Abstract
The promyelocytic leukaemia gene PML was originally identified at the t(15;17) translocation of acute promyelocytic leukaemia, which generates the oncogene PML-retinoic acid receptor α. PML epitomises a subnuclear structure called PML nuclear body. Current models propose that PML through its scaffold properties is able to control cell growth and survival at many different levels. Here we discuss the current literature and propose new avenues for investigation.
Keywords: Snail, HNF4α, miRs-200, miR-34a, stemness
Promyelocytic Leukaemia (PML) and Cancer
Haematological malignancies
The majority of acute promyelocytic leukaemia (APL) cases are characterised by the t(15;17) chromosomal translocation that juxtaposes the PML gene and the retinoic acid receptor α (RARα).1, 2, 3, 4, 5 This translocation is reciprocal and balanced, and produces two fusion genes, PML-RARα and RARα-PML. PML-RARα is the main oncogene of APL and is able to transform haemopoietic precursors.1, 2, 3 On one hand, PML-RARα suppresses RARα transcriptional function, thus blocking differentiation at the promyelocytic stage.1, 2, 3 On the other hand, PML-RARα disrupts the PML nuclear bodies (PML-NBs) through formation of PML-RARα/PML heterodimers.1, 2, 3 It is still debated whether this interaction is crucial for leukaemogenesis in vivo. At least in vitro, the PML moiety within the oncogenic fusion protein has been proposed to only function to promote formation of multimers that cause transcriptional repression and ultimately transformation.6 However, loss of PML increases the incidence and accelerates the onset of leukaemia in a mouse model of APL,7 thus suggesting that its inactivation is important for promoting neoplastic transformation in vivo. Interestingly, two mutations of the remaining PML allele have been found in APL, which generate a premature stop codon and are predicted to encode truncated cytoplasmic proteins.8, 9 PML has been found translocated to the PAX5 locus to generate a PAX5-PML fusion gene in childhood acute lymphoblastic leukaemia,10 which disrupts both PML and Pax5 function, thus suggesting that disruption of PML function could have a role in non-APL tumours. Indeed, it has been reported that PML expression is lost in other haematological malignancies, such as in 83% diffuse large cell lymphomas (DLCL) and 77% follicular lymphomas.11 It would be very important to address the impact of PML loss in non-APL tumours. Overall, these studies suggest that PML works as tumour suppressor in the haemopoietic system. However, chronic myeloid leukaemia (CML) represents a notable exception. PML expression has been shown to act as positive regulator of self-renewal in CML-initiating cells.12 As a result, PML loss leads to exhaustion of the leukaemic stem cell pool and reduced disease progression. Accordingly, PML expression correlates with poor overall survival in CML patients. This report constitutes a change in paradigm in the field of PML research, as it suggests a potential oncogenic role of PML. This is reminiscent of the role played by the growth suppressor p21 in leukaemic stem cells.13 Further research is needed to fully dissect the mechanisms underlying PML function in cancer stem cells.
Solid tumours
It is becoming clear that PML expression is altered in many solid tumours. In this respect, the study by Gurrieri et al.11 showed that PML expression is absent in 17% of colon adenocarcinomas, 21% of lung tumours, 27% of prostate adenocarcinomas, 31% of breast adenocarcinomas, 49% of CNS tumours (100% medulloblastomas and over 90% oligodendroglial tumours), 49% of germ cell tumours and 68% of non-Hodgkin's lymphomas (83% DLCL and 77% follicular lymphomas). Other studies have shown that PML expression is lost in breast carcinomas,14 gastric cancer,15 small cell lung carcinoma16 and in invasive epithelial tumours.17 Interestingly, Gurrieri et al.11 showed that loss of PML expression correlates with a higher tumour grading in breast adenocarcinomas and prostate carcinomas. It has to be noted that this study was conducted using an antibody against a PML epitope reported as fixation-sensitive,14 thus suggesting that some of the tumours could have been erroneously classified as negative. Despite the lack of expression at protein levels, there is little evidence that PML is regulated at the transcriptional level in cancer.11 In addition, mutations in non-APL tumours are extremely rare.11 As a result, research efforts have been focused on studying posttranslational regulation of PML stability. In this respect, PML has been shown to be degraded by the proteasome in lung, colon and gastric cancer cell lines.11, 18 In addition, recent studies have demonstrated that the E3 ligase RNF4 mediates PML ubiquitylation and degradation.19, 20 Interestingly, this process appears to rely on disulphide bonds formation and subsequent SUMOylation of PML in PML-NBs.21 This work opens a completely new field for the investigation of redox-dependent and SUMO-dependent degradation of nuclear proteins. Given the importance of posttranscriptional/translational regulation of gene expression in cancer, it is conceivable that PML translation could be regulated in cancer cells. In this respect, it is presently unclear whether PML translation is controlled by microRNAs or RNA-binding proteins in normal versus cancer cells. This warrants an urgent investigation.
PML Function(s)
PML has been proposed to transduce various growth suppressive signals. Several studies have implicated PML in the regulation of cellular senescence and programmed cell death. Most of them focused on PML nuclear splice variants,22 but it is becoming clear that cytoplasmic localisation of PML can affect growth suppression and cell death. This review will focus on the role of PML in cell death control and will discuss the impact of most recent discoveries in the field.
