ventral midbrain specimens were obtained during routine autopsy and analyzed as described previously (Bannon and Whitty, 1997). Medicolegal investigations were conducted by forensic pathologists. The cause and manner of death were determined after evaluating the circumstances of death, toxicology data, and autopsy results. All cases were evaluated for common drugs of abuse (including alcohol), and positive urine screens were confirmed by quantitative analysis of blood. Blood levels of cocaine and metabolites were determined as described before (Hernandez et al., 1994). The cocaine abuse cohort in the present study had a documented history of drug abuse, tested positive for cocaine and/or metabolites but negative for other drugs of abuse (except for 10B, diazepam and 13B, methadone), and were determined to have died as a result of cocaine abuse or cocaine intoxication. Control subjects had no documented history of drug abuse, were drug-free at time of death, and died as a result of cardiovascular disease, gunshot wounds, or pulmonary embolism. Pair-wise matches of cocaine abusers and matched drug-free controls for age, gender, and pH are summarized in Table 1. Ethnicity was defined by medical legal records. Ventral midbrain was cut on a cryostat and the DA cell enriched area of ventral midbrain was hand dissected from slides. There was no difference between cocaine abusers and matched controls in pH and no effect of pH on hDAT mRNA levels.
Table 1.
Demographic information on the 36 subjects.
| ID | Drug1 | Race2 | Gender3 | Age | pH |
|---|---|---|---|---|---|
| A1 | drug-free | AA | F | 40 | 6.41 |
| B1 | cocaine | AA | F | 34 | 6.42 |
| A2 | drug-free | AA | M | 33 | 6.32 |
| B2 | cocaine | AA | M | 35 | 6.53 |
| A3 | drug-free | AA | M | 35 | 6.35 |
| B3 | cocaine | AA | M | 35 | 6.70 |
| A5 | drug-free | AA | M | 45 | 6.32 |
| B5 | cocaine | AA | M | 46 | 6.57 |
| A6 | drug-free | AA | M | 49 | 6.79 |
| B6 | cocaine | AA | M | 49 | 6.40 |
| A7 | drug-free | AA | M | 47 | 6.48 |
| B7 | cocaine | AA | M | 52 | 6.47 |
| A8 | drug-free | AA | M | 52 | 6.32 |
| B8 | cocaine | AA | M | 52 | 6.30 |
| 10A | drug-free | AA | F | 45 | 6.24 |
| 10B | cocaine | AA | F | 42 | 6.26 |
| 11A | drug-free | AA | F | 51 | 6.46 |
| 11B | cocaine | AA | F | 52 | 6.30 |
| 12A | drug-free | AA | M | 47 | 6.60 |
| 12B | cocaine | AA | M | 51 | 6,65 |
| 13A | drug-free | AA | M | 56 | 6.67 |
| 13B | cocaine | AA | M | 57 | 6.70 |
| 15A | drug-free | AA | M | 34 | 6.62 |
| 15B | cocaine | AA | M | 34 | 6.40 |
| 16A | drug-free | AA | M | 45 | 6.43 |
| 16B | cocaine | AA | M | 45 | 6.90 |
| 19A | drug-free | AA | F | 47 | 6.62 |
| 19B | cocaine | AA | F | 50 | 6.30 |
| 20A | drug-free | AA | M | 53 | 6.31 |
| 20B | cocaine | AA | M | 59 | 6.60 |
| 21A | drug-free | AA | M | 51 | 6.70 |
| 21B | cocaine | AA | M | 54 | 6.60 |
| 22A | drug-free | AA | M | 66 | 6.74 |
| 22B | cocaine | AA | M | 64 | 6.70 |
| 23A | drug-free | AA | M | 46 | 6.78 |
| 23B | cocaine | AA | M | 52 | 6.60 |
drug-free control; cocaine: cocaine abuser
AA: African American (defined by medical legal records);
F: female; M: male.
Genomic DNA preparation
The Wizard® SV Genomic DNA Purification System (Promega, WI, USA) was used for genomic DNA isolation: briefly, 20 mg of tissue sample or 2–4×106 cells was digested in 275 μl of Digestion Solution Master Mix or 150 μl of Wizard® SV Lysis Buffer, followed by overnight incubation at 55°C. After the rest of the procedure, genomic DNA was dissolved in 500 μl Elution Buffer.
Genotyping
The polymerase chain reaction (PCR)-RFLP method was used for SNPs/VNTR genotyping. PCR followed standard protocols by using the Fast PCR Master Mix (Fermentas, MD, USA), associated instructions and primers listed in Table S1. For 5′VNTR, the PCR product was 426-bp, 486-bp, 546-bp, and 606-bp for 6-, 7-, 8-, and 9-repeat respectively. Int8VNTR showed 291-bp and 321-bp for 5- and 6-repeat; 3′VNTR, 441-bp and 481-bp for 9- and 10-repeat. These VNTR’s PCR products were subject to agarose gel electrophoresis directly for length polymorphism. PCR products were 174-bp and 300-bp for DNPi and rs6347. The B allele (GAGGAG), but not the A allele (GAGAGG), of DNPi (di-nucleotide polymorphisms in Intron 1) was BseRI-sensitive for 127-bp and 47-bp digests. 300-bp of rs6347 could be digested by BbvcI into 194-bp and 106-bp for the A allele but not for the G allele. NsiI digested the T alleles of both rs11564775 and rs6876890 and the genotypes were identified through electrophoresis-based length polymorphism as well.
