Background: Set2 catalyzes mono-, di-, and trimethylation of lysine 36 on histone H3 (H3K36).
Results: Phosphorylation of the C-terminal domain of RNA polymerase II (RNAPII CTD) regulates Set2 protein levels and H3K36 methylation.
Conclusion: Set2 protein is regulated to maintain balanced levels of H3K36 methylation.
Significance: These results suggest that different methylation states perform distinct biological functions.
Keywords: Chromatin Histone Modification, Histone Methylation, Histone Modification, Protein Stability, RNA Polymerase II
Abstract
Methylation of lysine 36 on histone H3 (H3K36) is catalyzed by the Set2 methyltransferase and is linked to transcriptional regulation. Previous studies have shown that trimethylation of H3K36 by Set2 is directed through its association with the phosphorylated repeats of the RNA polymerase C-terminal domain (RNAPII CTD). Here, we show that disruption of this interaction through the use of yeast mutants defective in CTD phosphorylation at serine 2 results in a destabilization of Set2 protein levels and H3K36 methylation. Consistent with this, we find that Set2 has a short half-life and is co-regulated, with RNAPII CTD phosphorylation levels, during logarithmic growth in yeast. To probe the functional consequence of uncoupling Set2-RNAPII regulation, we expressed a truncated and more stable form of Set2 that is capable of dimethylation but not trimethylation in vivo. Results of high throughput synthetic genetic analyses show that this Set2 variant has distinct genetics from either SET2 or set2Δ and is synthetically sick or lethal with a number of transcription elongation mutants. Collectively, these results provide molecular insight into the regulation of Set2 protein levels that influence H3K36 methylation states.
Introduction
The N-terminal tails of histones are richly decorated with post-translational modifications that regulate a variety of DNA-templated processes through the recruitment of additional factors to chromatin. These factors interact with modifications such as acetylation, methylation, and phosphorylation through specific protein domains such as bromo-, chromo-, PHD, and TUDOR domains (1). Furthermore, these domains are usually found in the context of multiprotein complexes that perform diverse functions in chromatin remodeling, replication, transcription, and DNA repair, among others (2–4). Thus, there is great interest in both identifying biological important post-translational modifications on histones and understanding how these modifications function within chromatin.
One such post-translational modification is methylation of lysine 36 on histone H3. In yeast, H3K363 is modified by the methyltransferase Set2, which can add one, two, or three methyl groups to the ϵ-amino group of the lysine side chain using S-adenosylmethionine as the methyl donor (5). Genome-wide chromatin immunoprecipitation studies have demonstrated that H3K36 methylation is generally associated with actively transcribed genes; however, dimethylation (H3K36me2) and trimethylation (H3K36me3) show different patterns of localization (6–8). H3K36me2 appears uniformly distributed across transcribed genes and is not correlated with transcription frequency, whereas H3K36me3 seems to be most prevalent at the 3′ end of coding regions and is positively correlated with the rate of transcription. These data suggest that the deposition of different levels of H3K36 methylation is controlled within the cell and that the different degrees of this modification likely direct distinct biological functions.
H3K36 methylation has been reported to be recognized by the Rpd3(S) deacetylase complex in Saccharomyces cerevisiae through the chromodomain of Eaf3 (a member of both the Rpd3(S) deacetylase and NuA4 histone acetyltransferase complexes) (9–11). Subsequent studies revealed that H3K36me2 is the preferred substrate for Rpd3(S) binding (12–14) and that a primary function for H3K36 methylation is to recruit the repressive activity of Rpd3(S) to genes to “reset” or reestablish chromatin structure between multiple rounds of transcription (15–17). Such resetting, or at least maintaining a more compact chromatin structure within gene bodies, also protects genes from spurious transcription from improper initiation sites (cryptic transcription) (15–18). To date, no separate function has been reported for H3K36me3. However, H3K36me3 can be recognized by both the chromodomain of Eaf3 and in vitro by the PHD domain of Nto1, a member of the NuA3 histone acetyltransferase complex, suggesting that H3K36me3 might have a positive role in transcription through the recruitment of histone acetyltransferase complexes (19). Consistent with this, NuA3 association to chromatin has been shown to be partially dependent on Set2 methylation (20).