PML and death receptors
PML has been shown to regulate apoptosis induced by FAS ligand (FASL) and tumour necrosis factor (TNF) α, which are key regulators of immunity and inflammation.23, 24, 25, 26, 27 In particular, PML-deficient lymphocytes show decreased cell death following treatment with FASL.28 Furthermore, bone marrow cells from PML−/− animals are resistant to TNFα treatment. In both cases, this correlates with decreased caspase activation. It is worth noting that PML has been shown to regulate cell death in a caspase-independent manner, thus suggesting that its role is not confined to caspase-dependent cell death.29 Finally, PML role is not limited to FASL and TNF, as it has been shown to potentiate interferon α-triggered cell death through induction of TRAIL,30 a death receptor expressed in cancer cells.31, 32, 33, 34, 35, 36, 37
PML and pro-apoptotic transcription factors
More recent studies have shown that PML regulates the tumour suppressor p53, a major regulator of apoptosis.38, 39, 40 In particular, it controls p53 degradation through the inhibition of Mdm2, which is the major p53 E3 ubiquitin ligase.41, 42, 43, 44, 45 This function appears to be in part PML-NB independent and occurs through sequestration of Mdm2 into nucleoli, thus promoting p53 activation upon DNA damage.41 The PML-interacting protein DAXX46, 47 has been shown to control p53 ubiquitylation by inhibiting MDM2 degradation.48, 49 PML can also control p53 by promoting its acetylation50 and phosphorylation at multiple residues.33, 51, 52, 53 cPML isoforms have been shown to negatively regulate PML function,54 thus suggesting that balance between nuclear and cytoplasmic isoforms could dictate the response to growth suppressive signals. PML can also regulate DNA damage response in a p53-independent manner. In this respect, PML is under the control of the ATM/Chk2 pathway for induction of cell death upon genotoxic stress.55, 56 Finally, PML has been shown to regulate cell death induced by HIV infection.57 In particular, PML transduces ATM/p53-dependent pro-apoptotic signals in HIV-induced syncytia.58, 59 Overall, these findings indicate that PML can regulate p53 function by acting at different levels of the p53 pathway. It remains to be established whether these different regulatory routes are stimulus- and/or tissue-specific.
PML regulates the function of other members of the p53 family. In this respect, PML has been shown to inhibit the degradation of the p53 family member p73.60, 61, 62, 63, 64, 65, 66, 67 More recent studies have further dissected the functional consequences of PML/p73 interaction.68, 69, 70, 71 Notably, the shorter, growth-promoting isoform ΔN-p73 is regulated by the APL oncogene PML-RARα, thus adding another level of complexity.72 Finally, PML is known to regulate the remaining member of the family, p63,73, 74, 75, 76, 77, 78, 79, 80 which has a key role in development and homeostasis of different epithelia.81
PML interacts also with c-Jun upon UV irradiation, and modulates its pro-apoptotic function through c-Jun-N-terminal kinase (JNK)-dependent phosphorylation,82 a pathway implicated in the regulation of apoptosis.67, 83, 84, 85 UV causes dramatic PML-NB reorganisation, which leads to formation of multiple microspeckles positive for both phosphorylated c-Jun and PML.82 Interestingly, DAXX has been shown to regulate JNK in human fibroblasts,46 thus suggesting that PML could regulate the JNK/c-Jun pathway via DAXX.
PML, PTEN and AKT
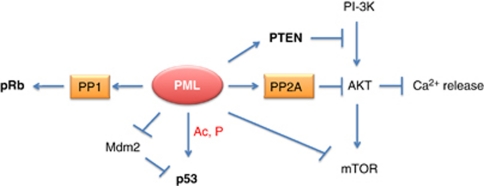
Recent evidence has implicated PML in the regulation of the PI-3K pathway at multiple levels.86 This work predominantly comes from the Pandolfi's group. First, PML has been shown to promote PTEN nuclear localisation by affecting its interaction with HAUSP and its ubiquitylation status.87 Second, PML is able to inhibit Akt function by promoting its PP2A-dependent dephosphorylation.88 Notably, we have shown that PML interacts with another phosphatase PP1, and promotes PP1-dependent dephosphorylation of retinoblastoma protein (pRb) in neural stem cells.89 Finally, PML directly interacts with mTOR and induces its localisation to the PML-NBs, thus inhibiting its function. Taken together, these findings indicate that PML has an important role in regulation of the PI-3K pathway. In this respect, PML has been shown to regulate the intracellular degradation mechanism autophagy,90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103 which is negatively regulated by mTOR, and has been implicated in cancer development and longevity. It is therefore possible that PML through inhibition of mTOR could promote induction of autophagy. As mentioned above, it is still unclear whether these regulatory nodes exist in the same cell or whether they vary depending on the cell type or extracellular environment.
PML and transforming growth factor (TGF) β
TGFβ is known to control key tumour suppressive functions in normal cells, whereas in cancer cells it has been proposed to bear pro-metastatic functions.104, 105 The group led by Pier Paolo Pandolfi has shown that in PML-deficient fibroblasts the response to TGFβ is blunted, with both senescence and apoptosis being severely impaired.106 Surprisingly, this effect was mainly caused by loss of PML cytoplasmic isoforms (cPML). In particular, cPML regulates endosomal trafficking of TGFβ receptors by promoting the association of Smad2/3 and Smad anchor for receptor activation. Interestingly, this pathway can be modulated by nuclear retention of cPML via a mechanism involving TG-interacting factor (TGIF-) and c-Jun.107, 108 In turn, TGIF is inhibited by PML competitor for TGIF association (PCTA), thus activating cPML tumour suppressive function.108 A recent study has demonstrated that the nuclear corepressor SnoN, a known regulator of TGFβ, controls p53 stabilisation via interaction with PML and PML-NBs and independent of Smads.109 This study suggests that nuclear PML is also involved in regulation of the TGFβ pathway. Further research efforts are needed to fully dissect the role of different PML isoforms in regulation of this pathway.
PML and the endoplasmic reticulum
A very recent study from Pandolfi's group has proposed a novel role for PML in the cytoplasm.110, 111 PML appears enriched at the endoplasmic reticulum and at the mitochondria-associated membranes, which constitute ER-to-mitochondria communication sites involved in transport of Ca2+ and induction of apoptosis.110, 111, 112 At these sites, PML interacts with the 1,4,5-triphosphate receptor (IP3R), AKT and PP2A. In the absence of PML, AKT-dependent phosphorylation of IP3R is increased, whereas Ca2+ release from the ER is impaired, resulting in blunted apoptosis. These data suggest that PML can affect both nuclear and cytoplasmic functions of AKT through its interaction with PP2A, thus promoting its inactivation and apoptosis induction. Mitochondria act as crucial regulators of cell death through a complex interplay of pro- and anti-apoptotic proteins associated with these organelles. The tumour suppressor p53 has been demonstrated to localise to mitochondria and promote apoptosis via regulation of BCL-2 family members.113, 114, 115, 116, 117, 118 It is conceivable that PML could regulate p53 not only in the nucleus but also in mitochondria. Further studies are needed to address a potential functional interaction between PML and p53 in mitochondria, and the impact of this interaction on BCL-2 family members and apoptosis induction.
