SUMMARY
MicroRNAs (miRNA) are implicated in brain development and function but the underlying mechanisms have been difficult to study in part due to the cellular heterogeneity in neural circuits. To systematically analyze miRNA expression in neurons, we have established a miRNA tagging and affinity purification (miRAP) method that is targeted to cell types through the Cre-loxP binary system in mice. Our studies of the neocortex and cerebellum reveal the expression of a large fraction of known miRNAs with distinct profiles in glutamatergic and GABAergic neurons, and subtypes of GABAergic neurons. We further detected putative novel miRNAs, tissue or cell type-specific strand selection of miRNAs, and miRNA editing. Our method thus will facilitate a systematic analysis of miRNA expression and regulation in specific neuron types in the context of neuronal development, physiology, plasticity, pathology and disease models, and is generally applicable to other cell types and tissues.
INTRODUCTION
In the mammalian brain, neural circuits often consist of diverse cells types characterized by their stereotyped location, connectivity patterns, and physiological properties. To a large extent, the identity and physiological state of neuron types are determined by their patterns of gene expression (Nelson et al., 2006, Hobert et al., 2010). Therefore, a comprehensive understanding of gene expression profiles in defined cell types not only provides a molecular explanation of cell phenotypes but also is necessary for establishing the link from gene function to neural circuit organization and dynamics. In addition to gene transcription which dictates mRNA production, the stability and translation of mRNAs are regulated by microRNAs (miRNAs), the class of 20~23nt small noncoding RNAs (He and Hannon, 2004, Bartel, 2004). miRNAs can also influence transcription by regulating the translation of transcriptional factors (Hobert, 2008). Recent studies begin to reveal diverse role of miRNAs in the brain, such as in neural patterning (Ronshaugen et al., 2005), neural stem cell differentiation (Kuwabara et al., 2004), cell type specification (Poole and Hobert, 2006), synaptic plasticity (Schratt et al., 2006), and also in neuropsychiatric disorders (Shafi et al., 2010, Xu et al., 2010a). However, the mechanism and logic by which miRNAs regulate neuronal development, function, and plasticity are not well understood. A necessary step is a comprehensive characterization of miRNA expression profiles at the level of distinct neuron types, because individual cell types are the building blocks of neural circuits as well as the basic units of gene regulation.
Analysis of gene expression, including miRNA expression, in the brain has posed a major challenge in genomics despite rapid advances in sequencing technology, because neuronal subtypes are highly heterogeneous and intermixed. Until recently, cell type based expression profiling in the brain has mainly relied on physical enrichment of target cell population such as laser capture microdissection (Meguro et al., 2004, Rossner et al., 2006), fluorescence-activated cell sorting (FACS) (Tomomura et al., 2001, Lobo et al., 2006), or manual sorting (Sugino et al., 2006). These methods are often labor intensive and of low yield. Furthermore, the physical damage and stress inherent to these procedures may alter the physiological state of cells and likely their gene expression. The recent invention of genetic tagging methods, such as TRAP (Heiman et al., 2008b) and Ribo-tag (Sanz et al., 2009), begin to overcome these obstacles, but these strategies have been limited to the analysis of mRNA expression.
Here we present a novel miRNA tagging and Affinity Purification method, miRAP, which can be applied to genetically defined cell types in any complex tissues in mice. This method is based on the fact that mature miRNAs are incorporated into RNA-induced silencing complex (RISC), in which the Argonaute protein AGO2 directly binds miRNAs and their mRNA targets (Hammond et al., 2001). We demonstrate that epitope tagging of Ago2 protein allows direct purification of miRNAs from tissue homogenates using antibodies against the engineered molecular tag. We further established a Cre-loxp binary expression system to deliver epitope-tagged AGO2 (tAGO2) to genetically defined cell types. To demonstrate the feasibility of this approach in the brain, we have analyzed miRNA profiles from five neuron types in mouse cerebral cortex and cerebellum by deep sequencing. Our study reveals the expression of a large fraction of known miRNAs (over 480) which show distinct profiles in glutamatergic and GABAergic neurons, and subtypes of GABAergic neurons. Our method further detects 23 putative novel miRNAs and also provides evidence for tissue-specific strand selection of miRNAs and miRNA editing in subset of neuron types. The miRAP method therefore enables a systematic analysis of miRNA expression and regulation in different neuron types in the brain and is generally applicable to other cell types and tissues in mice.
RESULTS
A genetically targeted microRNA tagging methodology
Our strategy for molecular tagging and affinity purification is based on the knowledge that mature miRNA is incorporated into the RNA-Induced Silencing Complex (RISC) where the miRNA and its mRNA target interact (Hammond et al., 2001). Argonaute (AGO) proteins are at the core of RISC complex and directly bind miRNAs. AGO immunoprecipitation has been successfully used to isolate miRNAs and their mRNA targets (Easow et al., 2007, Beitzinger et al., 2007, Hendrickson et al., 2008, Zhang et al., 2007, Hammell et al., 2008, Karginov et al., 2007). Among the four members in the AGO family in human and mouse, only AGO2 exhibits endoneuclase activity (Meister et al., 2004, Ikeda et al., 2006) and is indispensible for Dicer-independent miRNA biogenesis (Cheloufi et al., 2010). We therefore chose to tag Ago2 by fusing GFP and MYC at its N-terminus (tAGO2) (Figure 1B). GFP allows visualization of tAgo2 expression and MYC allows efficient affinity purification of associated miRNAs.
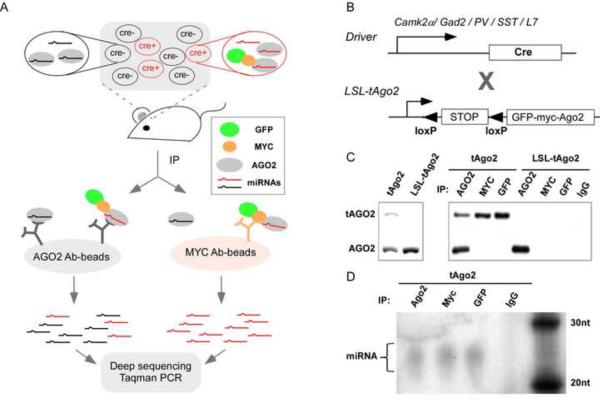
Figure 1.
The miRAP methodology. (A) The scheme of miRNA tagging and Affinity Purification (miRAP). tAGO2 (a GFP-MYC-AGO2 fusion protein) expression is activated by Cre recombinase in certain cell types in mouse brain. Using MYC antibody, miRNAs in Cre+ cells are co-precipitated with tAGO2. Using AGO2 antibody, miRNAs in all cells of the tissue are co-precipitated with AGO2 or tAGO2. RNAs prepared from immunoprecipitation (IP) product is subjected to deep sequencing or miRNA Taqman PCR. (B) Cre-loxP binary system to achieve cell type specific delivery of tAGO2. (C)Western analysis of tAGO2 expression and immunoprecipitation. (D) Autoradiogram of 32P-labelled RNA purified from tAgo2 brain lysate by immunoprecipitation. RNAs ranging between 20–30nt were observed, corresponding to the size of miRNAs. (see also Sppl Figure 1)
In addition to confirming the functionality of tAGO2, it is essential to reliably and systematically deliver tAGO2 to different cell types at consistent levels. We achieved this using the Cre/loxp binary expression system (Figure 1B). We generated a knockin allele at the Gt(ROSA)26Sor (Rosa26) locus which expresses tAGO2 upon Cre/loxp recombination (R26-LSL-tAGO2). Combined with an increasing inventory of cell type-restricted Cre driver lines (Taniguchi et al., 2011) this strategy would allow systematic analysis of miRNA expression in different cell types in mice.
To test the functionality of the LSL-tAgo2 allele, we first crossed the reporter mouse to a CMVCre line to activate tAgo2 expression in the germline. Offspring of CMV-Cre;LSL-tAgo2 were identified in which tAgo2 is ubiquitously expressed in all cells of the animal (the tAgo2 mouse) (Sppl Figure 1). Using antibody against AGO2, a 100kD endogenous band was detected in whole brain homogenates from both the tAgo2 and LSL-tAgo2 mouse, while the higher molecular weight tAGO2 band is only detected in tAgo2 sample (Figure 1C). This result demonstrates the tight Cre-dependent activation of tAgo2 expression from the LSL-STOP cassette.
