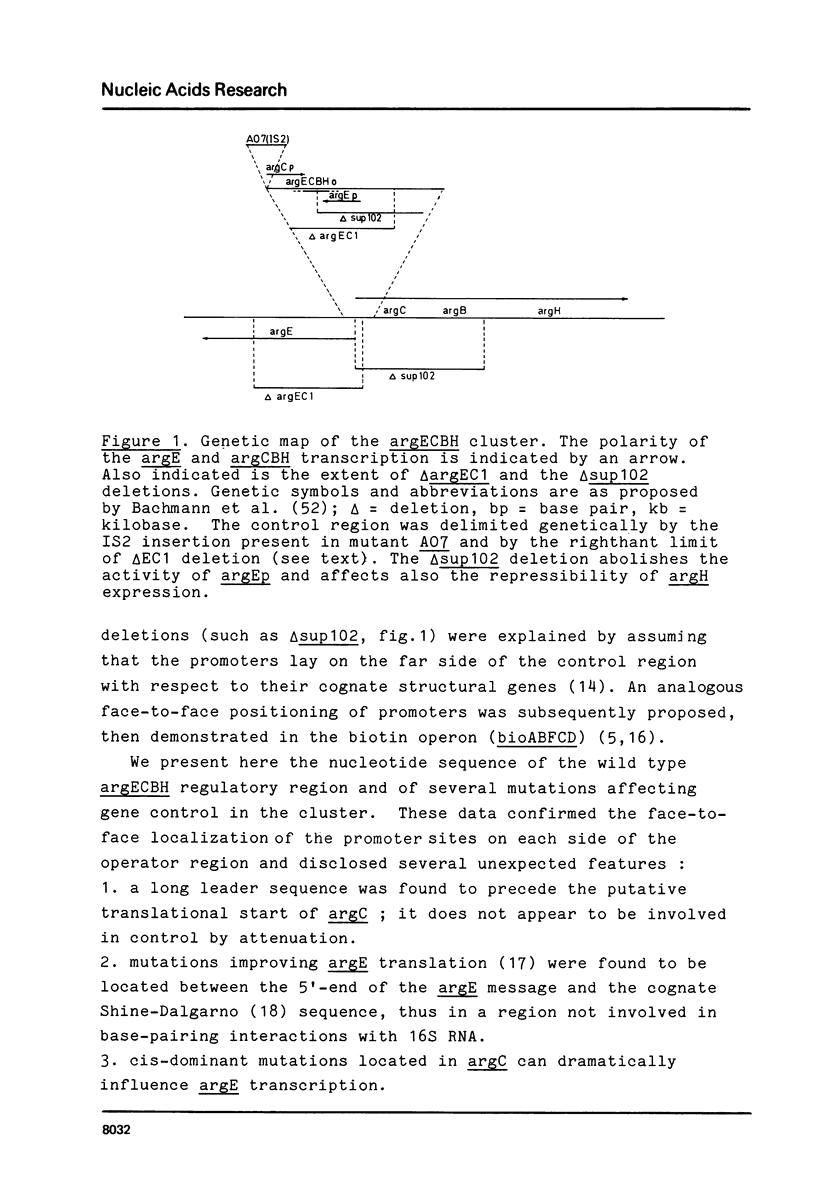
Abstract
The nucleotide sequence of the control region of the divergent argECBH operon has been established in the wild type and in mutants affecting expression of these genes. The argE and argCBH promoters face each other and overlap with an operator region containing two domains which may act as distinct repressor binding sites. A long leader sequence - not involved in attenuation - precedes argCBH. Overlapping of the argCBH promoter and the region involved in ribosome mobilization for argE translation explains the dual effect of some mutations. Mutations causing semi-constitutive expression of argE improve putative promoter sequences within argC. Implications of these results regarding control mechanisms in amino acid biosynthesis and their evolution are discussed.