Overall, studies in the last few years indicate that PML, through interaction with PP1 and PP2A phosphatases in the nucleus and the cytoplasm, could affect key tumour suppressive (pRb, see below) and oncogenic pathways (AKT). It remains to be established whether the function of p53 and c-Jun could also be modulated by PML-mediated regulation of PP1 and/or PP2A (Figure 1). Finally, it is conceivable that PML itself could be a target for PP1 and/or PP2A-mediated dephosphorylation as part of a positive or negative feedback loop.
Figure 1.
Multifaceted role of PML in regulation of apoptosis and growth suppression. PML activates pRb and inhibits AKT via interactions with PP1 and PP2A phosphatases, respectively. In addition, it negatively affects the PI-3K pathway by inhibiting mTOR and activating PTEN. cPML retains the ability to promote PP2A-dependent AKT inhibition, thus causing Ca2+ release at contact sites between the mitochondria and the ER. Finally, PML positively regulates p53 by acting at different levels (that is, acetylation, phosphorylation and Mdm2-dependent degradation). It is presently unclear whether sequestration of PP1 and PP2A into PML-NBs can inhibit their function
PML Function in Stem Cells: what is the Contribution of Cell Death Regulation?
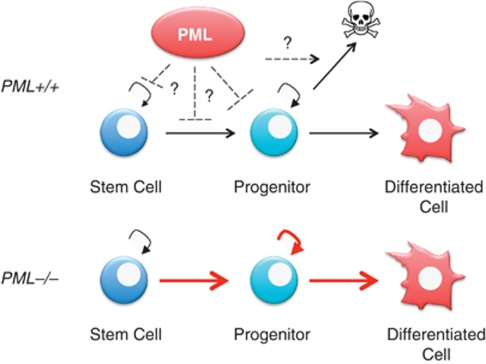
PML has emerged as an important factor regulating stem cell function within multiple tissues. In particular, our work has shown that PML regulates neural stem cell (NSC) function during corticogenesis by a mechanism involving PP1 and pRb.89 In the bone marrow, PML loss affects self-renewal in haemopoietic stem cells (HSCs) potentially through its action on the mTOR pathway.12 Finally, PML regulates mammary gland development and its loss results in skewing of mammary progenitor subtypes.119, 120 It is presently unclear whether the phenotypes caused by PML loss in these different tissues is in part due to alterations of cell death. For instance, increased NSC number in PML−/− cortices during development could be because of increased cycling as well as decreased cell death. Vice versa, reduction in neuronal numbers in PML-deficient cortices could be because of increased cell death following commitment of neural progenitors to neuronal fate. In the haemopoietic system, the increased proliferation of PML−/− committed progenitors could be caused by impaired cell death (Figure 2). Finally, altered cell death pathways could explain the increased generation of ERα+ luminal progenitors in PML-deficient mammary glands.108, 109
Figure 2.
Role of PML in stem/progenitor cells. PML regulates stem cell function in different tissues. In the haematopoietic system, PML loss leads to expansion of the progenitor pool and increased entry into differentiation, accompanied by reduction in the stem cell pool. This phenotype could be explained by increased transition to committed progenitors, augmented proliferation and/or decreased cell death. Further research is needed to determine whether PML can promote cell death in committed progenitors, and by which mechanism(s)
The last few years of PML research have produced fascinating results. However, the increasing complexity of PML function and its promiscuous interactions raise a number of key questions: (i) Are these interactions tissue- or context-specific? (ii) Does PML work differently in normal cells versus immortalised or transformed cells? (iii) How is the interplay between cPML and nuclear functions regulated? The field is in need of more refined mouse models, such as knockin and conditional knockouts, which will help addressing these important points.
Acknowledgments
We thank other members of Salomoni's lab for their support. Furthermore, we thank Pierluigi Nicotera (DZNE, Bonn, Germany) for scientific discussion and support. PS, MD and DM are supported by the Samantha Dickson Brain Tumour Trust and the Wellcome Trust.
Glossary
- APL
acute promyelocytic leukaemia
- CML
chronic myeloid leukaemia
- DLCL
diffuse large cell lymphomas
- ER
endoplasmic reticulum
- FL
follicular lymphomas
- HAUSP
herpesvirus-associated ubiquitin-specific protease
- HSCs
haemopoietic stem cells
- IP3R
inositol 1,4,5-trisphosphate receptor
- PI3K
phosphoinositide kinase-3
- PML
promyelocytic leukaemia gene
- PML-NB
PML nuclear body
- cPML
cytoplasmic PML
- PCTA
PML competitor for TGIF association
- PP1
protein phosphatase 1
- PP2A
protein phosphatase 2A
- PTEN
phosphatase and tensin homolog
- RARα
retinoic acid receptor alpha
- pRb
retinoblastoma protein
- SMAD
mothers against decapentaplegic homolog
- TGFα
transforming growth factor α
- TGIF
TG-–interacting factor
- TNF
tumour necrosis factor
- TRAIL
TNF-related apoptosis-inducing ligand
- mTOR
mammalian target of rapamycin
- SUMO
small ubiquitine-like modifier
The authors declare no conflict of interest.