Set2 interacts with the repeating C-terminal domain (CTD) of RNA polymerase II (RNAPII) (12, 21–25). In yeast, the CTD of RNAPII consists of 26 repeats of a heptapeptide repeat sequence (YSPTSPS) in which all three serine residues can be phosphorylated (26–28). Set2 preferentially recognizes the Ser2/Ser5-modified form of the CTD through a region at the C terminus of Set2 termed the SRI (Set2-Rpb1 interacting) domain (21, 29). Consistent with this, H3K36me3 is well correlated with Ser2- and Ser2/Ser5-modified forms of the CTD that occur within the bodies and 3′ ends of genes (6, 30–32).
We recently reported that deletion of the kinase responsible for Ser2 phosphorylation of the CTD, Ctk1, results not only in loss of H3K36me but also decreased Set2 protein levels (14). This finding suggested a novel mechanism for regulating histone methyltransferase activity. Here, we present a molecular basis for the regulation of Set2 protein levels. We find that events that alter or perturb Ser2 CTD phosphorylation levels result in reduced levels of Set2 protein and global loss of H3K36me3 without much change in H3K36me2. Furthermore, we demonstrate that expression of a stable truncation of Set2, incapable of H3K36me3, greatly compromises the growth of several transcription elongation mutants. Lastly, high throughput epistatic (E-MAP) analysis reveals different interactions for strains capable of catalyzing different levels of H3K36 methylation in vivo. Collectively, our data delineate a novel mechanism for Set2 regulation and support the model that different degrees of H3K36 methylation likely harbor distinct functions.
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmids
Strains used in this study are detailed in supplemental Table S1. Strains created for this study were constructed using heterologous gene replacement (33, 34) or through classical yeast mating in combination with tetrad dissection or high throughput strain construction techniques (35). Set2 expression plasmids (14, 21) were transformed using the lithium chloride technique (36), and shuffle strains (where important genes were genomically deleted but carried on a uracil-selectable plasmid) were maintained on selective media until the time of the appropriate experiment, and plasmid loss was instigated by plating on media containing 5-fluoroorotic acid (5-FOA) (37).
Western Blot Analysis
Yeast cultures were grown in either rich medium (YPD) or synthetic defined medium from a starting A600 of between 0.1 and 0.2 and grown to mid-log phase (A600 0.6–1.0) with the exception of cultures started from stationary phase. In this instance, 200-ml cultures were started from saturated cultures of yeast grown in synthetic defined medium for >7 days and inoculated at an initial A600 of 0.5. In all cases, five optical units of cells were harvested by centrifugation, and extracts were prepared using a protocol adapted from Ref. 38. Briefly, cell pellets were resuspended in 75 μl of SUMEB buffer (1% SDS, 8 m urea, 10 mm MOPS, pH 6.8, 10 mm EDTA, 0.01% bromphenol blue), mixed with 200 μl of glass beads, and vortexed for 3 min. To the beads was added 175 μl of additional buffer, and the total supernatant was collected and clarified to give a total volume of 250 μl of cell extract. Extracts were heated at 95 °C for 5 min and frozen at −80 °C until use. For Western blot analysis of histones, 5 μl of extract was separated by 17% SDS-PAGE. For Western analysis of Set2 and RNAPII, proteins were separated by 8% SDS-PAGE. Gels were transferred to PVDF for 90 min at 45 mA and dried in methanol to block according to the manufacturer's instructions (Millipore-Immobilon-P). Dried membranes were rehydrated briefly in methanol and incubated with primary antibodies overnight at 4 °C in PBST (phosphate-buffered saline including 0.05% Tween 20) containing 5% blocking (Pierce). Antibodies were used at the following dilutions: anti-H3K36me2 (Active Motif), 1:1,000; anti-H3K36me3 (Abcam ab9050), 1:2,500; anti-H3 1:5,000; anti-Set2 (1:8,000), anti-G6PDH (1:100,000), anti-Ser(P)2 CTD (gift of D. Eick), 1:200. Western blots were visualized using HRP-conjugated secondary antibodies and ECL Plus chemiluminescence (GE Healthcare).