RNA isolation
RNA was extracted from ventral midbrain tissues with TRIzol Reagent (Invitrogen, CA, USA). RNA samples were quantified using a NanoDrop (ThermoScientific, DE, USA) and the RNA integrity was assessed with a BioAnalyzer (Agilent Technologies, CA, USA). There was no difference in 260/280 ratio between the drug-free control (2.04 ± 0.01) and cocaine abuser groups (2.04 ± 0.01; P; 0.881) nor in the RNA integrity numbers (RINs; control 6.5 ± 0.2; cocaine abuser 6.9 ± 0.1; P: 0.134) as determined by t-tests.
cDNA synthesis
To prepare cDNA, 0.5–1 μg of total DNase-treated RNA was reverse-transcribed with Oligo(dT)20 primers by using a SuperScript cDNA synthesis system (Invitrogen). The cDNA was either used immediately for PCR or stored at −20°C for subsequent analysis.
BStop-PCR
To quantify allelic cDNA, a modification of hot-stop PCR (Uejima et al., 2000) was made by replacing an isotope-labeled primer with a biotin-labeled primer in the final quantification PCR cycle and we termed this modified method as BStop-PCR. Hot-start reactions (94°C for 5 min) were initiated with 0.4U Taq DNA polymerase (New England Biolabs, Ipswich, MA, USA) followed by 28 cycles of amplification. Each cycle consisted of 30 sec at 94°C, 30 sec at 63°C, and 1 min at 72°C. Elimination of heteroduplexes from the analysis of PCR products followed the hot-stop PCR procedure. The corresponding PCR products containing the “A” allele of rs6347 were subject to BbvcI digestion to produce 114-bp and 26-bp fragments. After capillary transfer of DNA to a positively charged nylon membrane, biotin-labeled DNA was visualized by using the BrightStar BioDetect Kit (Ambion), followed by quantification with TL100.
Quantification of mRNA levels
Quantitative real-time PCR (qRT-PCR) reactions used the SYBR Green PCR Master Mix (Applied Biosystems, CA, USA). Data were normalized with GAPDH because GAPDH transcript abundance did not differ between cocaine abusers and the matched controls. All primers used in this study, prepared by Integrated DNA Technology (IDT, IA, USA), are listed in Table S1.
Statistics
Data are expressed as mean ± SEM values. The statistical significance of differences among groups was determined by one-way ANOVA or by Mann-Whitney U tests. Analysis of Covariance was used to compare two line tests. Linkage disequilibrium (LD) parameters (D′ and r2) and haplotype frequencies were estimated with the Haploview 4.1 program (Barrett et al., 2005) and SHEsis softwares (Li et al., 2009). Dandelion Version 3.0 was for association analysis; PHASE (version 2.1) (Stephens et al., 2003) and PL-EM for haplotype inferences (Niu, 2004).
RESULTS
Selection and linkage disequilibrium (LD) of hDAT markers
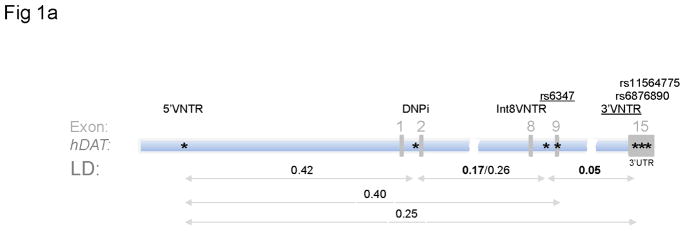
Five markers were selected to physically cover 90% (63.1 kb) of the 70 kb gene: a 5′VNTR, DNPi, Int8VNTR (a VNTR in Intron 8), rs6347 (a single nucleotide polymorphism or SNP in Exon 9), and the often-studied 3′VNTR in the last exon, Exon 15 or the 3′ untranslated region (3′UTR). Two additional SNPs were typed, too, due to their proximity to 3′VNTR: rs6876890 and rs11564775 (Figure 1). 5′VNTR and DNPi were selected because of their high heterogeneities for potential functionality because Tajima’s D statistic was positive for both: 2.44 (P < 0.05) and 1.26 (Drgon et al., 2006; Tajima 1989). Int8VNTR and 3′VNTR were selected for their reported in vitro functionalities and positive associations with diseases (Guindalini et al., 2006). rs6347 was included because of its exonic location as well as its MAF (minor allele frequency) of > 0.2 in many populations, based on dbSNP information. Allelic frequency information in the 36 subjects is summarized in Table 2 (genotype frequency information is provided in Table S2). Four alleles were found for 5′VNTR in this sample, two alleles for each of the two other VNTRs. All of them are highly informative based on their high MAF values. None of the markers failed the Hardy-Weinberg Equilibrium (HWE) tests (Table 2). Notice that 6-repeat of Int8VNTR was the minor allele in this cohort whereas 5-repeat was the minor allele in other populations (e.g. Guindalini et al., 2006), indicating ethnicity-related variation in allele frequency,
Figure 1.