Footnotes
Edited by G Melino
References
- de The H, Chen Z. Acute promyelocytic leukaemia: novel insights into the mechanisms of cure. Nat Rev Cancer. 2010;10:775–783. doi: 10.1038/nrc2943. [DOI] [PubMed] [Google Scholar]
- Piazza F, Gurrieri C, Pandolfi PP. The theory of APL. Oncogene. 2001;20:7216–7222. doi: 10.1038/sj.onc.1204855. [DOI] [PubMed] [Google Scholar]
- Salomoni P, Pandolfi PP. The role of PML in tumor suppression. Cell. 2002;108:165–170. doi: 10.1016/s0092-8674(02)00626-8. [DOI] [PubMed] [Google Scholar]
- Salomoni P, Ferguson BJ, Wyllie AH, Rich T. New insights into the role of PML in tumour suppression. Cell Res. 2008;18:622–640. doi: 10.1038/cr.2008.58. [DOI] [PubMed] [Google Scholar]
- Salomoni P. Stemming out of a new PML era. Cell Death Differ. 2009;16:1083–1092. doi: 10.1038/cdd.2009.63. [DOI] [PubMed] [Google Scholar]
- Minucci S, Maccarana M, Cioce M, De Luca P, Gelmetti V, Segalla S, et al. Oligomerization of RAR and AML1 transcription factors as a novel mechanism of oncogenic activation. Mol Cell. 2000;5:811–820. doi: 10.1016/s1097-2765(00)80321-4. [DOI] [PubMed] [Google Scholar]
- Rego EM, Wang ZG, Peruzzi D, He LZ, Cordon-Cardo C, Pandolfi PP. Role of promyelocytic leukemia (PML) protein in tumor suppression. J Exp Med. 2001;193:521–529. doi: 10.1084/jem.193.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellodi C, Kindle K, Bernassola F, Dinsdale D, Cossarizza A, Melino G, et al. Cytoplasmic function of mutant promyelocytic leukemia (PML) and PML-retinoic acid receptor-alpha. J Biol Chem. 2006;281:14465–14473. doi: 10.1074/jbc.M600457200. [DOI] [PubMed] [Google Scholar]
- Gurrieri C, Nafa K, Merghoub T, Bernardi R, Capodieci P, Biondi A, et al. Mutations of the PML tumor suppressor gene in acute promyelocytic leukemia. Blood. 2004;103:2358–2362. doi: 10.1182/blood-2003-07-2200. [DOI] [PubMed] [Google Scholar]
- Kurahashi S, Hayakawa F, Miyata Y, Yasuda T, Minami Y, Tsuzuki S, et al. PAX5-PML acts as a dual dominant-negative form of both PAX5 and PML. Oncogene. 2011;30:1822–1830. doi: 10.1038/onc.2010.554. [DOI] [PubMed] [Google Scholar]
- Gurrieri C, Capodieci P, Bernardi R, Scaglioni PP, Nafa K, Rush LJ, et al. Loss of the tumor suppressor PML in human cancers of multiple histologic origins. J Natl Cancer Inst. 2004;96:269–279. doi: 10.1093/jnci/djh043. [DOI] [PubMed] [Google Scholar]
- Ito K, Bernardi R, Morotti A, Matsuoka S, Saglio G, Ikeda Y, et al. PML targeting eradicates quiescent leukaemia-initiating cells. Nature. 2008;453:1072–1078. doi: 10.1038/nature07016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale A, De Franco F, Orleth A, Cambiaghi V, Giuliani V, Bossi D, et al. Cell-cycle restriction limits DNA damage and maintains self-renewal of leukaemia stem cells. Nature. 2009;457:51–56. doi: 10.1038/nature07618. [DOI] [PubMed] [Google Scholar]
- Gambacorta M, Flenghi L, Fagioli M, Pileri S, Leoncini L, Bigerna B, et al. Heterogeneous nuclear expression of the promyelocytic leukemia (PML) protein in normal and neoplastic human tissues. Am J Pathol. 1996;149:2023–2035. [PMC free article] [PubMed] [Google Scholar]
- Lee HE, Jee CD, Kim MA, Lee HS, Lee YM, Lee BL, et al. Loss of promyelocytic leukemia protein in human gastric cancers. Cancer Lett. 2007;247:103–109. doi: 10.1016/j.canlet.2006.03.034. [DOI] [PubMed] [Google Scholar]
- Zhang P, Chin W, Chow LT, Chan AS, Yim AP, Leung SF, et al. Lack of expression for the suppressor PML in human small cell lung carcinoma. Int J Cancer. 2000;85:599–605. doi: 10.1002/(sici)1097-0215(20000301)85:5<599::aid-ijc1>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Koken MH, Linares-Cruz G, Quignon F, Viron A, Chelbi-Alix MK, Sobczak-Thepot J, et al. The PML growth-suppressor has an altered expression in human oncogenesis. Oncogene. 1995;10:1315–1324. [PubMed] [Google Scholar]
- Scaglioni PP, Yung TM, Cai LF, Erdjument-Bromage H, Kaufman AJ, Singh B, et al. A CK2-dependent mechanism for degradation of the PML tumor suppressor. Cell. 2006;126:269–283. doi: 10.1016/j.cell.2006.05.041. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, et al. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- Jeanne M, Lallemand-Breitenbach V, Ferhi O, Koken M, Le Bras M, Duffort S, et al. PML/RARA oxidation and arsenic binding initiate the antileukemia response of As2O3. Cancer Cell. 2010;18:88–98. doi: 10.1016/j.ccr.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Condemine W, Takahashi Y, Zhu J, Puvion-Dutilleul F, Guegan S, Janin A, et al. Characterization of endogenous human promyelocytic leukemia isoforms. Cancer Res. 2006;66:6192–6198. doi: 10.1158/0008-5472.CAN-05-3792. [DOI] [PubMed] [Google Scholar]
- Debatin KM, Krammer PH. Death receptors in chemotherapy and cancer. Oncogene. 2004;23:2950–2966. doi: 10.1038/sj.onc.1207558. [DOI] [PubMed] [Google Scholar]
- Fas SC, Fritzsching B, Suri-Payer E, Krammer PH. Death receptor signaling and its function in the immune system. Curr Dir Autoimmun. 2006;9:1–17. doi: 10.1159/000090767. [DOI] [PubMed] [Google Scholar]