Protein Turnover Experiments
Turnover experiments were adapted from a protocol from Ref. 39. Briefly, 100-ml cultures of yeast in YPD were grown to an A600 of ∼0.6, and cycloheximide was added to a final concentration of 200 μg/ml. Five optical units of cells were taken at various time points, and 1 ml of 10 mm sodium azide and 1 ml of dimethyl sulfoxide were added. Cells were then immediately frozen in liquid nitrogen. Frozen samples were thawed simultaneously, and extracts were prepared using the SUMEB method described above.
Yeast Growth Assays
For phenotypic growth assays, yeast were grown overnight in appropriate media (YPD or synthetic complete medium with necessary additives). Saturated cultures were used to start fresh cultures in the same medium at an A600 of ∼0.2. Cells were allowed to double at least two times, and cells were harvested and resuspended to an A600 of 1.0 in sterile water in a 96-well plate. Cells were 5-fold serially diluted and then spotted onto plates containing necessary selections using a 48-pin replicating tool. Plates were incubated at indicated temperatures and imaged daily.
Epistatic Miniarray Analysis
E-MAP analysis was carried out as described (40, 41). Growth rates were assessed by automated image analysis of colony size (42).
RESULTS
Set2 and H3K36 Methylation Are Regulated Differentially during Cell Growth
It has been well established that methylation by Set2 is correlated with hyperphosphorylation of the CTD of RNAPII. More than a decade ago, West and Corden showed that CTD phosphorylation was highly correlated with log phase growth and the diauxic shift in yeast (43). We therefore measured Set2 protein and histone modifications in different stages of yeast growth. Starting from stationary phase cells, a 200-ml culture of yeast in YPD was inoculated at an initial A600 of 0.5, and time points/A600 measurements were taken at regular intervals (Fig. 1A). Extracts from each time point were separated by SDS-PAGE and probed for Set2 as well as RNAPII phosphorylation and histone modifications by Western blot analysis. As shown in Fig. 1B, and consistent with previous findings (43), RNAPII and CTD phosphorylation levels peaked at late log phase and during diauxic shift (t = 6–8 h). Significantly, we found that Set2 protein levels also changed under these same conditions and correlated strongly with the occurrence of RNAPII CTD phosphorylation (Fig. 1B). In examination of H3K36 methylation levels, we observed that H3K36me2 was most abundant at t = 0 and 2 h but decreased during mid- to late log phase (Fig. 1C). In contrast, H3K36me3 levels were lowest at t = 0–4 h but increased during mid- to late log phase at the same time where Set2 levels and CTD phosphorylation levels increased (Fig. 1C). Although Set2 and CTD phosphorylation levels decreased during late log phase, H3K36me3 levels remained constant. These data uncover that Set2 and H3K36me2 and H3K36me3 are dynamically regulated and that Set2 proteins levels and the occurrence of H3K36me3 are tightly coupled to CTD phosphorylation.
FIGURE 1.
Set2, H3K36 methylation, and RNAPII phosphorylation levels are dynamically regulated during cell growth. A, growth curve of wild-type BY4741 cells in complete synthetic defined medium with approximate growth stages. s.p., stationary phase; lag, lag phase; log, anaerobic fermentation of glucose, d.s., diauxic shift; and res., aerobic respiration of ethanol. B, Western blot analyses of extracts from the time course shown in A, indicating the presence of RNAPII as well as phosphorylated forms of the CTD and Set2. C, Western blot analyses from samples taken in A probing for the presence of various post-translational modifications (H3K36me2, H3K36me3, H3K4me3, and H3K27ac) on histone H3.