Using antibody to AGO2, GFP or MYC, tAGO2 can be efficiently immunoprecipitated (IP) from tAgo2 brain lysate (Figure 1C). To examine the identity of co-precipitated RNAs, they were extracted from IP product, radio-labeled and visualized by denaturing urea-PAGE. Enrichment of RNAs corresponding to miRNAs (e.g. 20–23 nucleotides) was observed using AGO2, MYC or GFP antibody-conjugated Dynabeads, but not with IgG control (Figure 1D). In addition, miRNA Taqman PCR detected miRNAs that are known to be brain specific in these RNA extracts (e.g. miR-124) at much higher levels than others that are known to be absent (e.g. miR-122, data not shown).
miRNA profiling in several neuron types of the neocortex and cerebellum
We used the LSL-tAgo2 strategy to profile miRNA expression in glutamatergic, GABAergic, and subclasses of GABAergic neurons in P56 mouse neocortex and cerebellum. Cortical excitatory pyramidal neurons and inhibitory interneurons differ in their embryonic origin, neurotransmitters, and physiological function (Sugino et al., 2006). GABAergic interneurons further consist of multiple subtypes characterized by distinct connectivity, physiological properties, and gene expression patterns (Markram et al., 2004, Ascoli et al., 2008). In particular, the Ca2+-binding protein parvalbumin (PV) and neuropeptide somatostatin (SST) mark two major, non-overlapping subtypes (Gonchar et al., 2007, Xu et al., 2010b, Rudy et al., 2011). Whereas the PV interneurons innervate the soma and proximal dendrites and control the output and synchrony of pyramidal neurons, the SST interneurons innervate the more distal dendrites and control the input and plasticity of pyramidal neurons (Somogyi et al., 1998, Di Cristo et al., 2004). Finally, Purkinje cells are GABAergic projection neurons which carry the sole output of cerebellar cortex.
We used well characterized transgenic or knockin Cre driver lines to activate tAgo2 in these cell types (Figure 1B, Sppl Table 1). The cell type specificity of tAGO2 was validated by dual immunostaining using antibodies against GFP and appropriate cell type markers (Figure 2, Sppl Figure 2, Sppl Table 1). Although not all neurons of a given classes expressed tAgo2, almost all GFP+ cells colocalized with corresponding cell type markers, proving the highly stringent specificity of our system. tAGO2 predominantly localized to the cytoplasm in neuronal somata, but was also detected in neurites, with a particularly prominent example in the dendritic tree of Purkinje cells (Figure 2). miRAP was performed in cortical or cerebellar homogenates. Tissues from multiple mice were pooled as one IP sample when necessary. RNAs were immunopurified using a MYC antibody, extracted for construction of small RNA libraries which were analyzed by deep sequencing (Figure 1A). As a reference for these cell type specific miRNA profiles, we also sequenced miRNAs immunopurified by Ago2 antibody from neocortex and cerebellum and generated tissue-wide miRNA profiles. Consistent with previous reports, we observed that a group of miRNAs known to be brain specific, such as miR-124, miR-29b and miR-9, were highly enriched in all the samples. (Bak et al., 2008, Landgraf et al., 2007).
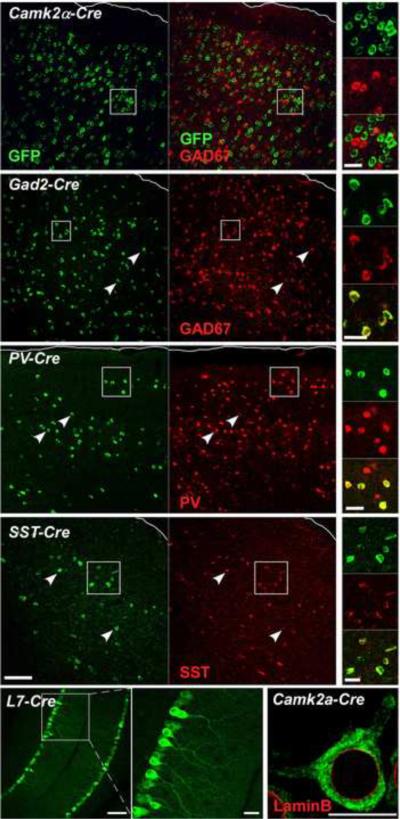
Figure 2.
Cell type specific tAgo2 expression in the neocortex and cerebellum activated by Cre drivers. Cre driver lines used are marked at top of each panel. tAgo2 expression was detected by GFP immunostaining. Pia of cortex is outlined. In the Camk2α -Cre line which labels pyramidal cells, GAD2 antibody was used due to the lack of appropriate Camk2α antibody; the lack of co-localization with GAD2 indicates tAgo2 expression in pyramidal neurons (also see sppl Figure 2 which demonstrates neuronal expression). In cortical GABA neuron drivers (Gad2, Pv, Sst), GFP+ cells colocalized with corresponding cell type markers. In the L7-cre line, Purkinje cells were identified by their characteristic morphology and position. Note the prominent GFP signal in Purkinje dendrites. tAGO2 is predominantly localized in cell somata but not the nucleus, surrounded by LaminB (red) which labels nuclear envelop. Scale bar: 100μm in low magnification images, 20μm in high magnification images, 10μm in LaminB image. (see also Sppl Figure 2)
In total, we generated19 libraries and 291,164,604 raw reads. After removal of low-quality reads and those lacking 3' adaptor, 68.2% of the filtered reads equal or longer than 18nt were perfectly mapped to the mouse genome (mm9). 99.5% of all mapped reads can be aligned to known miRNA hairpins (miRbase version 16). This percentage is higher than those from small RNA libraries constructed from size fractionated total RNA in most previous studies (commonly ranging 50%~80%), indicating that Ago2 immuno-precipitation is a more efficient way to enrich miRNAs while excluding other small RNAs such as degradation product of mRNAs (Sppl Table 1). In our experiments, the correlation coefficient of miRNA profiles from biological replicates within a group (the same cell or tissue type) were extremely high (>0.96, Sppl Table 1), indicating the high reproducibility of the miRAP method.
Hierarchical clustering based on the average linkage of Pearson Correlation (Eisen et al., 1998) of miRNA profiles revealed non-random partition of the samples into two major branches, one containing all five individual neuron types and the other containing the two tissue types (Figure 3). This result demonstrates the necessity and power of cell type based analysis. A common assumption is that brain specific or CNS specific miRNAs are likely to be neuron specific. Our findings suggest this is not always the case. For example, miR-143 was expressed at relatively high levels in both neocortex and cerebellum (ranking at 30 and 51 respectively, from highest to lowest, average normalized per million reads number > 10,000 ), but expressed at relatively low levels in all the examined neuron types (ranking after 160, average normalized per million reads number <500, pairwise fold-change > 20, P<10−84) (Sppl Table 2). It is likely that miR-143 and certain other brain enriched miRNAs may in fact be highly expressed in non-neuronal cells such as astrocytes. On the other hand, miRNAs that are enriched in specific but relatively rare neuron types are likely to be underrepresented and under-appreciated in miRNA profiles generated from the tissue homogenates. For example, miR-34a ranked at 14 in PV profiles (average normalized per million reads number>100,000), but only at 116 in neocortex profiles (average normalized per million reads number<5,000, pairwise logarithm fold-change > 200, P<10−40) (Sppl Table2). Together, these results highlight the critical importance of cell type-based approach in analyzing miRNA expression and function.
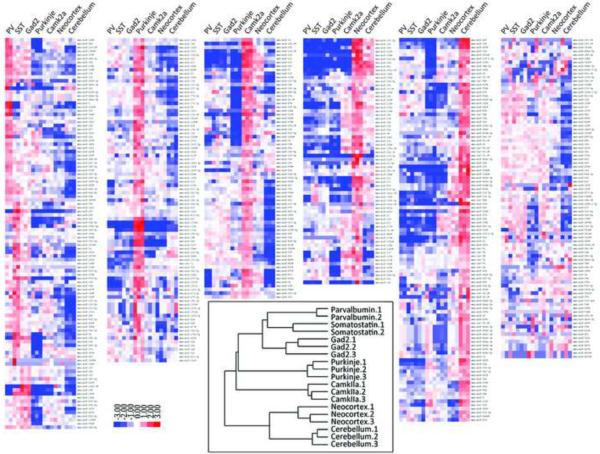
Figure 3.
Relative miRNA expression profiles and hierarchical clustering of samples. Relative expression profiles of mature miRNAs were constructed as follows: the raw reads counts of each miRNA are first normalized to the total number of reads that were mapped to all of the known mature miRNAs and miRNA* species, then log2 transformed, and mean centered. Inset: Hieratical clustering of miRNA expression of all the libraries. (see also Sppl Table 2)
Although more similar to each other than to individual neuron types, difference in miRNA expression is observed between neocortex and cerebellum (Sppl Figure 3A and Sppl Table 3). This is consistent with results from different brain regions using miRNA microarray. For example, miR-128 is expressed ~4 folds higher in neocortex than in cerebellum, while miR-195 and miR-497 are expressed at >10 folds higher in cerebellum than in neocortex. In total, 221 out of 527 detected miRNAs and miRNA* were identified as differentially expressed between these two brain regions in P56 mouse (P value <0.001, Sppl Figure 3A and Sppl Table 3), some of which exhibit differences as high as 40~55 fold (mmu-miR-141, mmu-miR-133b, mmu-miR-219-3p, mmu-miR-485).
Within the five neurons types, cortical Gad2, PV and SST neurons cluster together most closely, as expected from their common origin and transmitter phenotype. The miRNAs enriched in these groups, such as miR-34c and miR-130b, are likely to regulate functions that are common in all neocortical GABAergic interneurons. Others which show differential expression might serve as “finger prints” for subtypes of GABAergic neurons and regulate subtype specific functions. For example, both miR-133b and miR-187 are highly enriched in GABAergic neurons when compared to glutamatergic pyramidal neurons (Figure 4F, Sppl Table 4). However, miR-133b is significantly more enriched in PV cells, while miR-187 is more enriched in SST cells (Figure 4F, Sppl Table 5). Interestingly, neocortical GABAergic neurons cluster more closely with Purkinje cells from cerebellum than with cortical glutamatergic neurons. This suggests that neurotransmitter phenotype, a major aspect of neuronal identity, is a more significant determinant than brain regional location in neuronal miRNA expression.
