Abstract
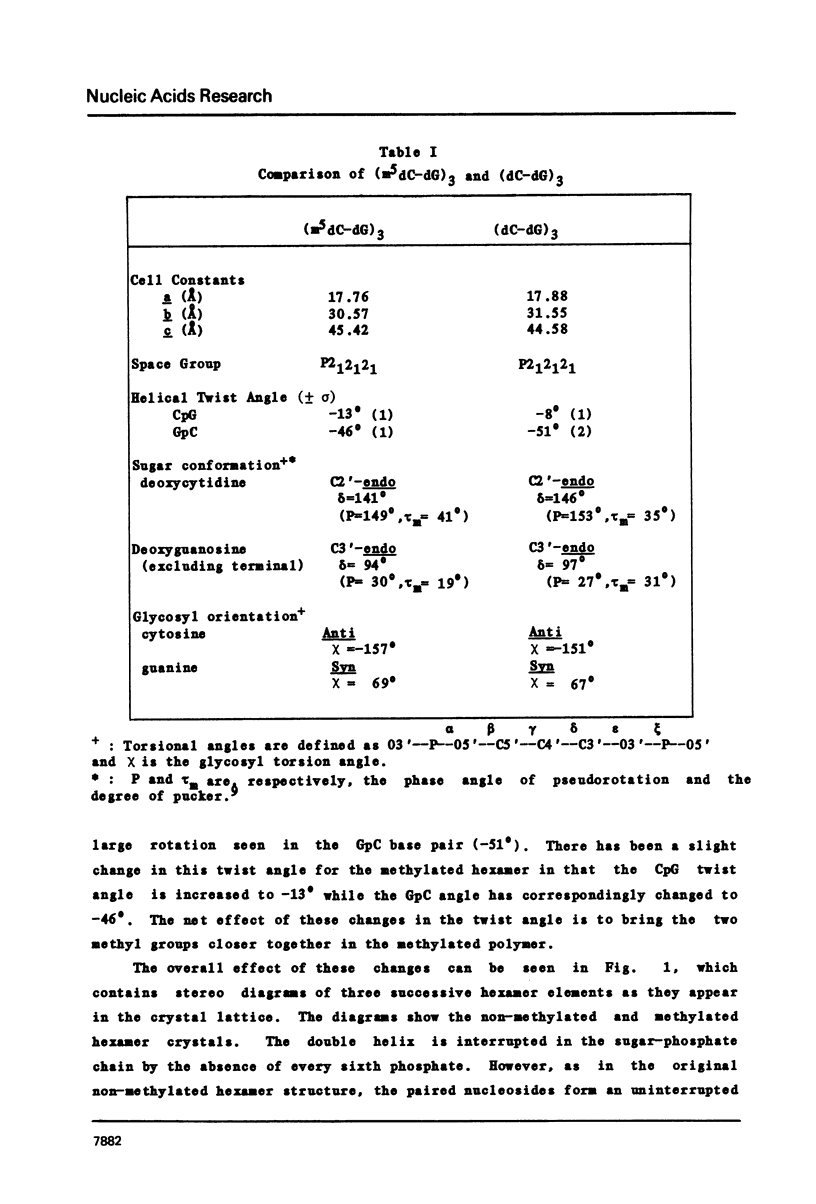
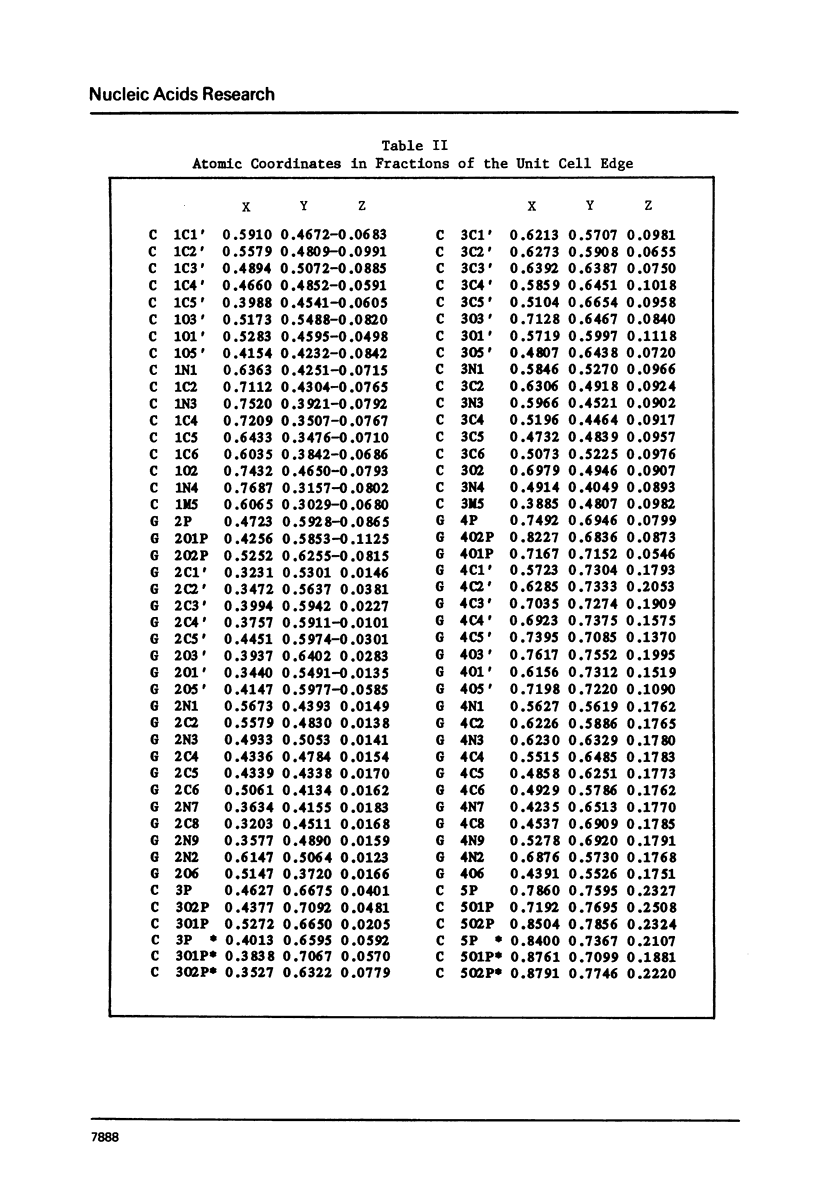
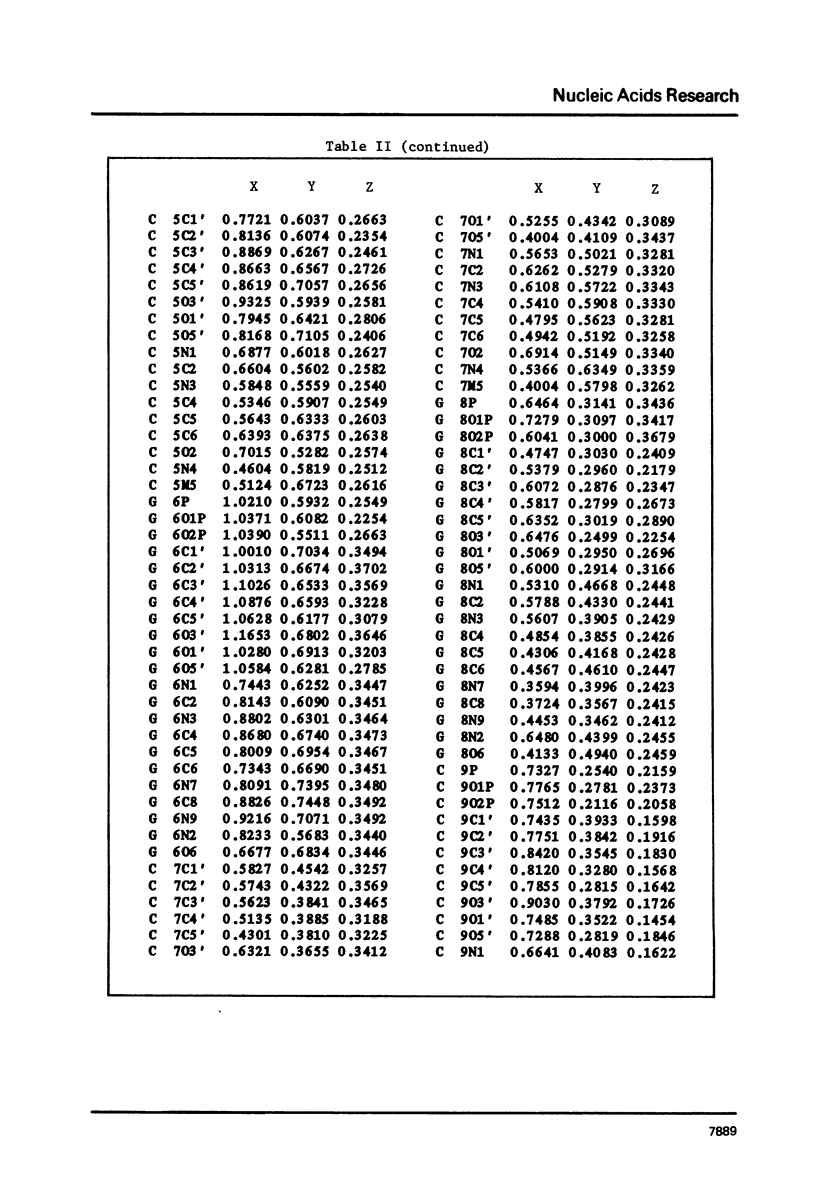
The hexamer (m5 dC-dG)3 has been synthesized and its three-dimensional structure determined by a single crystal X-ray diffraction analysis. The structure has been refined to a final R value of 15.6% at 1.3 A resolution. The molecule forms a left-handed Z-DNA helix which is similar to the unmethylated Z-DNA structure. The presence of the methyl group has resulted in slight changes in the twist angle between successive base pairs and modification of some of the interatomic contacts. Methylation of cytosine in the C5 position is associated with a relative destabilization of the B-DNA structure and a stabilization through hydrophobic bonding of the Z-DNA structure.
Full text
PDF










Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altona C., Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. A new description using the concept of pseudorotation. J Am Chem Soc. 1972 Nov 15;94(23):8205–8212. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