Full text
PDF


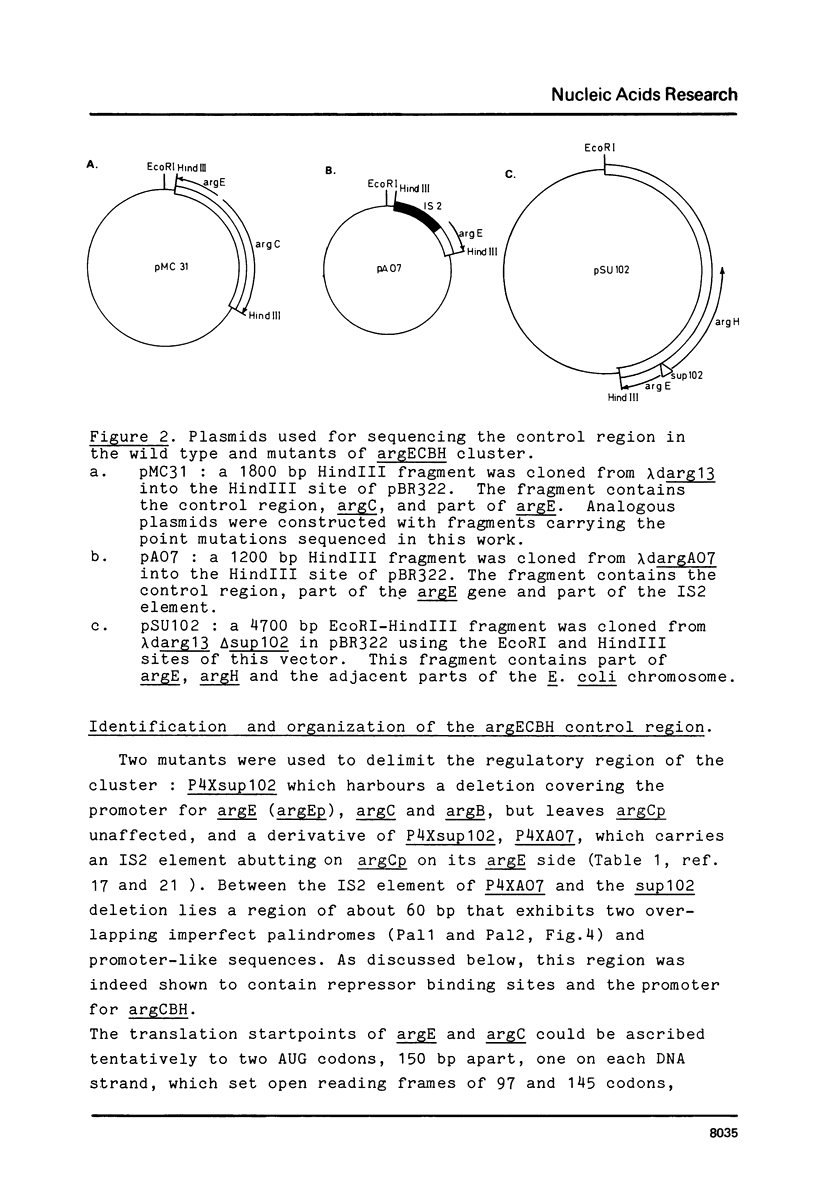


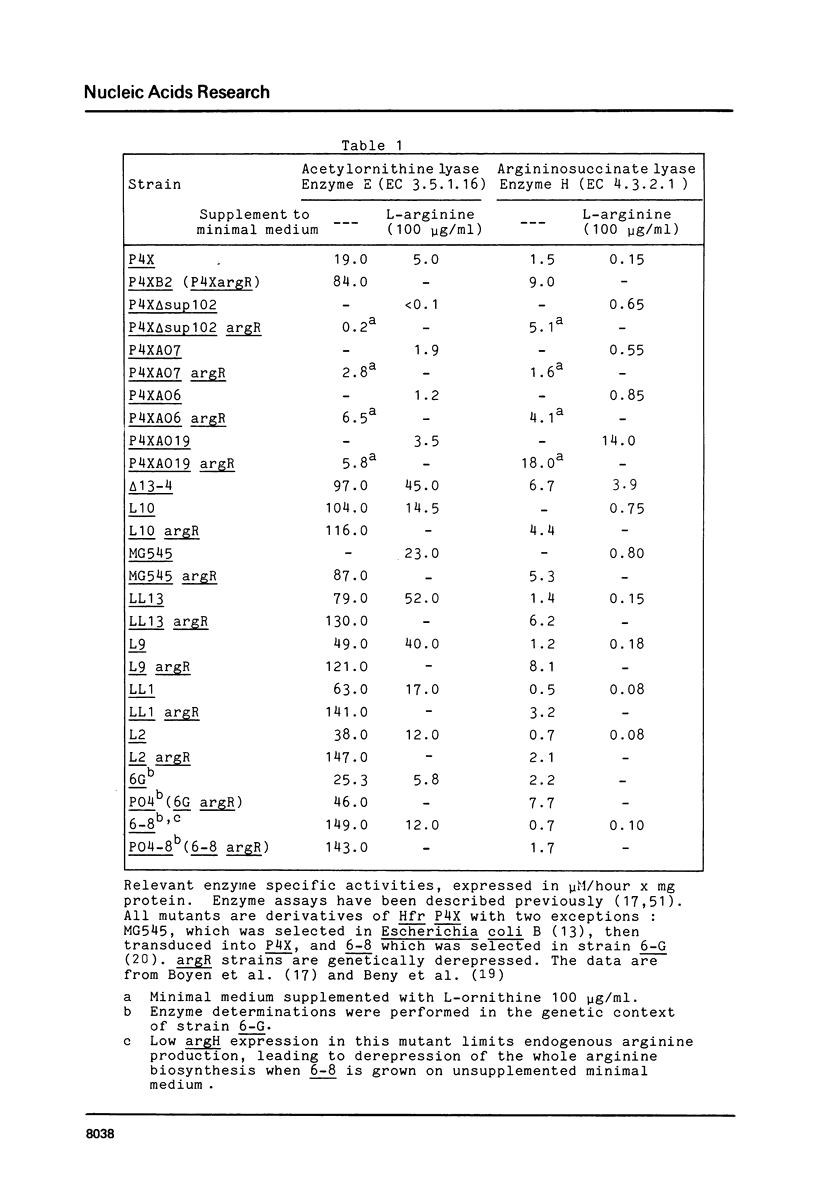









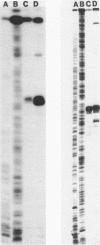
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkins J. F. Is UAA or UGA part of the recognition signal for ribosomal initiation? Nucleic Acids Res. 1979 Oct 25;7(4):1035–1041. doi: 10.1093/nar/7.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J., Low K. B., Taylor A. L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976 Mar;40(1):116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker D. F., Campbell A. M. Genetic and biochemical characterization of the birA gene and its product: evidence for a direct role of biotin holoenzyme synthetase in repression of the biotin operon in Escherichia coli. J Mol Biol. 1981 Mar 15;146(4):469–492. doi: 10.1016/0022-2836(81)90043-7. [DOI] [PubMed] [Google Scholar]
- Barker D. F., Kuhn J., Campbell A. M. Sequence and properties of operator mutations in the bio operon of Escherichia coli. Gene. 1981 Jan-Feb;13(1):89–102. doi: 10.1016/0378-1119(81)90046-9. [DOI] [PubMed] [Google Scholar]
- Barry G., Squires C., Squires C. L. Attenuation and processing of RNA from the rplJL--rpoBC transcription unit of Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3331–3335. doi: 10.1073/pnas.77.6.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedouelle H., Hofnung M. A DNA sequence containing the control regions of the malEFG and malK-lamB operons in Escherichia coli K12. Mol Gen Genet. 1982;185(1):82–87. doi: 10.1007/BF00333794. [DOI] [PubMed] [Google Scholar]
- Beny G., Boyen A., Charlier D., Lissens W., Feller A., Glansdorff N. Promoter mapping and selection of operator mutants by using insertion of bacteriophage Mu in the argECBH divergent operon of Escherichia coli K-12. J Bacteriol. 1982 Jul;151(1):62–67. doi: 10.1128/jb.151.1.62-67.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beny G., Cunin R., Glansdorff N., Boyen A., Charlier J., Kelker N. Transcription of Regions within the divergent argECBH operon of Escherichia coli: evidence for lack of an attenuation mechanism. J Bacteriol. 1982 Jul;151(1):58–61. doi: 10.1128/jb.151.1.58-61.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyen A., Charlier D., Crabeel M., Cunin R., Palchaudhuri S., Glansdorff N. Studies on the control region of the bipolar argECBH operon of Escherichia coli. I. Effect of regulatory mutations and IS2 insertions. Mol Gen Genet. 1978 May 3;161(2):185–196. doi: 10.1007/BF00274187. [DOI] [PubMed] [Google Scholar]