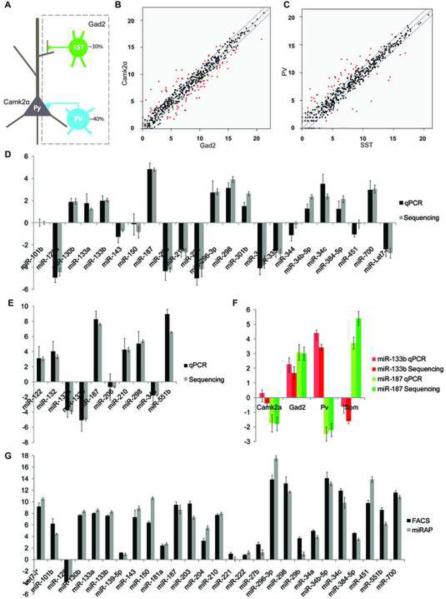
Figure 4.
miRNA expression profiling in neocortical neurons. (A) A simple schematic of neocortical neurons included in this study. The two major neuron types, glutamatergic projection neurons and GABAergic interneurons are targeted by Camk2α-cre and Gad2-cre lines, respectively. PV and SST interneurons are two non-overlapping subtypes of GABAergic neurons targeted by Pv-Cre and Sst-Cre, respectively. (B–C) Normalized expression values of miRNAs are plotted for Camk2α vs. Gad2 samples, and PV vs SST samples. The middle diagonal line represents equal expression, and lines to each side represent 2-fold enrichment in either cell population. The labels of axes are log 2 scaled. miRNAs with fold changes equal or larger than 2 and P values lower than 0.001 are represented in red. (D–E) miRNA Taqman PCR validation of deep sequencing. All readings were normalized to miRNA-124 using the ΔΔCt method. Deep sequencing reads number were log2 transformed and then normalized to miRNA-124. (D) The value of Camk2α sample was compared to Gad2 sample. Values above 0 represent higher expression in Gad2 than Camk2α, and vice versa. n=3 for deep sequencing; n=4 for Taqman PCR. (E) The value of PV sample was compared to SST sample. Values above 0 represent higher expression in SST than PV, and vice versa. n=2 for deep sequencing; n=4 for Taqman PCR. (F) miR-133b and miR-187 expression in four neocortical neuron types. The cell type specific data was normalized to data of neocortex homogenates in each case. Value above 0 indicates higher expression than neocortex sample, and vice versa. (G) Comparison between RNAs purified from FACS sorting and miRAP using Taqman PCR in Camk2α neurons. n=4.Error bar: standard deviation. (see also Sppl Figure 3)
To compare expression levels of each miRNA in different libraries, we constructed relative miRNA expression profiles and arranged the miRNA according to their expression pattern (Figure 3). Many miRNAs have substantially stronger expression in one or more neuron types or tissue types than in others. This selective expression pattern suggests specificity in target regulation, and thus specific miRNA function, in different cell types and anatomical regions.
Validation of deep sequencing results by miRNA Taqman PCR
To more closely examine differential miRNA expression in different cell types (Figure 4A), we performed pair wise comparison between cortical glutamatergic and GABAergic neurons (Figure 4B), and subtypes of GABAergic neurons (Figure 4C). To validate the cell type differences revealed by deep sequencing, we used Taqman PCR to assess subsets of miRNAs from independent sets of samples. As the miRAP method enriches miRNAs but depletes other RNA species by design, we cannot use house-keeping mRNAs as endogenous control for normalization. Instead, difference for each miRNA between two cell types was calculated using the ΔΔCt method, ie. first normalized to the Ct value of miR-124, then compared between each other. In order to directly compare deep sequencing result with Taqman PCR result, the per million reads number of individual miRNAs in deep sequencing profiles are log2 transformed and normalized to the value of miR-124 as well. As what we examined was the relative expression of miRNAs among samples, not their absolute abandunce, in theory we could choose any miRNA with consistent and detectable level of expression as normalization standard. miR-124 is chosen for practical reasons: it was sequenced with high reads number in all samples, and it can be consistently amplified with rather low amount of starting material by Taqman PCR.
We first examined the Camk2α and Gad2 group which represent the two cardinal neuron types in neocortex. 157 out of 523 detected miRNA or miRNA* were identified to have significant differential expression in deep sequencing profiles (P<0.001, Figure 4B and Sppl Table 4). We did Taqman PCR for 21 miRNAs, and found very high concordance between the two profiling techniques. Not only the trend of enrichment or depletion matched, but also the exact fold change value resembled closely (Figure 4D).
Next, we compared the PV and SST groups, which represent two major non-overlapping subtypes of cortical GABAergic interneurons. 125 out of 511 detected miRNA or miRNA* were identified to have significant differential expression in deep sequencing profiles (P<0.001, Figure 4C and Sppl Table 5). A set of 10 miRNAs were examined by Taqman PCR, which also validated the deep sequencing results very well (Figure 4E). Similarly, Taqman PCR validated the deep sequencing results in Purkinje cell vs cerebellum (Sppl Figure 3B and Sppl Table 6).
As an independent validation of the miRAP method, we performed FACS sorting to isolate Camk2α cells in neocortex and extracted RNA for Taqman PCR analysis. In order to label Camk2α neurons specifically, we generated a Rosa26-loxp-STOP-loxp-H2B-GFP reporter line which brightly labels cell nuclei upon Cre activation (Sppl Figure 3C). Breeding with the same Camk2α -Cre line used for miRAP, we were able to sort Camk2α cells at high purity. Most of miRNAs tested show very similar expression level between the two purification methods, proving miRAP as a reliable method to enrich cell type specific miRNAs (Figure 4F).
miRNA clusters and families
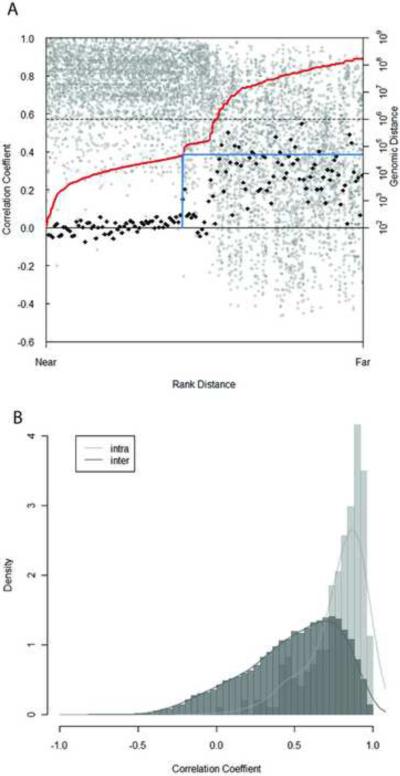
A large number of miRNAs reside in clusters in the genome (Altuvia et al., 2005), with miRNAs in the same cluster sharing the same promoter and polycistronic pri-miRNA transcript. Short distances between miRNA genes on the chromosome imply they may be located in a cluster, but it is not obvious what would be the appropriate distance to define a cluster and different standards have been used (Baskerville and Bartel, 2005, Leung et al., 2008, Altuvia et al., 2005). Because miRNAs in the same cluster are co-transcribed, their expression should be more consistent with each other than those which are located in different clusters (Tanzer and Stadler, 2004). We examined the relationship between genomic distance and pair-wise correlation coefficient of miRNAs on the same strand of the chromosome. The average correlation of paired miRNAs drops sharply at the genomic distance of 50kb (Figure 5), suggesting the average size of miRNA clusters may be approximately 50kb. This result agrees with those from Chiang et al which examined miRNA profiles in different tissue types (Chiang et al., 2010).
Figure 5.
Correlation among miRNA gene expression, genomic organization, and sequence similarity. (A) Correlation of the expression and genomic distance of miRNA genes. Each miRNA gene was paired with every other miRNA gene lying in the same orientation on the same chromosome. For each pair, the distance between the two loci was ranked, and the correlation coefficient for their expression was plotted according this rank (grey circle). The red line indicates the relationship between the rank and actual genomic distance. 50-point moving average of correlation coefficients was calculated. The difference of each moving average and the next was plotted according to the rank (black diamond). The blue line indicates the first point where this value deviates abruptly from the previous ones, i.e. where the average correlation coefficient beyond certain genomic distance changes abruptly. (B) Correlation of expression among miRNA genes within a family (intra-family) and those that do not belong to the same family (inter-family). miRNA genes within the same family tend to have higher correlation coefficient than those that belong to different families. The relative frequency counts (density) on Y axis are arbitrarily defined so that the sum of area in all bins is equal to 1. (see also Sppl Figure 4 and Sppl Table 3–6)
Based on primary sequence and secondary structure conservation, miRNAs can be grouped into different families. miRNAs from the same family evolved from a common ancestor and have high sequence homology. Therefore, family members may share mRNA targets and are involved in the similar aspects of biological function. As cell type is the basic unit of gene regulation in brain tissues, we examined whether miRNAs within a family have similar expression pattern. Information on miRNA families was obtained from miRBase. By examine the distribution of correlation coefficient, we observed higher correlation of expression across the 5 cell types and 2 tissue types for miRNAs within the same family than ones between families. This result indicates cooperation and co-regulation of homologous miRNAs (Figure 5B).