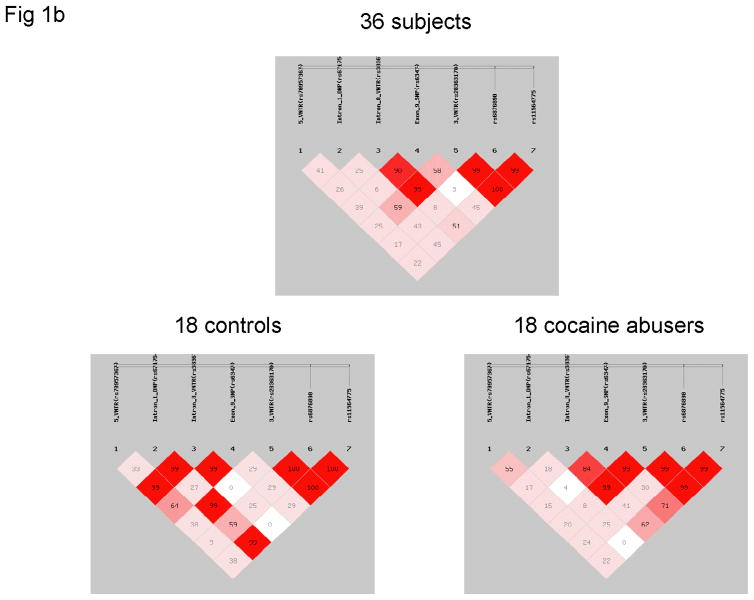
Seven hDAT markers typed in this study: location, frequency and linkage disequilibrium (LD). (a) Schematic location of seven markers (*) and approximate LD (D′, boldface value by Haploview, otherwise by SHEsis) for selected hDAT regions (horizontal bar). Underline, exonic markers used in monitoring mRNA levels; double-arrowed horizontal line, selected LD; exons in gray. (b) LD among seven hDAT markers. Upper panel: SHEsis; lower panels, Haploview. Strong LD is observed for 3′ end and LD differences observed between controls and cocaine abusers.
Table 2.
Summary of hDAT markers typed in 36 African American subjects.
| Variant number | Variant ID | Location
|
Variation | Allele frequency
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| on chr 5a | in hDAT | Total (n = 36) | cocaine (n = 18) | HWE P-value | drug-free (n = 18) | HWE P-value | |||
| 1 | 5′VNTR (rs70957367) | 1509887- 1509403 | −11kb in promoter | 60bp 6-7-8-9-repeatb | 0.014-0.361-0.208-0.417 | 0.007-0.410-0.278-0.306 | 0.295 | 0.007-0.326-0.139-0.528 | 0.179 |
| 2 | DNPic (rs67175440) | 1496605-1496604 | Intron 1 | A/B | 0.375/0.625 | 0.333-0.667 | 0.790 | 0.417-0.583 | 0.396 |
| 3 | Int8′VNTR (rs3836790) | 1465452-1464679 | Intron 8 | 30 bp 5-6-repeatd | 0.722-0.278 | 0.611-0.389 | 0.782 | 0.833-0.167 | 0.396 |
| 4 | rs6347 | 1464412 | Exon9 | A/G | 0.389/0.611 | 0.472-0.582 | 0.989 | 0.333-0.667 | 1.0 |
| 5 | 3′VNTR (rs28363170) | 1447128-1446635 | Exon15-(3′-UTR) | 40 bp 9-10-repeat e | 0.181-0.819 | 0.194-0.806 | 0.305 | 0.167-0.833 | 0.396 |
| 6 | rs6876890 | 1446878 | Exon15-(3′-UTR) | C/T | 0.333/0.667 | 0.306-0.694 | 0.142 | 0.382- 0.618 | 0.129 |
| 7 | rs11564775 | 1446753 | Exon15-(3′-UTR) | C/T | 0.611/0.389 | 0.650-0.350 | 0.271 | 0.577-0.423 | 0.396 |
Variant location from dbSNP database (NCBI), Genome Build 36.3.
5′VNTR: TCAAGGAACAGAGATAAGAACGACAGCCGACTTCTCTGCAGAAACTATGCAACCCAGACG
A, AG; B, GA.
Int8′VNTR: CACATACCATGCAACATACACACTCAGACA
3′VNTR: CCCCCACAGGAGCATGTCCTATCCCATGGACGCATGCAGGG
The 63.1 kb region from 5′VNTR to rs11564775 was not in high LD except those between Int8VNTR and rs6347 and among the last three markers in Exon 15 (see Figure 1b upper panel). LD was medium between 3′VNTR and DNPi (D′ = 0.12 by Haploview or 0.59 by SHEsis) or 5′VNTR (D′ = 0.25). 5′VNTR and DNPi displayed medium level of LD (D′ 0.42) while Int8VNTR and rs6347 displayed strong LD (D′ 0.57 by Haploview or 0.90 by SHEsis). LD in the matched controls differed slightly from that in the cocaine abusers (Figure 1b lower panels).
Reduced expression of a major hDAT haplotype in cocaine abusers
The 36 subjects used here were strictly paired between controls and cocaine abusers in race, age and gender (Table 1). The 18 cocaine abusers tended to have decreased expression (18.9%) without reaching any statistical significance, comparing to the controls. A total of 27 haplotypes were inferred statistically from the seven polymorphisms for the 36 subjects. Controls carried eight unique haplotypes and abusers carried ten unique haplotypes, sharing nine haplotypes.