- Arnott S., Chandrasekaran R., Birdsall D. L., Leslie A. G., Ratliff R. L. Left-handed DNA helices. Nature. 1980 Feb 21;283(5749):743–745. doi: 10.1038/283743a0. [DOI] [PubMed] [Google Scholar]
- Behe M., Felsenfeld G. Effects of methylation on a synthetic polynucleotide: the B--Z transition in poly(dG-m5dC).poly(dG-m5dC). Proc Natl Acad Sci U S A. 1981 Mar;78(3):1619–1623. doi: 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Dickerson R. E. Conformation and dynamics in a Z-DNA tetramer. J Mol Biol. 1981 Nov 15;152(4):723–736. doi: 10.1016/0022-2836(81)90124-8. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Wang R. Y. 5-Methylcytosine in eukaryotic DNA. Science. 1981 Jun 19;212(4501):1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- Möller A., Gabriels J. E., Lafer E. M., Nordheim A., Rich A., Stollar B. D. Monoclonal antibodies recognize different parts of Z-DNA. J Biol Chem. 1982 Oct 25;257(20):12081–12085. [PubMed] [Google Scholar]
- Nordheim A., Pardue M. L., Lafer E. M., Möller A., Stollar B. D., Rich A. Antibodies to left-handed Z-DNA bind to interband regions of Drosophila polytene chromosomes. Nature. 1981 Dec 3;294(5840):417–422. doi: 10.1038/294417a0. [DOI] [PubMed] [Google Scholar]
- Pohl F. M., Jovin T. M. Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly (dG-dC). J Mol Biol. 1972 Jun 28;67(3):375–396. doi: 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Thamann T. J., Lord R. C., Wang A. H., Rich A. The high salt form of poly(dG-dC).poly(dG-dC) is left-handed Z-DNA: Raman spectra of crystals and solutions. Nucleic Acids Res. 1981 Oct 24;9(20):5443–5457. doi: 10.1093/nar/9.20.5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wang A. J., Quigley G. J., Kolpak F. J., van der Marel G., van Boom J. H., Rich A. Left-handed double helical DNA: variations in the backbone conformation. Science. 1981 Jan 9;211(4478):171–176. doi: 10.1126/science.7444458. [DOI] [PubMed] [Google Scholar]