Tissue-specific strand selection of miRNAs
Mature miRNAs can be processed from either the 5' arm or 3' arm of the precursor miRNA hairpin but in most cases are preferentially processed from only one arm. Overall, the discrimination of preferred strand (miRNA) over the other strand (miRNA*) is very high (Hu et al., 2009). Indeed, in our libraries the preferred stand comprises >90% of all the reads mapped to miRNA or miRNA* (Sppl Figure 4). In our data set, the preferred arm largely remained consistent across the cell and tissue types. However, a few miRNAs switched dominant arms in different libraries. For example, miR-544-3p was sequenced more frequently in Purkinje cell and cerebellum samples, while miR-544-5p was sequenced more frequently in the four neocortical cell samples and neocortex tissue sample. Because the two strands are expected to regulate different in mRNA targets, this brain regional difference in strand selection may have functional relevance in cellular phenotypes and physiology. In the case of miR-299 vs miR-299*, there were more reads of miR-299* in all libraries except in the two from SST cells. The case of miR-485 is more complicated: there is higher reads number for miR-485* in Purkinje cells, Camk2α cells and cerebellum, but similar reads number for miR-485 vs miR-485* in other libraries (Table 1).
Table 1.
Summary of tissue/cell type specific strand selection
| Pv | Som | Gad2 | Purkinje | Camklla | Neocortex | Cerebellum | |
|---|---|---|---|---|---|---|---|
| mmu-mir-544 | 0.088 | 1.132 | 0.119 | −2.337 | 0.065 | 1.837 | −0.224 |
| mmu-mir-299 | 0.853 | −0.105 | 0.893 | 1.915 | 1.097 | 1.507 | 3.543 |
| mmu-mir-485 | 0.036 | 0.512 | −0.246 | −5.073 | −0.991 | 0.514 | −2.630 |
Value in the table is the log 2 transformed ratio of 5' and 3' arm reads after adding pseudocount of one. Negative value means more reads from 5' arm than 3' arm of the miRNA precursor and vice versa.
Analysis of miRNA editing
RNA editing is the alteration of RNA sequence post transcription through nucleotide insertion, deletion or modification (Brennicke et al., 1999). The most common type is adenosine (A) to inosine (I) base modification in dsRNA which is catalyzed by adenosine deaminases (ADAR). Pri-miRNAs and Pre-miRNAs are double stranded and can serve as ADAR substrate (Blow et al., 2006, Kawahara et al., 2008, Luciano et al., 2004). Such modification of miRNAs could affect their biogenesis and alter target specificity, thus affecting miRNA function (Yang et al., 2006, Nishikura, 2010).
Since the brain is a primary site of ADAR expression in mammals, we looked for evidence of miRNA editing in our samples. We first searched reads that have single nucleotide mismatches to miRNA and miRNA* but not perfectly matched to the genome. To avoid considering untemplated 3'-terminal addition, we focused on mismatches that occurred >2 nucleotides from the 3' end. We observed substantially higher A-to-G base change above any other types of single nucleotide changes, indicating A-to-I modifications in miRNAs (Sppl Figure 5A). To look for specific sites of A-to-I editing in individual miRNAs, we calculated the rate of A-to-G changes at every genomic position of the sequenced reads. If there are at least 10 raw reads supporting the editing event, and the fraction of A-to-G modification at certain position exceeded 5% in at least two libraries, it was considered as inferred A-to-I editing sites. Under these criteria, we discovered 18 editing sites in all the libraries. None of these sites corresponded to known SNPs. Most of them have been reported before, such as miR-381,miR-376b/c and miR-377, etc (Chiang et al., 2010, Kawahara et al., 2007, Linsen et al., 2010). (Table 2)
Table 2.
Inferred A-to-I editing sites in miRNAs
| miRNA | Editing position | Editing site | Editing fraction | Reference |
|---|---|---|---|---|
| mmu-miR-100 | 1 | AAA | 0.2% | N.A. |
| mmu-miR-1251 | 6 | UAG | 11.0% | (Chiang, Schoenfeld et al. 2010) |
| mmu-miR-3099 | 7 | UAG | 72.5% | (Chiang, Schoenfeld et al. 2010) |
| mmu-miR-337-3p | 11 | UAU | 6.5% | N.A. |
| mmu-miR-34b-5p | 11 | AAU | 4.5% | N.A. |
| mmu-miR-376b | 6 | UAG | 72.9% | (Kawahara, Megraw et al. 2008; Linsen, de Wit et al. 2010) |
| mmu-miR-376c | 6 | UAG | 56.0% | (Kawahara, Megraw et al. 2008; Chiang, Schoenfeld et al. 2010; Linsen, de Wit et al. 2010) |
| mmu-miR-377 | 10 | AAG | 12.6% | (Linsen, de Wit et al. 2010) |
| mmu-miR-378 | 16 | CAG | 10.2% | (Chiang, Schoenfeld et al. 2010) |
| mmu-miR-379 | 5 | UAG | 26.8% | (Chiang, Schoenfeld et al. 2010) |
| mmu-miR-381 | 4 | UAC | 23.0% | (Chiang, Schoenfeld et al. 2010) |
| mmu-miR-411 | 5 | UAG | 43.6% | (Chiang, Schoenfeld et al. 2010) |
| mmu-miR-421 | 14 | UAA | 9.7% | (Chiang, Schoenfeld et al. 2010) |
| mmu-miR-467c | 3 | AAG | 10.2% | N.A. |
| mmu-miR-467e | 4 | AAG | 0.5% | N.A. |
| mmu-miR-497 | 2 | CAG | 0.9% | (Chiang, Schoenfeld et al. 2010) |
| mmu-miR-99a | 1 | AAA | 6.1% | (Kawahara, Megraw et al. 2008) |
The 5' end of mature miRNA is counted as position 1. The edited fraction for each miRNA is calculated by dividing combined edited reads number with total reads number (perfectly matched reads + edited reads) from all the libraries. Red indicates editing in UAG triplets.
As a control, we examined the background error rate of single mismatch in the two synthetic RNA oligos (M19 and M24) that we used during library construction. The total percentage of single mismatch is significantly lower than that from miRNAs, as is the rate of mismatch at each position of the oligos compared to the 5% filter criteria we set. In addition, A-to-G mismatch is not the highest kind of mismatches in the 12 possible single nucleotide mismatches found in the reads of control oligos (Sppl Figure 5B). This result indicates that the A-to-I editing events we observed in miRNA reads are most likely to be biological.
Discovery of novel miRNAs
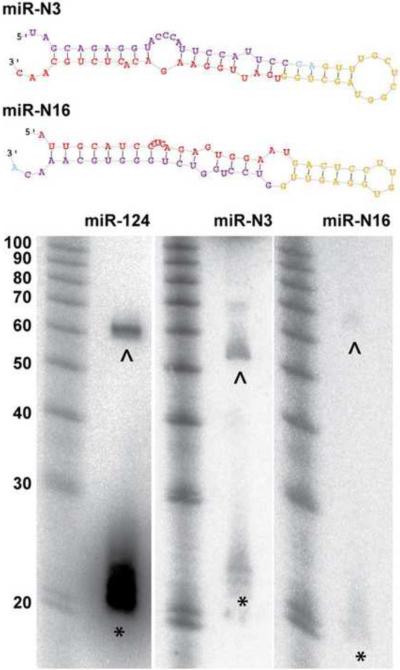
We sought to identify novel miRNAs from our deep sequencing data using a miRNA-discovery algorithm, miRDeep2 (Friedlander et al., 2008). We did prediction for each of the 19 libraries, and combine the results according to genomic position of the prediction. In total, 26 candidates passed the score cutoff of 6, which provided the highest signal-to-noise ratio (7.36) of the prediction, and had normalized read number larger than 1 per million in at least two libraries. These candidates were manually curated to remove highly palindromic precursors or redundant sequences resulting in a refined set of 24 miRNA candidates (Sppl Tabe 7). One of them has recently been deposited into miRbase V17 (mmu-mir-3572) (Spierings et al., 2011). Most of these predicted novel miRNAs have the same seed sequence as known miRNAs in other species, supporting the hypothesis that they are bona fide miRNAs. Among these, 15 predicted novel miRNAs reside in introns of other genes and 4 derive from small nucleolar RNAs(snoRNAs). To validate the prediction, we performed miRNA northern blotting. We were able to detect positive signals corresponding to putative miRNA precursors and/or mature miRNAs for 11 candidates using total RNA extracted from P56 mouse neocortex (Figure 6 and Sppl Figure 6A). Because some candidate miRNAs were only sequenced in libraries made from cell type specific miRAP samples, it is possible they were undetectable by northern blot due to extremely low expression levels. We further performed Taqman PCR for a set of 11 candidate novel miRNAs, 7 of which overlapped with the northern blotting validated set. All of them were well amplified from whole brain total RNA. Whenever the candidates were detected at sufficient levels (data not shown), highly consistent level of expression for each candidate were observed not only among biological replicates of whole brain total RNA (Sppl Figure 6B) but also among those of cell type specific miRAP samples..