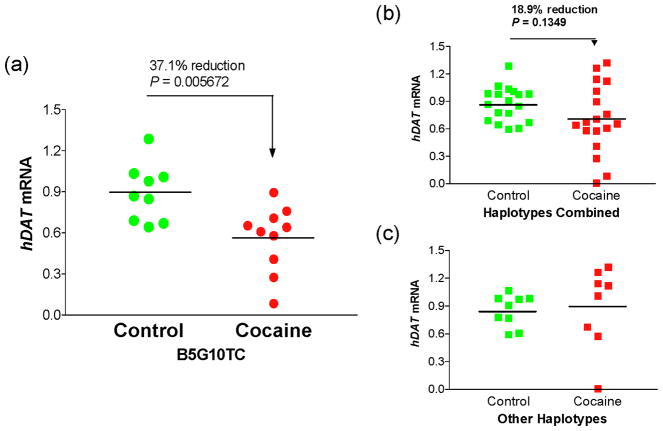
Analysis of correlation between haplotypes and expression levels revealed that one of the haplotypes, B5G10TC that covered the last six polymorphisms (frequency: 0.33 in drug-free controls versus 0.31 cocaine abusers) displayed an average expression level 37.1% less in cocaine abuser carriers than in control carriers (P = 0.005672 by Mann-Whitney U tests, Figure 2a). This B5G10TC-mediated reduction contributed to the overall small decrease (Figure 2b) because the rest of the haplotypes did not display any reduced expression in the abusers (Figure 2c). In drug-free controls, the B5G10TC activity remained the averages of other haplotypes combined.
Figure 2.
Identification of a cocaine abuse-related haplotype B5G10TC. (a) Down-regulation of hDAT mRNA levels in B5G10TC-carriers. (b) hDAT mRNA levels in all subjects. (c) hDAT mRNA levels in other-haplotype (non-B5G10TC)-carriers where P value by Mann–Whitney U tests is 0.3704 (all P values two-tailed).
Haplotype-dependent hDAT expression in human brain: lack of correlation between 3′ markers and mRNA levels
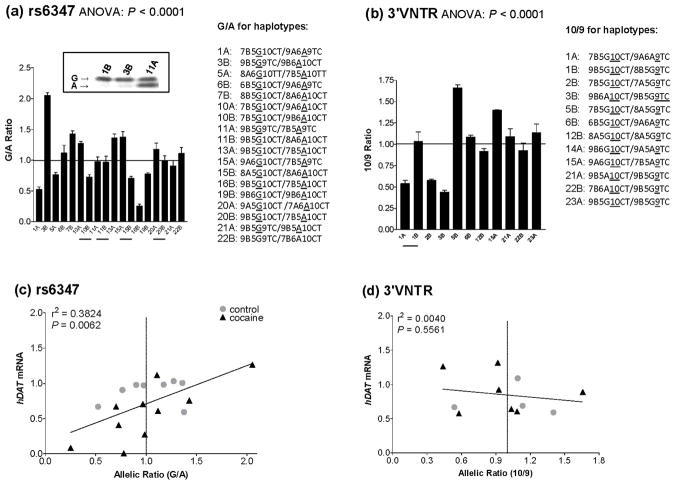
We next went on to use both exonic markers rs6347 and 3′VNTR to assess allelic hDAT expression in human midbrain samples by BStop-PCR, making sure of high degree of confidence in the expression data and increasing the use of available samples. BStop-PCR used biotin as the labeling signal, which displayed good dynamics (Figure S1). Among the 36 subjects, 18 carried informative (or both alleles) rs6347, 11 carried informative 3′VNTR and 6 carried both. Among the 18 rs6347-carriers, 13 (61.1%) displayed allelic dependence (> 1.2-fold allelic difference for statistical significance) of hDAT expression. The average ratio of G/A is 1.027 ± 0.095-fold for all 18 subjects and 1.023 ± 0.156-fold among the 11 subjects (Figure 3a), suggesting that neither of the alleles are consistently associated with higher expression. Similarly among the 11 3′VNTR-carriers, five (45.5%) displayed allelic dependence (> 1.2-fold allelic difference). The average ratio of 10/9 is 0.983 ± 0.111-fold for all 11 samples and 0.924 ± 0.252-fold among the five subjects that displayed allelic dependence (Figure 3b), again suggesting that neither of the alleles are consistently associated with higher expression. The corresponding haplotypes in each subject are indicated on the right side of both graphs, showing that none of the alleles of typed markers are consistently associated with a high or low ratio.
Figure 3.