- Fulda S, Debatin KM. Signaling through death receptors in cancer therapy. Curr Opin Pharmacol. 2004;4:327–332. doi: 10.1016/j.coph.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Guardiola-Serrano F, Rossin A, Cahuzac N, Luckerath K, Melzer I, Mailfert S, et al. Palmitoylation of human FasL modulates its cell death-inducing function. Cell Death Disease. 2010;1:e88. doi: 10.1038/cddis.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JP, Betin VM, Weir H, Shelmani GM, Moss DK, Lane JD. Caspase cleavage of the Golgi stacking factor GRASP65 is required for Fas/CD95-mediated apoptosis. Cell Death Disease. 2010;1:e82. doi: 10.1038/cddis.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZG, Ruggero D, Ronchetti S, Zhong S, Gaboli M, Rivi R, et al. PML is essential for multiple apoptotic pathways. Nat Genet. 1998;20:266–272. doi: 10.1038/3073. [DOI] [PubMed] [Google Scholar]
- Quignon F, De Bels F, Koken M, Feunteun J, Ameisen JC, de The H. PML induces a novel caspase-independent death process. Nat Genet. 1998;20:259–265. doi: 10.1038/3068. [DOI] [PubMed] [Google Scholar]
- Crowder C, Dahle O, Davis RE, Gabrielsen OS, Rudikoff S. PML mediates IFN-alpha-induced apoptosis in myeloma by regulating TRAIL induction. Blood. 2005;105:1280–1287. doi: 10.1182/blood-2004-04-1614. [DOI] [PubMed] [Google Scholar]
- Falschlehner C, Schaefer U, Walczak H. Following TRAIL's path in the immune system. Immunology. 2009;127:145–154. doi: 10.1111/j.1365-2567.2009.03058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazi A. Directing cancer cells to self-destruct with pro-apoptotic receptor agonists. Nat Rev Drug Discov. 2008;7:1001–1012. doi: 10.1038/nrd2637. [DOI] [PubMed] [Google Scholar]
- Schneider-Jakob S, Corazza N, Badmann A, Sidler D, Stuber-Roos R, Keogh A, et al. Synergistic induction of cell death in liver tumor cells by TRAIL and chemotherapeutic drugs via the BH3-only proteins Bim and Bid. Cell Death Disease. 2010;1:e86. doi: 10.1038/cddis.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis CR, van der Sloot AM, Natoni A, Szegezdi E, Setroikromo R, Meijer M, et al. Rapid and efficient cancer cell killing mediated by high-affinity death receptor homotrimerizing TRAIL variants. Cell Death Disease. 2010;1:e83. doi: 10.1038/cddis.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider B, Munkel S, Krippner-Heidenreich A, Grunwald I, Wels WS, Wajant H, et al. Potent antitumoral activity of TRAIL through generation of tumor-targeted single-chain fusion proteins. Cell Death Disease. 2010;1:e68. doi: 10.1038/cddis.2010.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFarlane M, Inoue S, Kohlhaas SL, Majid A, Harper N, Kennedy DB, et al. Chronic lymphocytic leukemic cells exhibit apoptotic signaling via TRAIL-R1. Cell Death Differ. 2005;12:773–782. doi: 10.1038/sj.cdd.4401649. [DOI] [PubMed] [Google Scholar]
- Harper N, MacFarlane M. Recombinant TRAIL and TRAIL receptor analysis. Methods Enzymol. 2008;446:293–313. doi: 10.1016/S0076-6879(08)01618-2. [DOI] [PubMed] [Google Scholar]
- Meley D, Spiller DG, White MR, McDowell H, Pizer B, See V. p53-mediated delayed NF-kappaB activity enhances etoposide-induced cell death in medulloblastoma. Cell Death Disease. 2010;1:e41. doi: 10.1038/cddis.2010.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, Green DR. Dissecting p53-dependent apoptosis. Cell Death Differ. 2006;13:994–1002. doi: 10.1038/sj.cdd.4401908. [DOI] [PubMed] [Google Scholar]
- Fricker M, O'Prey J, Tolkovsky AM, Ryan KM. Phosphorylation of Puma modulates its apoptotic function by regulating protein stability. Cell Death Disease. 2010;1:e59. doi: 10.1038/cddis.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat Cell Biol. 2004;6:665–672. doi: 10.1038/ncb1147. [DOI] [PubMed] [Google Scholar]
- Kurki S, Latonen L, Laiho M. Cellular stress and DNA damage invoke temporally distinct Mdm2, p53 and PML complexes and damage-specific nuclear relocalization. J Cell Sci. 2003;116:3917–3925. doi: 10.1242/jcs.00714. [DOI] [PubMed] [Google Scholar]
- Louria-Hayon I, Grossman T, Sionov RV, Alsheich O, Pandolfi PP, Haupt Y. The promyelocytic leukemia protein protects p53 from Mdm2-mediated inhibition and degradation. J Biol Chem. 2003;278:33134–33141. doi: 10.1074/jbc.M301264200. [DOI] [PubMed] [Google Scholar]
- Wei X, Yu ZK, Ramalingam A, Grossman SR, Yu JH, Bloch DB, et al. Physical and functional interactions between PML and MDM2. J Biol Chem. 2003;278:29288–29297. doi: 10.1074/jbc.M212215200. [DOI] [PubMed] [Google Scholar]
- Zhu H, Wu L, Maki CG. MDM2 and promyelocytic leukemia antagonize each other through their direct interaction with p53. J Biol Chem. 2003;278:49286–49292. doi: 10.1074/jbc.M308302200. [DOI] [PubMed] [Google Scholar]
- Khelifi AF, D'Alcontres MS, Salomoni P. Daxx is required for stress-induced cell death and JNK activation. Cell Death Differ. 2005;12:724–733. doi: 10.1038/sj.cdd.4401559. [DOI] [PubMed] [Google Scholar]
- Salomoni P, Khelifi AF. Daxx: death or survival protein. Trends Cell Biol. 2006;16:97–104. doi: 10.1016/j.tcb.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Song MS, Song SJ, Kim SY, Oh HJ, Lim DS. The tumour suppressor RASSF1A promotes MDM2 self-ubiquitination by disrupting the MDM2-DAXX-HAUSP complex. EMBO J. 2008;27:1863–1874. doi: 10.1038/emboj.2008.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Qu LK, Zhang J, Wang W, Michaelson JS, Degenhardt YY, et al. Critical role for Daxx in regulating Mdm2. Nat Cell Biol. 2006;8:855–862. doi: 10.1038/ncb1442. [DOI] [PubMed] [Google Scholar]
- Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, et al. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature. 2000;406:207–210. doi: 10.1038/35018127. [DOI] [PubMed] [Google Scholar]
- Hofmann TG, Will H. Body language: the function of PML nuclear bodies in apoptosis regulation. Cell Death Differ. 2003;10:1290–1299. doi: 10.1038/sj.cdd.4401313. [DOI] [PubMed] [Google Scholar]
- Moller A, Sirma H, Hofmann TG, Rueffer S, Klimczak E, Droge W, et al. PML is required for homeodomain-interacting protein kinase 2 (HIPK2)-mediated p53 phosphorylation and cell cycle arrest but is dispensable for the formation of HIPK domains. Cancer Res. 2003;63:4310–4314. [PubMed] [Google Scholar]
- Sung KS, Lee YA, Kim ET, Lee SR, Ahn JH, Choi CY. Role of the SUMO-interacting motif in HIPK2 targeting to the PML nuclear bodies and regulation of p53. Exp Cell Res. 2011;317:1060–1070. doi: 10.1016/j.yexcr.2010.12.016. [DOI] [PubMed] [Google Scholar]
- Bellodi C, Kindle K, Bernassola F, Cossarizza A, Dinsdale D, Melino G, et al. A cytoplasmic PML mutant inhibits p53 function. Cell Cycle. 2006;5:2688–2692. doi: 10.4161/cc.5.22.3504. [DOI] [PubMed] [Google Scholar]
- Yang S, Kuo C, Bisi JE, Kim MK. PML-dependent apoptosis after DNA damage is regulated by the checkpoint kinase hCds1/Chk2. Nat Cell Biol. 2002;4:865–870. doi: 10.1038/ncb869. [DOI] [PubMed] [Google Scholar]
- Meltser V, Ben-Yehoyada M, Reuven N, Shaul Y. c-Abl downregulates the slow phase of double-strand break repair. Cell Death Disease. 2010;1:e20. doi: 10.1038/cddis.2009.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins NW, Badley AD. Mechanisms of HIV-associated lymphocyte apoptosis: 2010. Cell Death Disease. 2010;1:e99. doi: 10.1038/cddis.2010.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfettini JL, Nardacci R, Seror C, Bourouba M, Subra F, Gros L, et al. The tumor suppressor protein PML controls apoptosis induced by the HIV-1 envelope. Cell Death Differ. 2009;16:298–311. doi: 10.1038/cdd.2008.158. [DOI] [PubMed] [Google Scholar]
- Perfettini JL, Nardacci R, Seror C, Raza SQ, Sepe S, Saidi H, et al. 53BP1 represses mitotic catastrophe in syncytia elicited by the HIV-1 envelope. Cell Death Differ. 2010;17:811–820. doi: 10.1038/cdd.2009.159. [DOI] [PubMed] [Google Scholar]
- Agostini M, Tucci P, Chen H, Knight RA, Bano D, Nicotera P, et al. p73 regulates maintenance of neural stem cell. Biochem Biophys Res Commun. 2010;403:13–17. doi: 10.1016/j.bbrc.2010.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernassola F, Salomoni P, Oberst A, Di Como CJ, Pagano M, Melino G, et al. Ubiquitin-dependent degradation of p73 is inhibited by PML. J Exp Med. 2004;199:1545–1557. doi: 10.1084/jem.20031943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotsch V, Bernassola F, Coutandin D, Candi E, Melino G. p63 and p73, the ancestors of p53. Cold Spring Harb Perspect Biol. 2010;2:a004887. doi: 10.1101/cshperspect.a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Cano L, Herreros-Villanueva M, Fernandez-Alonso R, Ayuso-Sacido A, Meyer G, Garcia-Verdugo JM, et al. p73 deficiency results in impaired self renewal and premature neuronal differentiation of mouse neural progenitors independently of p53. Cell Death Disease. 2010;1:e109. doi: 10.1038/cddis.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killick R, Niklison-Chirou M, Tomasini R, Bano D, Rufini A, Grespi F, et al. p73: a multifunctional protein in neurobiology. Mol Neurobiol. 2011;43:139–146. doi: 10.1007/s12035-011-8172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufini A, Agostini M, Grespi F, Tomasini R, Sayan BS, Niklison-Chirou MV, et al. p73 in cancer. Genes Cancer. 2011;2:491–502. doi: 10.1177/1947601911408890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini R, Mak TW, Melino G. The impact of p53 and p73 on aneuploidy and cancer. Trends Cell Biol. 2008;18:244–252. doi: 10.1016/j.tcb.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Tomlinson V, Gudmundsdottir K, Luong P, Leung KY, Knebel A, Basu S. JNK phosphorylates Yes-associated protein (YAP) to regulate apoptosis. Cell Death Disease. 2010;1:e29. doi: 10.1038/cddis.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strano S, Monti O, Pediconi N, Baccarini A, Fontemaggi G, Lapi E, et al. The transcriptional coactivator Yes-associated protein drives p73 gene-target specificity in response to DNA Damage. Mol Cell. 2005;18:447–459. doi: 10.1016/j.molcel.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Downward J, Basu S. YAP and p73: a complex affair. Mol Cell. 2008;32:749–750. doi: 10.1016/j.molcel.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Lapi E, Di Agostino S, Donzelli S, Gal H, Domany E, Rechavi G, et al. PML, YAP, and p73 are components of a proapoptotic autoregulatory feedback loop. Mol Cell. 2008;32:803–814. doi: 10.1016/j.molcel.2008.11.019. [DOI] [PubMed] [Google Scholar]
- Klanrit P, Taebunpakul P, Flinterman MB, Odell EW, Riaz MA, Melino G, et al. PML involvement in the p73-mediated E1A-induced suppression of EGFR and induction of apoptosis in head and neck cancers. Oncogene. 2009;28:3499–3512. doi: 10.1038/onc.2009.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainardi S, Pelosi A, Palescandolo E, Riccioni R, Fontemaggi G, Diverio D, et al. deltaN-p73 is a transcriptional target of the PML/RARalpha oncogene in myeloid differentiation. Cell Death Differ. 2007;14:1968–1971. doi: 10.1038/sj.cdd.4402210. [DOI] [PubMed] [Google Scholar]