Figure 6.
Secondary structures and northern blot validation of two candidate novel miRNAs, miR-N3 and miR-N16. miR-124 was included as positive control. Red region: mature miRNA; yellow region: loop; purple region: miRNA*. * :signals corresponding to putative mature miRNA. ^ : signals corresponding to putative miRNA precursor. (see also Sppl Figure 6 and Sppl Table 7)
DISCUSSION
A necessary step towards understanding the regulatory function of miRNAs in the brain is gaining a comprehensive knowledge on their expression in the relevant cell types of the neural circuits and in the relevant physiological and developmental processes. However, to date, analysis of miRNA expression profiles in the brain has relied on tissue homogenates which completely erase cell type distinction. This approach not only cannot detect and quantify miRNAs in rare (and yet important) cell types but also makes it nearly impossible to interpret results in the context of neural circuits that underlie the function under investigation. Physical enrichment of target cell population can significantly improve cell type resolution but suffer from several limitations. FACs (Arlotta et al., 2005) or manual sorting (Sugino et al., 2006) of fluorescence labeled cells and laser captured microdissection (Rossner et al., 2006) are often laborious and of low yield. The extensive manipulation of tissue causes cell damage and stress which alter gene expression. Here we establish a genetically targeted and affinity-based miRNA profiling method, miRAP, which largely overcomes these obstacles. miRAP is conceptually similar to TRAP and Ribo-Tag, which are designed for mRNA profiling, and presents several major advantages. First, miRAP captures miRNAs during their biogenesis and function within the cell in situ, thus bypassing physical enrichment and can be applied to tissues that are difficult to dissociate. Second, miRAP avoids physical damage and stress of cells. Third, miRAP procedure is simple and sensitive. Indeed, when estimated by the Ct value of miR-124 from two methods for the same cell type, the yield of RNA from miRAP samples is 70–400 times higher than FACS from the same amount of starting material, likely due to loss of fluorescence and cell death during FACS preparation and process. Together, these features of miRAP make it ideal for deep sequencing and Taqman PCR analysis in rare cell types when sample pooling is necessary.
Our implementation of miRAP using the Cre/loxP binary system is similar to Ribo-Tag (Sanz et al., 2009) and has several advantages over bacTRAP (Heiman et al., 2008a). First, we can make use of well characterized Cre drivers which allow reliable and consistent expression of tAGO2 in genetically defines cell types in any tissue of interest, thus avoiding concerns about random insertion of BAC transgenes in the genome and ectopic expression in different transgenic lines.
Second, Cre-dependent tAGO2 expression is well suited to study the effect of cell specific gene manipulations when combined with various floxed alleles. Third, because miRAP captures miRNA in situ under the physiological state of the cell, it allows meaningful assessment of miRNA profiles in the context of neuronal development, function, plasticity, pathology, and in mouse models of brain disorders. Finally, combined with other Cre dependent genetic tagging system, such as the Ribo-Tag, miRAP allows an integrated analysis of different molecular profiles in the same cell type using a defined drive line; this will facilitate a deeper understanding of the multi-level and multi-faceted gene regulatory mechanisms, such as those involving miRNA-mRNA interactions.
The Cre-activated expression of tAGO2 in our miRAP method is unlikely to significantly alter miRNA profile in the cells for the following reasons. In the tAgo2 mouse line in which tAGO2 is expressed in all cells of the animal, the expression level of tAGO2 is significantly lower than the endogenous AGO2, assayed by western blotting from whole brain lysate (Figure 1C), or neocortex and cerebellum lysate (Sppl Figure 1D, E). Interestingly, the combined level of tAGO2 and AGO2 in tAgo2 brain is comparable to, if not less than, the level of AGO2 in LSL-tAgo2 mouse brain (with no tAGO2 expression), suggesting a feedback regulation of Ago2 expression. Similar phenomena was observed in Drosophila S2 cell lines, where the Flag-Ago2 stable cell line expresses less total AGO2 than do naïve S2 cells (Czech et al., 2009). When we perform miRNA Taqman PCR using total RNA extracted from neocortex and cerebellum of P56 LSL-tAgo2 vs. tAgo2 mice, same pattern was observed in a set of 28 miRNAs (Sppl Figure 1G). None of the Cre activated LSL-tAgo2 mouse lines show any notable phenotype in development and behavior. The fact that the expression of cell type-specific markers (e.g. PV, SOM) appears unaltered also suggests that there is no major change of cell identity due to tAgo2 expression. Epitope tagged Ago2 has been widely used to study RISC function and to immunopurify miRNAs (Liu et al., 2004, Karginov et al., 2007), and no change of Ago2 function has been reported due to fusion with an epitope tag. In addition, in the validation experiment for Camk2α -Cre, the expression of miRNAs in two mouse lines harboring different transgenic allele, i.e. LSL-tAgo2 and LSL-H2BGFP, showed the same expression level for the miRNAs examined. All together, these results indicate that the miRAP system is unlikely to affect the native miRNA profiles.
When comparing expression data obtained from miRAP and FACS, we detected discrepancy in expression levels of a few miRNAs (Figure 4E). This is likely due to the following factors. First, physical damage and stress during FACS sorting may alter miRNA profiles, because expression of certain miRNAs are sensitive to neuronal activity or respond to cellular stress. Second, FACS sorted neurons only retain cell body whereas most of their neuronal processes are lost, along with the miRNAs that are localized in dendrites (Tai and Schuman, 2006) and synapses (Schratt, 2009, Lugli et al., 2008). miRAP, on the other hand, should capture miRNAs in neurites since AGO2 has been shown to localize in dendtrites (Cougot et al., 2008, Lugli et al., 2005) and tAGO2 signal can be detected in dendrites (Figure 3). Third, not all mature miRNAs are incorporated into RISC complex. Profiles from miRAP likely represent “active” miRNAs which are associated with Ago2, while miRNA extraction from sorted cells harvests steady state miRNAs regardless of their functional status. Finally, within each major GLU and GABAergic neurons in our study, subtypes likely express tAgo2 at different levels and show different miRNA expression and regulation, including their response to stress and physical damage during FACS. The compounding effect of these factors will affect the miRNAs profiles obtained from these two methods. Another common method to validate RNA expression is in situ hybridization using LNA probes. Unfortunately, our extensive effort did not yield consistent and interpretable results, probably due to the relatively low expression of cell type specific miRNAs.
A potential caveat in a molecular tagging strategy to nucleic acid purification is the redistribution of the affinity tag to the un-tagged pool during homogenization and IP. This is more concerning when the tag is of low affinity and requires chemical cross linking. This issue is inherent to the tag and IP procedure itself and thus is common to all datasets; it would reduce the observed magnitude of differential expression among cell types. This issue is less of a concern for miRAP because AGO2-miRNA interaction is very stable and of high affinity (Tang et al., 2008). In addition, comparison of FACS with miRAP in the Camk2α-Cre line suggests that our method faithfully represent the miRNA profiles in this cell type.
Discovered less than two decades ago, miRNAs have since been implicated in the regulation of almost all aspects of cellular processes. Despite their prominent expression in the mammalian brain, the role of miRNAs in brain development, function, and plasticity remains poorly understood. A major challenge is to link miRNA activity in defined neuron types to specific aspects of neuronal specification, development and physiology; characterizing miRNA profiles in specific cell types is the first step. Using miRAP, we have obtained the first set of miRNA expression profiles in defined neuron types in mouse brain. Our study reveals the expression of a large fraction of known miRNAs with distinct profiles in glutamatergic and GABAergic neurons, and subtypes of GABAergic neurons. We have further detected putative novel miRNAs, tissue or cell type- specific strand selection of miRNAs, and miRNA editing. This generally applicable miRAP method will facilitate a systematic analysis of miRNA expression and regulation in specific neuron types in the context of neuronal specification, development, physiology, plasticity, pathology and disease models. Targeting of rare cell types may further reveal novel, low abundant miRNAs, and link novel regulatory mechanisms such as miRNA editing to specific neuronal and circuitry function.
Identification of mRNA targets in defined cell types is key to understanding the biological function of miRNAs. As miRNA activity requires base-pairing with only 6–8 nucleotides of mRNA, target prediction through bioinformatics has proven to be challenging. Recently, Ago HITS-CLIP has been used to profile miRNA-mRNA targets in the mouse brain (Chi et al., 2009). Genetically targeted miRAP provides a possibility for cell type specific Ago2 HITS-CLIP using the MYC or GFP antibody.
EXPERIMENTAL PROCEDURES
Generation of mouse lines
To generate mouse line that conditionally express GFP –myc-Ago2, a cassette containing LoxP-STOP -LoxP-GFP-myc-Ago2 was cloned in to a Rosa26-CAG targeting vector which contains DTA negative selection marker. GFP-myc-Ago2 is a gift from Dr. Richard M. Schultz in University of Pennsylvania. The STOP cassette contains Neo gene which confers G418 resistance. The targeting vector was linearized by PacI digestion and transfected into C57BL/6 mouse ES cell line. G418-resistant ES clones were screened by PCR first using a forward primer upstream of 5' homologous arm and a reverse primer in the transgene promoter region, then confirmed by southern blot of EcoRV digested DNA, which was probed by a 134bp genomic fragment upstream of the 5' targeting arm. All PCR positive clones were also positive for southern blot.