Correlation between 3′ markers (rs6347 in panels a and c or 3′VNTR in panels b and d) and mRNA levels of hDAT in human ventral midbrain. Variable allele-dependence of hDAT mRNA levels, based on rs6347 (18 subjects in panel a) and 3′VNTR (11 subjects in panel b). Horizontal line, G/A or 10-repeat/9-repeat ratio = 1. Subject label: A for drug-free control, B for cocaine abuser; underline, control-case pair (see Table 1); corresponding haplotypes are statistically inferred and indicated on the right hand side. Data were obtained from three independent experiments. ANOVA P values of < 0.0001 indicate significant individual-dependence. Insert, representative BStop-PCR data. Correlation between allelic ratio of rs6347 (c) or 3′VNTR (d) and overall levels of hDAT mRNA in human midbrain tissues. rs6347 was carried by eight drug-free controls and ten cocaine abusers; 3′VNTR by four drug-free controls and seven cocaine abusers. Vertical line, allelic ratio as 1. r2, goodness of linear fit; P, significance of non-zero slope.
Approximately 42% of the controls (3/8 by rs6347 and 2/4 by 3′VNTR) and 47% of the abusers (4/10 by rs6347 and 4/7 by 3′VNTR) did not show any haplotype-dependence of expression (ratio ~1). Overall the allelic or haplotype-dependence of hDAT expression was very significant, as the ANOVA P value is < 0.0001 based on both markers. The data from the two markers are generally consistent with each other with a linear regression coefficient (r2) of 0.9468 (P: 0.0011) in six individuals that carried both markers.
The next question was whether the allelic or haplotype-dependence is correlated with overall hDAT expression in 24 informative individuals (those carried at least one of the two exonic markers). To address this question, we have analyzed the hDAT mRNA levels by qRT-PCR. The results show that allelic dependence of rs6347 correlates positively with the mRNA levels (r2: 0.3824 for goodness of fit, P: 0.0062 for non-zero linearity, see Figure 3c). We failed to find such correlation for the 3′VNTR marker (Figure 3d). We postulate that rs6347 was on the same background as other functional polymorphisms such as the nearby Int8VNTR and haplotypes of the upstream promoter (Greenwood and Kelsoe 2003; Kelada et al, 2005; Hill et al, 2010). However, 3′VNTR is located further away from those functional variants with weak LD (Figure 1a).
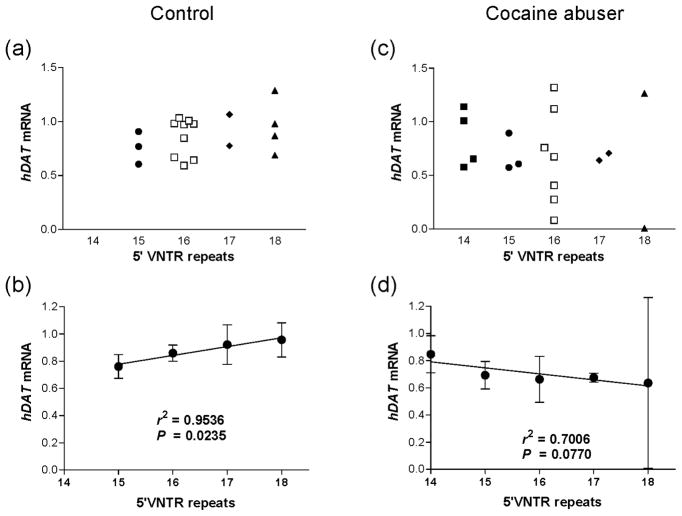
Positive correlation of 5′VNTR with hDAT mRNA levels
Delineation of allelic expression may lead to identification of new markers for association study or generating functionality hypotheses to test. We first looked at 5′VNTR because this could be a new risk locus. The subjects were stratified by drug phenotypes because cocaine abuse could confound a potential correlation. The total repeat number in each drug-free control can be positively correlated with the average hDAT mRNA levels (Figure 4a, b). However, this positive correlation was not observed, not even for the averages, in the cocaine abusers (Figure 4c, d). We summed the repeats on both chromosomes and correlated the sum with the mRNA level attributed to both chromosomes because if a transcription factor binds to a repeat sequence for functional effect, multiple bindings would increase the additive effect.
Figure 4.
5′VNTR-based positive allelic correlation with hDAT expression in midbrain tissues from drug-free controls (a, b) but not from cocaine abusers (c, d). Repeats: sum of both alleles. Panels a and c, distribution of individuals; panels b and d, averages of mRNA levels where r2, is goodness of linear fit and P, significance of non-zero slope.
By contrast, alleles for Int8VNTR (Figure S2) and 3′VNTR (Figure S3) or the VNTR haplotype 6–10 (Figure S4) display no significant correlation with the expression levels. No correlation was found for other markers either.
DISCUSSION
This study assessed the correlation of hDAT haplotypes with corresponding transcript abundance in human midbrain. We demonstrated that hDAT expression is clearly haplotype-dependent and furthermore that the dependence varies by a large range in human midbrain tissues. In particular, this study discovered that B5G10TC, a main African American haplotype with the same frequency in drug-free controls versus cocaine abusers, displays reduced expression in cocaine abusers, comparing to drug-free controls. Among the examined markers, 5′VNTR is the only marker that correlates positively with mRNA levels in drug-free controls but not in cocaine abusers. These preliminary but intriguing findings could imply hDAT epigenetic effects that warrant further investigation.