- Barton CE, Johnson KN, Mays DM, Boehnke K, Shyr Y, Boukamp P, et al. Novel p63 target genes involved in paracrine signaling and keratinocyte differentiation. Cell Death Disease. 2010;1:e74. doi: 10.1038/cddis.2010.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Luong P, Hudson C, Gudmundsdottir K, Basu S. c-Abl phosphorylation of DeltaNp63alpha is critical for cell viability. Cell Death Disease. 2010;1:e16. doi: 10.1038/cddis.2009.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melino G. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ. 2011;18:1487–1499. doi: 10.1038/cdd.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalom-Feuerstein R, Lena AM, Zhou H, De La Forest Divonne S, Van Bokhoven H, Candi E, et al. DeltaNp63 is an ectodermal gatekeeper of epidermal morphogenesis. Cell Death Differ. 2011;18:887–896. doi: 10.1038/cdd.2010.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanbokhoven H, Melino G, Candi E, Declercq W. p63, a story of mice and men. J Invest Dermatol. 2011;131:1196–1207. doi: 10.1038/jid.2011.84. [DOI] [PubMed] [Google Scholar]
- Aylon Y, Oren M. New plays in the p53 theater. Curr Opin Genet Dev. 2011;21:86–92. doi: 10.1016/j.gde.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell G, Fillinger J, Sittadjody S, Avila J, Burd R, Limesand K. IGF1 activates cell cycle arrest following irradiation by reducing binding of DeltaNp63 to the p21 promoter. Cell Death Disease. 2010;2010:e50. doi: 10.1038/cddis.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub WE, Weber TA, Schafer B, Candi E, Durst F, Ou HD, et al. The C-terminus of p63 contains multiple regulatory elements with different functions. Cell Death Disease. 2010;1:e5. doi: 10.1038/cddis.2009.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernassola F, Oberst A, Melino G, Pandolfi PP. The promyelocytic leukaemia protein tumour suppressor functions as a transcriptional regulator of p63. Oncogene. 2005;24:6982–6986. doi: 10.1038/sj.onc.1208843. [DOI] [PubMed] [Google Scholar]
- Salomoni P, Bernardi R, Bergmann S, Changou A, Tuttle S, Pandolfi PP. The promyelocytic leukemia protein PML regulates c-Jun function in response to DNA damage. Blood. 2005;105:3686–3690. doi: 10.1182/blood-2004-09-3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam D, Shah S, de Castro IP, Loh SH, Martins LM. Drosophila happyhour modulates JNK-dependent apoptosis. Cell Death Disease. 2010;1:e66. doi: 10.1038/cddis.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt B, Abou-Eladab EF, Tiedge M, Walzel H. Role of the JNK/c-Jun/AP-1 signaling pathway in galectin-1-induced T-cell death. Cell Death Disease. 2010;1:e23. doi: 10.1038/cddis.2010.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanasekaran DN, Reddy EP. JNK signaling in apoptosis. Oncogene. 2008;27:6245–6251. doi: 10.1038/onc.2008.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Bernardi R, Pandolfi PP. A novel signaling network as a critical rheostat for the biology and maintenance of the normal stem cell and the cancer-initiating cell. Curr Opin Genet Dev. 2009;19:51–59. doi: 10.1016/j.gde.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455:813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman LC, Wang X, Alimonti A, Chen Z, Teruya-Feldstein J, Yang H, et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128:141–156. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regad T, Bellodi C, Nicotera P, Salomoni P. The tumor suppressor Pml regulates cell fate in the developing neocortex. Nat Neurosci. 2009;12:132–140. doi: 10.1038/nn.2251. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV. Linking calorie restriction to longevity through sirtuins and autophagy: any role for TOR. Cell Death Disease. 2010;1:e12. doi: 10.1038/cddis.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EY, Longatti A, McKnight NC, Tooze SA. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol. 2009;29:157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Feng J, Broaddus RR. The novel estrogen-induced gene EIG121 regulates autophagy and promotes cell survival under stress. Cell Death Disease. 2010;1:e32. doi: 10.1038/cddis.2010.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg-Lerner A, Bialik S, Simon HU, Kimchi A. Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009;16:966–975. doi: 10.1038/cdd.2009.33. [DOI] [PubMed] [Google Scholar]
- Grander D, Kharaziha P, Laane E, Pokrovskaja K, Panaretakis T. Autophagy as the main means of cytotoxicity by glucocorticoids in hematological malignancies. Autophagy. 2009;5:1198–1200. doi: 10.4161/auto.5.8.10122. [DOI] [PubMed] [Google Scholar]
- Heidari N, Hicks MA, Harada H. GX15-070 (obatoclax) overcomes glucocorticoid resistance in acute lymphoblastic leukemia through induction of apoptosis and autophagy. Cell Death Disease. 2010;1:e76. doi: 10.1038/cddis.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laane E, Tamm KP, Buentke E, Ito K, Kharaziha P, Oscarsson J, et al. Cell death induced by dexamethasone in lymphoid leukemia is mediated through initiation of autophagy. Cell Death Differ. 2009;16:1018–1029. doi: 10.1038/cdd.2009.46. [DOI] [PubMed] [Google Scholar]