Positive ES cell clones were injected into albino C57BL/6 blastocysts to obtain chimeric mice following standard procedures. Chimeric mice were bred with albino C57BL/6 mice to obtain germline transmission. To generate mouse line that conditionally express H2B-GFP, ZsGreen gene in Ai6 vector (Madisen et al., 2010) targeting vector was replaced with H2B-GFP gene. Ai6 vector is a gift from Dr. Hongkui Zeng from Allen Institute for Brain Science. C57BL/6;129 hybrid ES cell line was transfected and screened using same strategy as Ai6 mice. Positive ES cell clones were used for tetroploid complementation to obtain male heterozygous mice following standard procedures. Heterozygous mice were bred with each other to obtain homozygotes. Homozygotes were bred with Cre driver lines for experiment.
CMV-cre (Stock NO 003465), Camk2α –Cre (Stock NO 005359) and two lines of L7-Cre mice(Stock NO 006207 and 004146; the first one was used for miRAP, the second one was used for immunostaining to show tAGO2 localization in Purkinje cells) were purchased from Jax laboratory. Pv-ires-Cre mice were gift from Dr Silvia Arber. Gad2-ires-Cre and Som-ires-Cre were generated in the Huang lab as described previously (Taniguchi et al., 2011). ES cell transfections, blastocyst injections and tetroploid complementation were performed by the gene targeting shared resource center in Cold Spring Harbor Laboratory.
Immunohistochemistry and confocal microscopy
Postnatal animals were anaesthetized (avertin) and perfused with 4% paraformaldehyde (PFA) in 0.1 M PB. The brains were removed and post-fixed overnight at 4°C. Brain sections (50μm) were cut with a vibratome. Sections were blocked with 10% normal goat serum (NGS) and 0.1% Triton in PBS and then incubated with the following primary antibodies in the blocking solution at 4°C overnight: GFP (rabbit polyclonal antibody; 1:800; Rockland), parvalbumin (Pv, mouse monoclonal antibody; 1:1000; Sigma), somatostatin (SOM; rat monoclonal antibody; 1:300; Millipore); lamin B(goat polyclonal antibody;1:100; Santa Cruz Biotechnology). Sections were then incubated with appropriate Alexa fluor dye-conjugated IgG secondary antibodies (1: 400; Molecular Probes) in blocking solution and mounted in Fluoromount-G (SouthernBiotech). For immunostaining against Gad67 (mouse monoclonal antibody; 1:800; Millipore), no detergent was added in any step, and incubation was done at room temperature for 2 days at the primary antibody step. In some experiments sections were incubated with TOTO-3 (1:3000; Molecular Probes) together with secondary antibodies to visualize nuclei. Sections were imaged with confocal microscopy (Zeiss LSM510 and Zeiss LSM710).
miRAP
Neocortex and cerebellum of P56 mouse brain were dissected on ice and flash frozen in liquid nitrogen, ground to a fine powder, and resuspend in 10 volume of ice-cold lysis buffer (10mM HEPES pH7.4, 100mM KCl, 5mM MaCl2, 0.5%NP-40, 1mM DTT, 100U/ml RNasin) containing Roche Complete proteinase inhibitors, EDTA-free. Tissue suspension was homogenized using glass douncer. Homogenates were centrifuged for 30min at 13,000g, 4°C to pellet cell debris and unsolubilized material. Mouse-anti-c-Myc(sc-40, Santa Cruz Biotechnology), mouse-anti-Ago2(2E12-1C9, Abnova) or mouse IgG1(Molipore) conjugated protein G Dynabeads(Invitrogen) were added into supernatant, and the mixture was incubated in 4°C with end-over-end rotation for 4 hours. Beads were washed twice with low salt NT2 buffer(50mM Tris.Hcl pH 7.5, 150mM NaCl, 1mM MgCl2, 0.5%NP-40, 1mM DTT, 100U/ml RNasin) and twice with high salt NT2 buffer(50mM Tris.Hcl pH 7.5, 600mM NaCl, 1mM MgCl2, 0.5%NP-40, 1mM DTT, 100U/ml RNasin) and treated with 0.6mg/ml proteinase K for 20min at 55 °C. RNA was extracted by acid phenochloroform(Ambion), followed by chloroform, and precipitated with sodium acetate and glycoblue(Ambion) in ethanol overnight −80°C. RNA pellet was washed once in 75% ethanol and resuspend in water for further application.
FACS sorting and RNA extraction
Neocortex and cerebellum were dissected and cut into small pieces on ice before dissociation. Tissue dissociation was performed using Papain Dissociation System (Worthington Biochemical Corporation, Lakewood, NJ) according to manufacturer's instruction. Cells were washed once with FACS buffer (1%BSA in PBS, 50U/ml RNasin, 12.5U/ml DNase), resuspended in 2ml FACS buffer with 1μg/ml RNase-free propidium iodide for dead-cell discrimination. GFP positive PI negative single cells were FACS-sorted directly into Trizol-LS(Invitrogen) for RNA extraction according to manufacturer's instruction.
miRNA Taqman PCR
Real-time RT-PCR analyses of RNA purified by miRAP or from FACS sorted cells were carried out using Taqman MicroRNA Assays(Applied Biosystem) on 7900HT real time PCR machine (Applied Biosystem) according to the manufacturer's instruction. All reactions were run in triplicate. Data were normalized to miRNA-124. When deep sequencing data was compared with RT-q-PCR data, the per million reads number for each miRNA was log2 transformed and normalized to miRNA-124.
Small RNA library generation and sequencing
Libraries for deep sequencing were prepared from RNAs extracted from immonuprecipitation products following standard protocol. Briefly, RNA was successively ligated to 3' and 5' adaptors, gel purified after each ligation, reverse transcribed and PCR amplified using Solexa sequencing primers. PCR product was gel purified, quantified, and sequenced for 36 cycles on Illumina Genome Analyzer II. Radiolabeled synthetic RNA oligos (M19: CGUACGGUUUAAACUUCGA and M24: CGUACGGUUUAAACUUCGAAAUGU) were spiked in to trace RNA on UREA-PAGE during library preparation, and were depleted by PmelI digestion after PCR amplification. Significant amount of oligos were retained in the libraries, and were used as spike-in oligo control for RNA editing analysis.
Sequence processing, mapping and annotation
Raw Illumina sequencing reads were trimmed from 3' linker, filtered for low-quality reads, and collapsed to unique sequences retaining their individual read count information. Unique sequences 18nt or longer in length were mapped to the University of California at Santa Cruz mm9 assembly of the mouse genome using bowtie allowing no mismatch. miRNA annotations were made according to miRbase version 16. Raw data and annotated sequences of the small RNA libraries are uploaded in the GEO database (accession number GSE30286).
Data normalization and comparison
To quantify and compare miRNA expression across data sets, we used edgeR package developed by Robinson and Smyth (Robinson et al., 2010). Briefly, we used “calcNormFactors” funtion which calculated the sample whose 75%-ile (of library-scale-scaled counts) is closest to the mean of 75%-iles as the reference to get the effective library size for normalization (TMM(trimmed mean of M values)-normalization). To detect pair-wise differential expression of miRNAs in different cell/tissue types, we used “exact test” which is based on negative binomial models and the qCML method(Robinson and Smyth, 2008, Robinson and Smyth, 2007). The results of the “exact test” was accessed by the function “topTags” to get the p-value, fold change and the false discovery rate (FDR) for error control (Benjamini.Y. and Hochberg, 1995). The same data sets were randomly shuffled 10,000 times, and then processed under the same procedure. According to this result, P-value for the actual data set was set to 0.001 as the cutoff to report differentia expression of miRNAs.(Robinson and Oshlack, 2010, Robinson et al., 2010). To generate the heatmap of miRNA expression across data set, we used the mean centered expression of each miRNA and miRNA*. For hierarchical clustering, the average linkage of Pearson Correlation was employed (Eisen et al., 1998).
Identification of arm switching miRNAs
To classify reads from 5' and 3' arms, we grouped reads from each library according to alignment with miRNA precursors. For each miRNA, we summarized the reads in libraries prepared from the same cell type or tissue type. The fold enrichment was calculated as the log2 ratio of 5' and 3' arm reads after adding pseudocounts of one. Only miRNAs with unique precursor and five or more reads on either arm in at least 10 libraries were considered and reported.
RNA editing analysis
Each sequencing library was filtered for sequences that uniquely aligned to the genome with one mismatch >2nt from the 3' end of miRNA or miRNA*. The 12 possible mismatch types were then quantified at each position covered by the filtered reads. The individual editing fraction in each library was calculated as the number of reads containing a certain mismatch at a particular position divided by the number of filtered reads covering that position. To screen inferred A-to-I editing sites, A-to-G mismatches were filtered for editing fraction>5% at a particular position and reads number>10 for each library respectively, and then combined together to calculate the editing fraction in all libraries. None of the inferred A-to-I editing sites was found to correspond to known SNPs by checking in the Perlegen SNP database and dbSNP.