- Brent R., Ptashne M. Mechanism of action of the lexA gene product. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4204–4208. doi: 10.1073/pnas.78.7.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. P., Baumberg S. Divergent transcription of the argECBH cluster of escherichia coli k12. Mutations which alter the control of enzyme synthesis. J Mol Biol. 1976 Apr 5;102(2):205–220. doi: 10.1016/s0022-2836(76)80049-6. [DOI] [PubMed] [Google Scholar]
- Busby S., Aiba H., de Crombrugghe B. Mutations in the Escherichia coli operon that define two promoters and the binding site of the cyclic AMP receptor protein. J Mol Biol. 1982 Jan 15;154(2):211–227. doi: 10.1016/0022-2836(82)90061-4. [DOI] [PubMed] [Google Scholar]
- Charlier D., Crabeel M., Cunin R., Glansdorff N. Tandem and inverted repeats of arginine genes in Escherichia coli: structural and evolutionary considerations. Mol Gen Genet. 1979 Jul 2;174(1):75–88. doi: 10.1007/BF00433308. [DOI] [PubMed] [Google Scholar]
- Chou J., Lemaux P. G., Casadaban M. J., Cohen S. N. Transposition protein of Tn3: identification and characterisation of an essential repressor-controlled gene product. Nature. 1979 Dec 20;282(5741):801–806. doi: 10.1038/282801a0. [DOI] [PubMed] [Google Scholar]
- Cleary P. P., Campbell A., Chang R. Location of promoter and operator sites in the biotin gene cluster of Escherichia coli. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2219–2223. doi: 10.1073/pnas.69.8.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabeel M., Charlier D., Cunin R., Glansdorff N. Cloning and endonuclease restriction analysis of argF and of the control region of the argECBH bipolar operon in Escherichia coli. Gene. 1979 Mar;5(3):207–231. doi: 10.1016/0378-1119(79)90079-9. [DOI] [PubMed] [Google Scholar]
- Cunin R., Elseviers D., Sand G., Freundlich G., Glandsdorff N. On the functional organization of the arg ECBH cluster of genes in Escherichia coli K-12. Mol Gen Genet. 1969;106(1):32–47. doi: 10.1007/BF00332819. [DOI] [PubMed] [Google Scholar]
- Debarbouille M., Cossart P., Raibaud O. A DNA sequence containing the control sites for gene malT and for the malPQ operon. Mol Gen Genet. 1982;185(1):88–92. doi: 10.1007/BF00333795. [DOI] [PubMed] [Google Scholar]