Markers for future association study
The observed low LD between the major regulatory regions such as 5′ promoter, Intron 8 and 3′UTR was consistent with previous findings especially in American samples (Guindalini et al., 2006; Greenwood et al., 2006). Such LD information suggests that genotype data with the Int8VNTR or 3′VNTR would not be able to reflect the 5′promoter activity variation and vice versa, as postulated in the results. For cocaine abuse, both 5′ and Intron 8-3′ ends could confer risk. Although previous studies have shown that 3′VNTR and Int8VNTR can regulate expression of a reporter gene in vitro (Inoue-Murayama et al., 2002; Miller and Madras, 2002; Hill et al., 2010), neither correlate with mRNA levels in human brain tissues in this study.
Because of the low LD, all three VNTRs should be evaluated for substance abuse. This notion is supported by findings for other diseases. For example, markers in upstream regions displayed more consistent positive signals than 3′VNTR for associations with schizophrenia and bipolar disorders (Khodayari et al., 2004; Stöber et al., 2006; Serretti and Mandelli, 2008; Talkowski et al., 2008).
Regulatory haplotypes in cocaine abusers
B5G10TC is a major African American haplotype of the transcribed region and has same frequency in drug-free controls versus cocaine abusers. Based on reported findings that low activity-associated alleles such as 6-repeat of Int8VNTR and 10-repeat of 3′VNTR are risk alleles (Inoue-Murayama et al., 2002; Miller and Madras 2002; Hill et al., 2010), expressional reduction of B5G10TC in cocaine abusers suggests two possibilities: B5G10TC is sensitive to cocaine abuse or this haplotype is genomically and functionally modified in individuals with the liability for cocaine abuse. There is no experimental data on the two Exon 15 SNPs rs6876890 and rs11564775 yet but MFOLD analysis shows that in the rs6347 G allele background, order of free energy is 10TC (−1453.74 kcal/mol) < 10TT (−1452.94 kcal/mol) < 10CT (−1448.91 kcal/mol), suggesting that TC of rs6876890 and rs11564775 might contribute to greater stability of this main mRNA species. However, the mechanism for the down-regulation merits further investigation also at an epigenetic level (Hawi et al., 2010; Shumay et al., 2010). Other regions including the 5′ end and the 3′ end represent risks and deserve further functional clarifications as well.
Variation of haplotype-dependence
Individual-dependent correlation between alleles and hDAT mRNA levels was observed for both exonic markers rs6347 and 3′VNTR independently of drug phenotypes (Figure 3), representing a flip-flop correlation. Two relevant questions are how the flip-flop correlation could happen and what this means to association studies. Among other possibilities, one potential answer to the first question is that the underlying regulatory regions lie in upstream regions such as the promoter and have low LD with these markers. This possibility is supported by the fact that the correlation between allelic ratio and overall mRNA levels is weaker for 3′VNTR than for rs6347 (Figure 3c vs. d). Another possibility is that these markers are functional but allelic expression varies among the individuals. No functional study on rs6347 has been done. Since both rs6347 and 3′VNTR are exonic and the polymorphisms may affect mRNA activity, we have carried out MFOLD modeling to see whether the polymorphisms might influence mRNA secondary structure or stability. The mRNA structures with the major alleles display lower free energy for greater stability than ones with the minor alleles. In the background of 10 repeat of 3′VNTR, the molecule with G (the major allele of rs6347) has a free energy of −1450.15 kcal/mol, comparing to −1448.91 kcal/mol in the molecule with A, or comparing to −1449.70 kcal/mol in the molecule with G but 9-repeat. In other words, the variation from G into A in rs6347 increases the free energy by 1.24 kcal/mol, 2.8-fold of the increase (0.45 kcal/mol) caused by the variation of 10-repeat into 9-repeat in 3′VNTR. These modeling results suggest that rs6347 is more likely to affect the mRNA stability than 3′VNTR. Consistent with this notion, the G allele that confers higher structural stability tends to correlate with higher expression levels than A in controls only (A/A-carriers had 30.2% reduction comparing to G/G-carriers, P = 0.0485 by Mann-Whitney U tests) but 3′VNTR lacks such correlation tendency. Despite the consistency between free energy modeling and experimental data, the small differences in free energy may suggest that other factors have contributed to the 30.2% reduction and these modeling data require future investigations for verification.
If expression variation of hDAT contributes to genetic etiologies of related brain disorders, the flip-flop allelic expression indicates the importance of marker selection in association study. Meanwhile, it is critical to continuously map the underlying genomic loci that may serve the best markers.
CONCLUSIONS
hDAT has many regulatory haplotypes and its activity can be down-regulated in a haplotype- and phenotype-dependent manner. Neither of the two most used markers, Int8VNTR and 3′VNTR, are correlated with the hDAT mRNA levels in the human midbrain. 5′VNTR may represent a novel marker based on its positive correlation with the expression levels.
Supplementary Material
Acknowledgments
This work was supported by US National Institutes of Health grants R01DA021409 (to ZL) and R01DA006470 (to MJB). The authors are grateful to Drs. Garrett Fitzmaurice and Joel Gelernter for helpful suggestions during preparation and revision of the manuscript.
Footnotes
DISCLOSURE
The authors declared no conflicts of interest.
Authors Contributions
ZL and MJB were responsible for study concept, design and interpretation of findings. CJS and MJB contributed to acquisition, dissection and characterization of brain tissues. SKM contributed to RNA preparation and characterization. YZ and ZL contributed to haplotyping, expressional and data analysis. JSL contributed to data analysis and interpretation. ZL drafted the manuscript and all authors critically reviewed content, revised the manuscript and approved final version for publication.