Set2 Protein Levels Are Regulated in Vivo
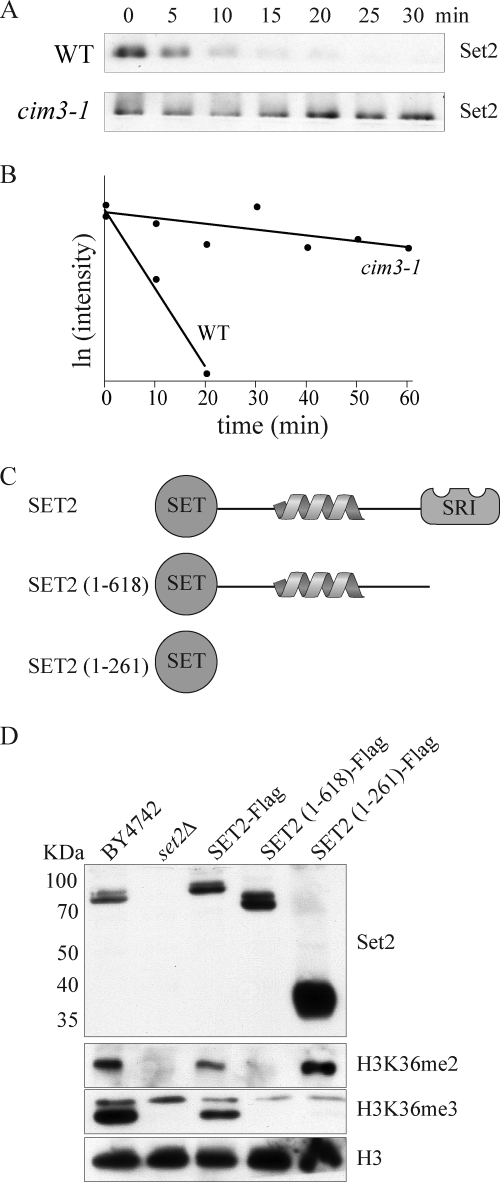
We previously noted that Set2 protein was lost in CTK1 delete cells (ctk1Δ), and this was not the result of changes in SET2 gene expression (14, 25). These data, combined with our observation that Set2 protein levels correlate with RNAPII CTD phosphorylation levels, prompted us to explore the regulation of the Set2 protein further. Accordingly, we examined the abundance of Set2 protein upon treatment with the translational inhibitor cycloheximide (Fig. 2A). Results showed in a time course experiment that Set2 exhibited a rapid turnover with a half-life of ∼9 min (Fig. 2, A and B). In contrast, performing the same experiment in a temperature-sensitive proteasome-deficient strain (cim3-1) at the nonpermissive temperature showed increased stability of Set2 (Fig. 2, A and B).
FIGURE 2.
Set2 is a highly unstable protein. A, Western blot analysis of a time course experiment probing for Set2 protein levels in wild-type and proteasome-deficient (cim3-1) cells following addition of the translation inhibitor cycloheximide (200 μg/ml). B, quantification of Set2 protein levels as a function of time following cycloheximide treatment. Protein abundance was calculated by densitometry of Western blot analysis and plotted as the natural log of the intensity. C, schematic of Set2 domain structure and truncation mutants used in D. D, Western blot analyses showing the relative abundance of Set2 and the corresponding H3K36 methylation levels of several Set2 truncation mutants.