- Dickson R. C., Abelson J., Barnes W. M., Reznikoff W. S. Genetic regulation: the Lac control region. Science. 1975 Jan 10;187(4171):27–35. doi: 10.1126/science.1088926. [DOI] [PubMed] [Google Scholar]
- Elseviers D., Cunin R., Glansdorff N. Control regions within the argECBH gene cluster of Escherichia coli K12. Mol Gen Genet. 1972;117(4):349–366. doi: 10.1007/BF00333028. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal D., Sommer H., Saedler H. Nucleotide sequence of the transposable DNA-element IS2. Nucleic Acids Res. 1979 Mar;6(3):1111–1122. doi: 10.1093/nar/6.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield L., Boone T., Wilcox G. DNA sequence of the araBAD promoter in Escherichia coli B/r. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4724–4728. doi: 10.1073/pnas.75.10.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofnung M. Divergent operons and the genetic structure of the maltose B region in Escherichia coli K12. Genetics. 1974 Feb;76(2):169–184. doi: 10.1093/genetics/76.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G. A. Control of the argECBH cluster in Escherichia coli. Mol Gen Genet. 1972;117(4):337–348. doi: 10.1007/BF00333027. [DOI] [PubMed] [Google Scholar]
- Ketner G., Campbel A. Operator and promoter mutations affecting divergent transcription in the bio gene cluster of Escherichia coli. J Mol Biol. 1975 Jul 25;96(1):13–27. doi: 10.1016/0022-2836(75)90179-5. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S. K., Garvin R. T., James E. Nucleotide sequence of the argF regulatory region of Escherichia coli K-12. Gene. 1981 Dec;16(1-3):119–132. doi: 10.1016/0378-1119(81)90068-8. [DOI] [PubMed] [Google Scholar]
- Otsuka A., Abelson J. The regulatory region of the biotin operon in Escherichia coli. Nature. 1978 Dec 14;276(5689):689–694. doi: 10.1038/276689a0. [DOI] [PubMed] [Google Scholar]
- Panchal C. J., Bagchee S. N., Guha A. Divergent orientation of transcription from the arginine gene ECBH cluster of Escherichia coli. J Bacteriol. 1974 Feb;117(2):675–680. doi: 10.1128/jb.117.2.675-680.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piette J., Cunin R., Van Vliet F., Charlier D., Crabeel M., Ota Y., Glansdorff N. Homologous control sites and DNA transcription starts in the related argF and argI genes of Escherichia coli K12. EMBO J. 1982;1(7):853–857. doi: 10.1002/j.1460-2075.1982.tb01259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouwels P. H., Cunin R., Glansdorff N. Letter: Divergent transcription in the argECBH cluster of genes in Escherichia coli K12. J Mol Biol. 1974 Mar;83(3):421–424. doi: 10.1016/0022-2836(74)90288-5. [DOI] [PubMed] [Google Scholar]