References
- Bannon MJ, Whitty CJ. Age-related and regional differences in dopamine transporter mRNA expression in human midbrain. Neurology. 1997;48:969–977. doi: 10.1212/wnl.48.4.969. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bezard E, Gross CE, Fournier MC, Dovero S, Bloch B, Jaber M. Absence of MPTP-induced neuronal death in mice lacking the dopamine transporter. Exp Neurol. 1999;155:268–273. doi: 10.1006/exnr.1998.6995. [DOI] [PubMed] [Google Scholar]
- Brookes KJ, Mill J, Guindalini C, Curran S, Xu X, Knight J, et al. A common haplotype of the dopamine transporter gene associated with attention-deficit/hyperactivity disorder and interacting with maternal use of alcohol during pregnancy. Arch Gen Psychiatry. 2006;63:74–81. doi: 10.1001/archpsyc.63.1.74. [DOI] [PubMed] [Google Scholar]
- Drgon T, Lin Z, Wang GJ, Fowler J, Pablo J, Mash DC, Volkow N, Uhl GR. Common human 5′ dopamine transporter (slc6a3) haplotypes yield varying expression levels in vivo. Cell Mol Neurobiol. 2006;26:875–889. doi: 10.1007/s10571-006-9014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraone SV, Mick E. Molecular genetics of attention deficit hyperactivity disorder. Psychiatr Clin North Am. 2010;33:159–180. doi: 10.1016/j.psc.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giros B, Jaber M, Jones SR, Wightman RM, Caron MG. Hyperlocomotion and indifference to cocaine and amphetamine in mice lacking the dopamine transporter. Nature. 1996;379:606–612. doi: 10.1038/379606a0. [DOI] [PubMed] [Google Scholar]
- Gainetdinov RR, Fumagalli F, Jones SR, Caron MG. Dopamine transporter is required for in vivo MPTP neurotoxicity: evidence from mice lacking the transporter. J Neurochem. 1997;69:1322–1325. doi: 10.1046/j.1471-4159.1997.69031322.x. [DOI] [PubMed] [Google Scholar]
- Greenwood TA, Kelsoe JR. Promoter and intronic variants affect the transcriptional regulation of the human dopamine transporter gene. Genomics. 2003;82:511–520. doi: 10.1016/s0888-7543(03)00142-3. [DOI] [PubMed] [Google Scholar]
- Greenwood TA, Schork NJ, Eskin E, Kelsoe JR. Identification of additional variants within the human dopamine transporter gene provides further evidence for an association with bipolar disorder in two independent samples. Mol Psychiatry. 2006;11:125–133. doi: 10.1038/sj.mp.4001764. [DOI] [PubMed] [Google Scholar]
- Guindalini C, Howard M, Haddley K, Laranjeira R, Collier D, Ammar N, Craig I, O’Gara C, Bubb VJ, Greenwood T, Kelsoe J, Asherson P, Murray RM, Castelo A, Quinn JP, Vallada H, Breen G. A dopamine transporter gene functional variant associated with cocaine abuse in a Brazilian sample. Proc Natl Acad Sci U S A. 2006;103:4552–4557. doi: 10.1073/pnas.0504789103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawi Z, Kent L, Hill M, Anney RJ, Brookes KJ, Barry E, Franke B, Banaschewski T, Buitelaar J, Ebstein R, Miranda A, Oades RD, Roeyers H, Rothenberger A, Sergeant J, Sonuga-Barke E, Steinhausen HC, Faraone SV, Asherson P, Gill M. ADHD and DAT1: further evidence of paternal over-transmission of risk alleles and haplotype. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:97–102. doi: 10.1002/ajmg.b.30960. [DOI] [PubMed] [Google Scholar]
- Hernandez A, Andollo W, Hearn WL. Analysis of cocaine and metabolites in brain using solid phase extraction and full-scanning gas chromatography/ion trap mass spectrometry. Forensic Sci Int. 1994;65:149–156. doi: 10.1016/0379-0738(94)90270-4. [DOI] [PubMed] [Google Scholar]
- Hill M, Anney RJ, Gill M, Hawi Z. Functional analysis of intron 8 and 3′ UTR variable number of tandem repeats of SLC6A3: differential activity of intron 8 variants. Pharmacogenomics J. 2010;10:442–447. doi: 10.1038/tpj.2009.66. [DOI] [PubMed] [Google Scholar]
- Inoue-Murayama M, Adachi S, Mishima N, Mitani H, Takenaka O, Terao K, Hayasaka I, Ito S, Murayama Y. Variation of variable number of tandem repeat sequences in the 3′-untranslated region of primate dopamine transporter genes that affects reporter gene expression. Neurosci Lett. 2002;334:206–210. doi: 10.1016/s0304-3940(02)01125-4. [DOI] [PubMed] [Google Scholar]
- Joyce JN, Smutzer G, Whitty CJ, Myers A, Bannon MJ. Differential modification of dopamine transporter and tyrosine hydroxylase mRNAs in midbrain of subjects with Parkinson’s, Alzheimer’s with Parkinsonism, and Alzheimer’s disease. Mov Disord. 1997;12:885–897. doi: 10.1002/mds.870120609. [DOI] [PubMed] [Google Scholar]