Novel miRNA prediction
Novel miRNAs were predicted by miReep2 which uses a probabilistic model of miRNA biogenesis to score compatibility of the position and frequency of sequenced RNA with the secondary structure of the miRNA precursor (Friedlander et al., 2008). Prediction was performed according to the manual of miRDeep2. Each library was processed separately, and the results were combined together according to genomic location. The signal-to-noise ration of the prediction was calculated according to the manual of miRDeep2.
miRNA northern blotting
miRNA northern blotting was performed following standard protocol (Pall and Hamilton, 2008). Briefly, total RNA was extracted from p56 mouse neocortex using Trizol-LS Reagent (Invitrogen) according to the manufacturer's instructions. 30–50 micrograms of total RNA were resolved on 15% denaturing polyacrylamide gels and transferred onto Hybond NX membrane (Amerhsam) with a Trans-Blot SD semi-dry transfer cell (Bio-Rad. RNA was crosslinked to the membrane using EDC method at 60°C for 1hour, pre-hybridized for at least 2h in Ultrahyb-Oligo (Ambion) at 37°C and hybridized overnight with 32P-labeled DNA probe. Membrane was washed 3–4 times in 0.1× SSC, 0.1% SDS 37°C and exposed to phosphor screen for 1 hour to 3 days.
Supplementary Material
HIGHLIGHTS
established a novel miRNA tagging and affinity purification method (miRAP)
miRAP enables cell type specific miRNA profiling from complex tissues in mice.
revealed distinct miRNA profiles in neuron types of cortex and cerebellum.
detected putative novel miRNAs and miRNA editing by deep sequencing.
ACKNOWLEDGEMENTS
We are grateful to Ingrid Ibarra, Astrid Desiree Haase, and Assaf Gordon for help with small RNA library preparation and deep sequencing processing, Benjamin Czech and Bing Zhang for help with miRNA northern blotting, Keerthi Krishnan for help with FACS sorting protocol optimization, Sang Yong Kim for help with generation of knock in mice. This work was supported in part by NIH MH088661 to M.Z, RC1 MH088661 to ZJH, Roberston Neuroscience Fund of CSHL to ZJH, and National Natural Science Foundation of China (60905013, 91019016, 31061160497) to XW, M.Z and YL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- ALTUVIA Y, LANDGRAF P, LITHWICK G, ELEFANT N, PFEFFER S, ARAVIN A, BROWNSTEIN MJ, TUSCHL T, MARGALIT H. Clustering and conservation patterns of human microRNAs. Nucleic acids research. 2005;33:2697–706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ARLOTTA P, MOLYNEAUX BJ, CHEN J, INOUE J, KOMINAMI R, MACKLIS JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–21. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- ASCOLI GA, ALONSO-NANCLARES L, ANDERSON SA, BARRIONUEVO G, BENAVIDES-PICCIONE R, BURKHALTER A, BUZSAKI G, CAULI B, DEFELIPE J, FAIREN A, FELDMEYER D, FISHELL G, FREGNAC Y, FREUND TF, GARDNER D, GARDNER EP, GOLDBERG JH, HELMSTAEDTER M, HESTRIN S, KARUBE F, KISVARDAY ZF, LAMBOLEZ B, LEWIS DA, MARIN O, MARKRAM H, MUNOZ A, PACKER A, PETERSEN CC, ROCKLAND KS, ROSSIER J, RUDY B, SOMOGYI P, STAIGER JF, TAMAS G, THOMSON AM, TOLEDO-RODRIGUEZ M, WANG Y, WEST DC, YUSTE R. Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat Rev Neurosci. 2008;9:557–68. doi: 10.1038/nrn2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAK M, SILAHTAROGLU A, MOLLER M, CHRISTENSEN M, RATH MF, SKRYABIN B, TOMMERUP N, KAUPPINEN S. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14:432–44. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARTEL DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- BASKERVILLE S, BARTEL DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. Rna. 2005;11:241–7. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BEITZINGER M, PETERS L, ZHU JY, KREMMER E, MEISTER G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007;4:76–84. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- BENJAMINI Y, HOCHBERG . Controlling the false discovery rate: a practical and powerful approach to multiple testing. ROYAUME-UNI, Royal Statistical Society; London: 1995. [Google Scholar]
- BLOW MJ, GROCOCK RJ, VAN DONGEN S, ENRIGHT AJ, DICKS E, FUTREAL PA, WOOSTER R, STRATTON MR. RNA editing of human microRNAs. Genome biology. 2006;7:R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRENNICKE A, MARCHFELDER A, BINDER S. RNA editing. FEMS microbiology reviews. 1999;23:297–316. doi: 10.1111/j.1574-6976.1999.tb00401.x. [DOI] [PubMed] [Google Scholar]
- CHELOUFI S, DOS SANTOS CO, CHONG MM, HANNON GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–9. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHI SW, ZANG JB, MELE A, DARNELL RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009 doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHIANG HR, SCHOENFELD LW, RUBY JG, AUYEUNG VC, SPIES N, BAEK D, JOHNSTON WK, RUSS C, LUO S, BABIARZ JE, BLELLOCH R, SCHROTH GP, NUSBAUM C, BARTEL DP. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes & development. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COUGOT N, BHATTACHARYYA SN, TAPIA-ARANCIBIA L, BORDONNE R, FILIPOWICZ W, BERTRAND E, RAGE F. Dendrites of mammalian neurons contain specialized P-body-like structures that respond to neuronal activation. J Neurosci. 2008;28:13793–804. doi: 10.1523/JNEUROSCI.4155-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CZECH B, ZHOU R, ERLICH Y, BRENNECKE J, BINARI R, VILLALTA C, GORDON A, PERRIMON N, HANNON GJ. Hierarchical rules for Argonaute loading in Drosophila. Mol Cell. 2009;36:445–56. doi: 10.1016/j.molcel.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DI CRISTO G, WU C, CHATTOPADHYAYA B, ANGO F, KNOTT G, WELKER E, SVOBODA K, HUANG ZJ. Subcellular domain-restricted GABAergic innervation in primary visual cortex in the absence of sensory and thalamic inputs. Nat Neurosci. 2004;7:1184–6. doi: 10.1038/nn1334. [DOI] [PubMed] [Google Scholar]
- EASOW G, TELEMAN AA, COHEN SM. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007;13:1198–204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EISEN MB, SPELLMAN PT, BROWN PO, BOTSTEIN D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRIEDLANDER MR, CHEN W, ADAMIDI C, MAASKOLA J, EINSPANIER R, KNESPEL S, RAJEWSKY N. Discovering microRNAs from deep sequencing data using miRDeep. Nature biotechnology. 2008;26:407–15. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- GONCHAR Y, WANG Q, BURKHALTER A. Multiple distinct subtypes of GABAergic neurons in mouse visual cortex identified by triple immunostaining. Front Neuroanat. 2007;1:3. doi: 10.3389/neuro.05.003.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAMMELL M, LONG D, ZHANG L, LEE A, CARMACK CS, HAN M, DING Y, AMBROS V. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat Methods. 2008;5:813–9. doi: 10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAMMOND SM, BOETTCHER S, CAUDY AA, KOBAYASHI R, HANNON GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–50. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- HE L, HANNON GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- HEIMAN M, SCHAEFER A, GONG S, PETERSON JD, DAY M, RAMSEY KE, SUÁREZ-FARIÑAS M, SCHWARZ C, STEPHAN DA, SURMEIER DJ. A Translational Profiling Approach for the Molecular Characterization of CNS Cell Types. Cell. 2008a;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEIMAN M, SCHAEFER A, GONG S, PETERSON JD, DAY M, RAMSEY KE, SUAREZ-FARINAS M, SCHWARZ C, STEPHAN DA, SURMEIER DJ, GREENGARD P, HEINTZ N. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008b;135:738–48. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HENDRICKSON DG, HOGAN DJ, HERSCHLAG D, FERRELL JE, BROWN PO. Systematic identification of mRNAs recruited to argonaute 2 by specific microRNAs and corresponding changes in transcript abundance. PLoS One. 2008;3:e2126. doi: 10.1371/journal.pone.0002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOBERT O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–6. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- HOBERT O, CARRERA I, STEFANAKIS N. The molecular and gene regulatory signature of a neuron. Trends in neurosciences. 2010;33:435–45. doi: 10.1016/j.tins.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HU HY, YAN Z, XU Y, HU H, MENZEL C, ZHOU YH, CHEN W, KHAITOVICH P. Sequence features associated with microRNA strand selection in humans and flies. BMC Genomics. 2009;10:413. doi: 10.1186/1471-2164-10-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IKEDA K, SATOH M, PAULEY KM, FRITZLER MJ, REEVES WH, CHAN EK. Detection of the argonaute protein Ago2 and microRNAs in the RNA induced silencing complex (RISC) using a monoclonal antibody. J Immunol Methods. 2006;317:38–44. doi: 10.1016/j.jim.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KARGINOV FV, CONACO C, XUAN Z, SCHMIDT BH, PARKER JS, MANDEL G, HANNON GJ. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci U S A. 2007;104:19291–6. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAWAHARA Y, MEGRAW M, KREIDER E, IIZASA H, VALENTE L, HATZIGEORGIOU AG, NISHIKURA K. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008;36:5270–80. doi: 10.1093/nar/gkn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAWAHARA Y, ZINSHTEYN B, SETHUPATHY P, IIZASA H, HATZIGEORGIOU AG, NISHIKURA K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–40. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUWABARA T, HSIEH J, NAKASHIMA K, TAIRA K, GAGE FH. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell. 2004;116:779–93. doi: 10.1016/s0092-8674(04)00248-x. [DOI] [PubMed] [Google Scholar]