- Pribnow D. Nucleotide sequence of an RNA polymerase binding site at an early T7 promoter. Proc Natl Acad Sci U S A. 1975 Mar;72(3):784–788. doi: 10.1073/pnas.72.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt L., Kaiser A. D. Control of lambda repressor synthesis. Proc Natl Acad Sci U S A. 1971 Sep;68(9):2185–2189. doi: 10.1073/pnas.68.9.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Saedler H., Reif H. J., Hu S., Davidson N. IS2, a genetic element for turn-off and turn-on of gene activity in E. coli. Mol Gen Genet. 1974;132(4):265–289. doi: 10.1007/BF00268569. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Tabak H. F., Flavell R. A. A method for the recovery of DNA from agarose gels. Nucleic Acids Res. 1978 Jul;5(7):2321–2332. doi: 10.1093/nar/5.7.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C. P., Cohen S. N. 3'-end labeling of DNA with [alpha-32P]cordycepin-5'-triphosphate. Gene. 1980 Jul;10(2):177–183. doi: 10.1016/0378-1119(80)90135-3. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Hardies S. C., Horn G. T., Klein B., Larson J. E., Neuendorf S. K., Panayotatos N., Patient R. K., Selsing E. RPC-5 column chromatography for the isolation of DNA fragments. Methods Enzymol. 1980;65(1):327–347. doi: 10.1016/s0076-6879(80)65043-5. [DOI] [PubMed] [Google Scholar]
- Wray L. V., Jr, Jorgensen R. A., Reznikoff W. S. Identification of the tetracycline resistance promoter and repressor in transposon Tn10. J Bacteriol. 1981 Aug;147(2):297–304. doi: 10.1128/jb.147.2.297-304.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981 Feb 26;289(5800):751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- Zurawski G., Gunsalus R. P., Brown K. D., Yanofsky C. Structure and regulation of aroH, the structural gene for the tryptophan-repressible 3-deoxy-D-arabino-heptulosonic acid-7-phosphate synthetase of Escherichia coli. J Mol Biol. 1981 Jan 5;145(1):47–73. doi: 10.1016/0022-2836(81)90334-x. [DOI] [PubMed] [Google Scholar]
- van de Sande J. H., Kleppe K., Khorana H. G. Reversal of bacteriophage T4 induced polynucleotide kinase action. Biochemistry. 1973 Dec 4;12(25):5050–5055. doi: 10.1021/bi00749a004. [DOI] [PubMed] [Google Scholar]