- Kelada SN, Costa-Mallen P, Checkoway H, Carlson CS, Weller TS, Swanson PD, Franklin GM, Longstreth WT, Jr, Afsharinejad Z, Costa LG. Dopamine transporter (SLC6A3) 5′ region haplotypes significantly affect transcriptional activity in vitro but are not associated with Parkinson’s disease. Pharmacogenet Genomics. 2005;15:659–668. doi: 10.1097/01.fpc.0000170917.04275.d6. [DOI] [PubMed] [Google Scholar]
- Khodayari N, Garshasbi M, Fadai F, Rahimi A, Hafizi L, Ebrahimi A, Najmabadi H, Ohadi M. Association of the dopamine transporter gene (DAT1) core promoter polymorphism -67T variant with schizophrenia. Am J Med Genet. 2004;129B:10–12. doi: 10.1002/ajmg.b.30067. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang Z, He Z, Tang W, Li T, Zeng Z, Lin H, Shi Y. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis. Cell Res. 2009;19:519–523. doi: 10.1038/cr.2009.33. http://analysis.bio-x.cn. [DOI] [PubMed] [Google Scholar]
- Lohoff FW, Bloch PJ, Hodge R, Nall AH, Ferraro TN, Kampman KM, Dackis CA, O’Brien CP, Pettinati HM, Oslin DW. Association analysis between polymorphisms in the dopamine D2 receptor (DRD2) and dopamine transporter (DAT1) genes with cocaine dependence. Neurosci Lett. 2010;473:87–91. doi: 10.1016/j.neulet.2010.02.021. [DOI] [PubMed] [Google Scholar]
- Miller GM, Madras BK. Polymorphisms in the 3′-untranslated region of human and monkey dopamine transporter genes affect reporter gene expression. Mol Psychiatry. 2002;7:44–55. doi: 10.1038/sj.mp.4000921. [DOI] [PubMed] [Google Scholar]
- Niu T. Algorithms for inferring haplotypes. Genet Epidemiol. 2004;27:334–347. doi: 10.1002/gepi.20024. [DOI] [PubMed] [Google Scholar]
- O’Gara C, Stapleton J, Sutherland G, Guindalini C, Neale B, Breen G, Ball D. Dopamine transporter polymorphisms are associated with short-term response to smoking cessation treatment. Pharmacogenet Genomics. 2007;17:61–67. doi: 10.1097/01.fpc.0000236328.18928.4c. [DOI] [PubMed] [Google Scholar]
- Schwartz K, Nachman R, Yossifoff M, Sapir R, Weizman A, Rehavi M. Cocaine, but not amphetamine, short term treatment elevates the density of rat brain vesicular monoamine transporter 2. J Neural Transm. 2007;114:427–430. doi: 10.1007/s00702-006-0549-8. [DOI] [PubMed] [Google Scholar]
- Serretti A, Mandelli L. The genetics of bipolar disorder: genome ‘hot regions,’ genes, new potential candidates and future directions. Mol Psychiatry. 2008;13:742–771. doi: 10.1038/mp.2008.29. [DOI] [PubMed] [Google Scholar]
- Shumay E, Fowler JS, Volkow ND. Genomic features of the human dopamine transporter gene and its potential epigenetic States: implications for phenotypic diversity. PLoS One. 2010;5:e11067. doi: 10.1371/journal.pone.0011067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sora I, Hall FS, Andrews AM, Itokawa M, Li XF, Wei HB, Wichems C, Lesch KP, Murphy DL, Uhl GR. Molecular mechanisms of cocaine reward: combined dopamine and serotonin transporter knockouts eliminate cocaine place preference. Proc Natl Acad Sci U S A. 2001;98:5300–5305. doi: 10.1073/pnas.091039298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöber G, Sprandel J, Jabs B, Pfuhlmann B, Moller-Ehrlich K, Knapp M. Family-based study of markers at the 5′-flanking region of the human dopamine transporter gene reveals potential association with schizophrenic psychoses. Eur Arch Psychiatry Clin Neurosci. 2006;256:422–427. doi: 10.1007/s00406-006-0657-3. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkowski ME, Kirov G, Bamne M, Georgieva L, Torres G, Mansour H, Chowdari KV, Milanova V, Wood J, McClain L, Prasad K, Shirts B, Zhang J, O’Donovan MC, Owen MJ, Devlin B, Nimgaonkar VL. A network of dopaminergic gene variations implicated as risk factors for schizophrenia. Hum Mol Genet. 2008;17:747–758. doi: 10.1093/hmg/ddm347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uejima H, Lee MP, Cui H, Feinberg AP. Hot-stop PCR: a simple and general assay for linear quantitation of allele ratios. Nat Genet. 2000;25:375–376. doi: 10.1038/78040. [DOI] [PubMed] [Google Scholar]
- van der Zwaluw CS, Engels RC, Buitelaar J, Verkes RJ, Franke B, Scholte RH. Polymorphisms in the dopamine transporter gene (SLC6A3/DAT1) and alcohol dependence in humans: a systematic review. Pharmacogenomics. 2009;10:853–866. doi: 10.2217/pgs.09.24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.