- LANDGRAF P, RUSU M, SHERIDAN R, SEWER A, IOVINO N, ARAVIN A, PFEFFER S, RICE A, KAMPHORST AO, LANDTHALER M, LIN C, SOCCI ND, HERMIDA L, FULCI V, CHIARETTI S, FOA R, SCHLIWKA J, FUCHS U, NOVOSEL A, MULLER RU, SCHERMER B, BISSELS U, INMAN J, PHAN Q, CHIEN M, WEIR DB, CHOKSI R, DE VITA G, FREZZETTI D, TROMPETER HI, HORNUNG V, TENG G, HARTMANN G, PALKOVITS M, DI LAURO R, WERNET P, MACINO G, ROGLER CE, NAGLE JW, JU J, PAPAVASILIOU FN, BENZING T, LICHTER P, TAM W, BROWNSTEIN MJ, BOSIO A, BORKHARDT A, RUSSO JJ, SANDER C, ZAVOLAN M, TUSCHL T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEUNG WS, LIN MC, CHEUNG DW, YIU SM. Filtering of false positive microRNA candidates by a clustering-based approach. BMC Bioinformatics. 2008;9(Suppl 12):S3. doi: 10.1186/1471-2105-9-S12-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LINSEN SE, DE WIT E, DE BRUIJN E, CUPPEN E. Small RNA expression and strain specificity in the rat. BMC Genomics. 2010;11:249. doi: 10.1186/1471-2164-11-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU J, CARMELL MA, RIVAS FV, MARSDEN CG, THOMSON JM, SONG JJ, HAMMOND SM, JOSHUA-TOR L, HANNON GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–41. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- LOBO MK, KARSTEN SL, GRAY M, GESCHWIND DH, YANG XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–52. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- LUCIANO DJ, MIRSKY H, VENDETTI NJ, MAAS S. RNA editing of a miRNA precursor. RNA. 2004;10:1174–7. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUGLI G, LARSON J, MARTONE ME, JONES Y, SMALHEISER NR. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J Neurochem. 2005;94:896–905. doi: 10.1111/j.1471-4159.2005.03224.x. [DOI] [PubMed] [Google Scholar]
- LUGLI G, TORVIK VI, LARSON J, SMALHEISER NR. Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J Neurochem. 2008;106:650–61. doi: 10.1111/j.1471-4159.2008.05413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MADISEN L, ZWINGMAN TA, SUNKIN SM, OH SW, ZARIWALA HA, GU H, NG LL, PALMITER RD, HAWRYLYCZ MJ, JONES AR, LEIN ES, ZENG H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–40. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARKRAM H, TOLEDO-RODRIGUEZ M, WANG Y, GUPTA A, SILBERBERG G, WU C. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- MEGURO R, LU J, GAVRILOVICI C, POULTER MO. Static, transient and permanent organization of GABA receptor expression in calbindin-positive interneurons in response to amygdala kindled seizures. J Neurochem. 2004;91:144–54. doi: 10.1111/j.1471-4159.2004.02701.x. [DOI] [PubMed] [Google Scholar]
- MEISTER G, LANDTHALER M, PATKANIOWSKA A, DORSETT Y, TENG G, TUSCHL T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–97. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- NELSON SB, HEMPEL C, SUGINO K. Probing the transcriptome of neuronal cell types. Curr Opin Neurobiol. 2006;16:571–6. doi: 10.1016/j.conb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- NISHIKURA K. Functions and regulation of RNA editing by ADAR deaminases. Annual review of biochemistry. 2010;79:321–49. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PALL GS, HAMILTON AJ. Improved northern blot method for enhanced detection of small RNA. Nat Protoc. 2008;3:1077–84. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- POOLE RJ, HOBERT O. Early embryonic programming of neuronal left/right asymmetry in C. elegans. Current biology : CB. 2006;16:2279–92. doi: 10.1016/j.cub.2006.09.041. [DOI] [PubMed] [Google Scholar]
- ROBINSON MD, MCCARTHY DJ, SMYTH GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBINSON MD, OSHLACK A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBINSON MD, SMYTH GK. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics. 2007;23:2881–7. doi: 10.1093/bioinformatics/btm453. [DOI] [PubMed] [Google Scholar]
- ROBINSON MD, SMYTH GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008;9:321–32. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]
- RONSHAUGEN M, BIEMAR F, PIEL J, LEVINE M, LAI EC. The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes Dev. 2005;19:2947–52. doi: 10.1101/gad.1372505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROSSNER MJ, HIRRLINGER J, WICHERT SP, BOEHM C, NEWRZELLA D, HIEMISCH H, EISENHARDT G, STUENKEL C, VON AHSEN O, NAVE KA. Global transcriptome analysis of genetically identified neurons in the adult cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2006;26:9956–66. doi: 10.1523/JNEUROSCI.0468-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RUDY B, FISHELL G, LEE S, HJERLING-LEFFLER J. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Developmental neurobiology. 2011;71:45–61. doi: 10.1002/dneu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANZ E, YANG L, SU T, MORRIS DR, MCKNIGHT GS, AMIEUX PS. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proceedings of the National Academy of Sciences. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHRATT G. microRNAs at the synapse. Nature reviews. Neuroscience. 2009;10:842–9. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- SCHRATT GM, TUEBING F, NIGH EA, KANE CG, SABATINI ME, KIEBLER M, GREENBERG ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–9. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- SHAFI G, ALIYA N, MUNSHI A. MicroRNA signatures in neurological disorders. Can J Neurol Sci. 2010;37:177–85. doi: 10.1017/s0317167100009902. [DOI] [PubMed] [Google Scholar]
- SOMOGYI P, TAMAS G, LUJAN R, BUHL EH. Salient features of synaptic organisation in the cerebral cortex. Brain Res Brain Res Rev. 1998;26:113–35. doi: 10.1016/s0165-0173(97)00061-1. [DOI] [PubMed] [Google Scholar]
- SPIERINGS DC, MCGOLDRICK D, HAMILTON-EASTON AM, NEALE G, MURCHISON EP, HANNON GJ, GREEN DR, WITHOFF S. Ordered progression of stage specific miRNA profiles in the mouse B2 B cell lineage. Blood. 2011 doi: 10.1182/blood-2010-10-316034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUGINO K, HEMPEL CM, MILLER MN, HATTOX AM, SHAPIRO P, WU C, HUANG ZJ, NELSON SB. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- TAI HC, SCHUMAN EM. MicroRNA: microRNAs reach out into dendrites. Curr Biol. 2006;16:R121–3. doi: 10.1016/j.cub.2006.02.006. [DOI] [PubMed] [Google Scholar]
- TANG F, HAJKOVA P, O'CARROLL D, LEE C, TARAKHOVSKY A, LAO K, SURANI MA. MicroRNAs are tightly associated with RNA-induced gene silencing complexes in vivo. Biochemical and biophysical research communications. 2008;372:24–9. doi: 10.1016/j.bbrc.2008.04.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TANIGUCHI H, HE M, WU P, KIM S, PAIK R, SUGINO K, KVITSANI D, FU Y, LU J, LIN Y, FISHELL G, NELSON S, HUANG ZJ. A Resource of Cre Driver Lines for Genetic Targeting of GABAergic Neurons in Cerebral Cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TANZER A, STADLER PF. Molecular evolution of a microRNA cluster. J Mol Biol. 2004;339:327–35. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- TOMOMURA M, RICE DS, MORGAN JI, YUZAKI M. Purification of Purkinje cells by fluorescence-activated cell sorting from transgenic mice that express green fluorescent protein. Eur J Neurosci. 2001;14:57–63. doi: 10.1046/j.0953-816x.2001.01624.x. [DOI] [PubMed] [Google Scholar]
- XU B, KARAYIORGOU M, GOGOS JA. MicroRNAs in psychiatric and neurodevelopmental disorders. Brain Res. 2010a;1338:78–88. doi: 10.1016/j.brainres.2010.03.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XU XM, ROBY KD, CALLAWAY EM. Immunochemical Characterization of Inhibitory Mouse Cortical Neurons: Three Chemically Distinct Classes of Inhibitory Cells. Journal of Comparative Neurology. 2010b;518:389–404. doi: 10.1002/cne.22229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG W, CHENDRIMADA TP, WANG Q, HIGUCHI M, SEEBURG PH, SHIEKHATTAR R, NISHIKURA K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG L, DING L, CHEUNG TH, DONG MQ, CHEN J, SEWELL AK, LIU X, YATES JR, 3RD, HAN M. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol Cell. 2007;28:598–613. doi: 10.1016/j.molcel.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.