- Li Q, Li H, Roughton K, Wang X, Kroemer G, Blomgren K, et al. Lithium reduces apoptosis and autophagy after neonatal hypoxia-ischemia. Cell Death Disease. 2010;1:e56. doi: 10.1038/cddis.2010.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C. Negative regulation of autophagy. Cell Death Differ. 2010;17:1807–1815. doi: 10.1038/cdd.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri MC, Tasdemir E, Criollo A, Morselli E, Vicencio JM, Carnuccio R, et al. Control of autophagy by oncogenes and tumor suppressor genes. Cell Death Differ. 2009;16:87–93. doi: 10.1038/cdd.2008.131. [DOI] [PubMed] [Google Scholar]
- Marino G, Madeo F, Kroemer G. Autophagy for tissue homeostasis and neuroprotection. Curr Opin Cell Biol. 2011;23:198–206. doi: 10.1016/j.ceb.2010.10.001. [DOI] [PubMed] [Google Scholar]
- McCoy F, Hurwitz J, McTavish N, Paul I, Barnes C, O'Hagan B, et al. Obatoclax induces Atg7-dependent autophagy independent of beclin-1 and BAX/BAK. Cell Death Disease. 2010;1:e108. doi: 10.1038/cddis.2010.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, et al. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Disease. 2010;1:e10. doi: 10.1038/cddis.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver N, Proctor GB, Arno M, Carpenter GH. Activation of mTOR coincides with autophagy during ligation-induced atrophy in the rat submandibular gland. Cell Death Disease. 2010;1:e14. doi: 10.1038/cddis.2009.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Sayeed A, Meng Z, Luciani G, Chen LC, Bennington JL, Dairkee SH. Negative regulation of UCP2 by TGFbeta signaling characterizes low and intermediate-grade primary breast cancer. Cell Death Disease. 2010;1:e53. doi: 10.1038/cddis.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HK, Bergmann S, Pandolfi PP. Cytoplasmic PML function in TGF-beta signalling. Nature. 2004;431:205–211. doi: 10.1038/nature02783. [DOI] [PubMed] [Google Scholar]
- Seo SR, Ferrand N, Faresse N, Prunier C, Abecassis L, Pessah M, et al. Nuclear retention of the tumor suppressor cPML by the homeodomain protein TGIF restricts TGF-beta signaling. Mol Cell. 2006;23:547–559. doi: 10.1016/j.molcel.2006.06.018. [DOI] [PubMed] [Google Scholar]
- Faresse N, Colland F, Ferrand N, Prunier C, Bourgeade MF, Atfi A. Identification of PCTA, a TGIF antagonist that promotes PML function in TGF-beta signalling. EMBO J. 2008;27:1804–1815. doi: 10.1038/emboj.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Zhu Q, Luo K. SnoN functions as a tumour suppressor by inducing premature senescence. EMBO J. 2009;28:3500–3513. doi: 10.1038/emboj.2009.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi C, Ito K, Lin HK, Santangelo C, Wieckowski MR, Lebiedzinska M, et al. PML regulates apoptosis at endoplasmic reticulum by modulating calcium release. Science. 2010;330:1247–1251. doi: 10.1126/science.1189157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinton P, Giorgi C, Pandolfi PP. The role of PML in the control of apoptotic cell fate: a new key player at ER-mitochondria sites. Cell Death Differ. 2011;18:1450–1456. doi: 10.1038/cdd.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A, Matute C, Alberdi E. Intracellular Ca2+ release through ryanodine receptors contributes to AMPA receptor-mediated mitochondrial dysfunction and ER stress in oligodendrocytes. Cell Death Disease. 2010;1:e54. doi: 10.1038/cddis.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomiyama A, Tachibana K, Suzuki K, Seino S, Sunayama J, Matsuda KI, et al. MEK-ERK-dependent multiple caspase activation by mitochondrial proapoptotic Bcl-2 family proteins is essential for heavy ion irradiation-induced glioma cell death. Cell Death Disease. 2010;1:e60. doi: 10.1038/cddis.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wabnitz GH, Goursot C, Jahraus B, Kirchgessner H, Hellwig A, Klemke M, et al. Mitochondrial translocation of oxidized cofilin induces caspase-independent necrotic-like programmed cell death of T cells. Cell Death Disease. 2010;1:e58. doi: 10.1038/cddis.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan L, Sebastia J, Tuffy LP, Spring A, Lichawska A, Devocelle M, et al. XIAP impairs Smac release from the mitochondria during apoptosis. Cell Death Disease. 2010;1:e49. doi: 10.1038/cddis.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlberg M, Ekoff M, Labi V, Strasser A, Huang D, Nilsson G. Pro-apoptotic Bax is the major and Bak an auxiliary effector in cytokine deprivation-induced mast cell apoptosis. Cell Death Disease. 2010;1:e43. doi: 10.1038/cddis.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposti MD. Bcl-2 antagonists and cancer: from the clinic, back to the bench. Cell Death Disease. 2010;1:e37. doi: 10.1038/cddis.2010.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Ferguson BJ, Khaled WT, Tevendale M, Stingl J, Poli V, et al. PML depletion disrupts normal mammary gland development and skews the composition of the mammary luminal cell progenitor pool. Proc Natl Acad Sci USA. 2009;106:4725–4730. doi: 10.1073/pnas.0807640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Rich T, Watson CJ. PML: a tumor suppressor that regulates cell fate in mammary gland. Cell Cycle. 2009;8:2711–2717. doi: 10.4161/cc.8.17.9462. [DOI] [PubMed] [Google Scholar]