We next sought to identify the region responsible for regulating Set2 stability. Using strains harboring truncation mutants of Set2 (Fig. 2C), we were able to map the region responsible for Set2 instability to several hundred amino acids of unknown function between the Set2 domain and the SRI domain responsible for interaction with RNAPII (Fig. 2D). Interestingly, analysis of Set2 and histone methylation levels in these strains revealed unexpected behavior of the different Set2 truncations. Deleting solely the SRI domain of Set2 (Set2(1–618)) resulted in no significant change in the abundance of Set2 protein but also no observable H3K36me2 or H3K36me3 (Fig. 2D). However, expression of just the SET domain (1–261) of Set2 resulted in a marked increase in Set2 protein and H3K36me2 but no detectable H3K36me3 (Fig. 2D).
Set2 Protein Levels Are Decreased in Mutants That Alter RNAPII CTD Phosphorylation
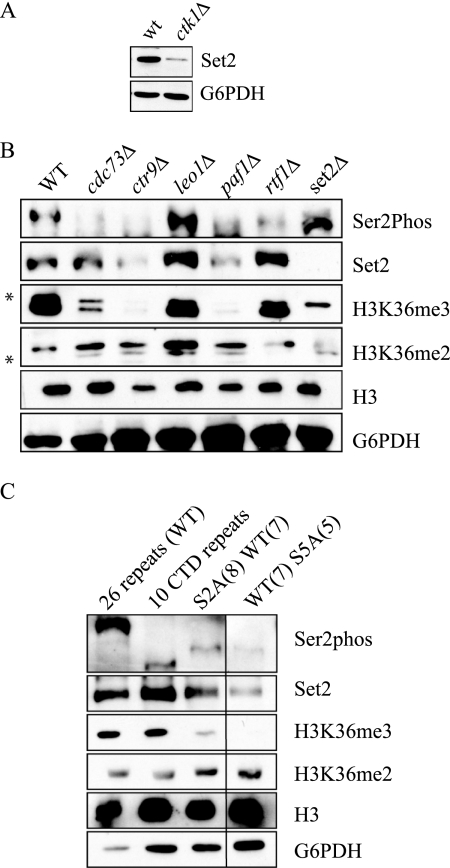
Set2 is known to bind to the Ser2/Ser5-phosphorylated form of RNAPII. Because Ctk1 is the major kinase responsible for Ser2 phosphorylation, we next wanted to determine whether the lower Set2 protein levels found in ctk1Δ strains (Fig. 3A) were related to the ability of Set2 to bind to the CTD of RNAPII. We therefore examined the levels of Set2 protein in a number of yeast mutants known to alter the phosphorylation state of the CTD. Set2 protein levels were noticeably decreased in mutants of the PAF complex, which is known to play an important role in transcription elongation and affect CTD Ser2 phosphorylation (Fig. 3B) (44). Significantly, Set2 protein levels were only decreased in PAF mutants that also exhibited changes in CTD Ser2 phosphorylation, cdc73Δ, paf1Δ, and ctr9Δ. Decreased levels of Set2 correlated with decreased levels of H3K36me3 but not H3K36me2, consistent with the model that CTD association is necessary for H3K36me3 by Set2. We also note that overall histone H3 levels are reduced in both the ctr9Δ and paf1Δ strains, likely also resulting in the lower levels of even the cross-reacting band in these two strains (Fig. 3B). Furthermore, Set2 levels were decreased in a temperature-sensitive allele of Kin28, as well as mutant alleles of the phosphatases Ssu72 (ssu72-2), Fcp1 (fcp1-1), and the cyclin-like protein Bur2 (supplemental Fig. S1). Because many of the mutants we tested were enzymes themselves, we wanted to confirm that the phenotype of decreased Set2 protein was directly due to a loss changes in its ability to recognize the CTD rather than other potential activities of these enzymes. Accordingly, we looked at Set2 protein levels in cells expressing mutant forms of the RNAPII CTD itself (Fig. 3C). Each strain carries an RNAPII gene with a CTD comprising wild-type repeats (WT) or mutations at either Ser2 or Ser5 within the given number of repeats. Again, we observed a decrease in Set2 protein levels when a number of Ser2 or Ser5 residues within the CTD repeat were mutated to alanine. However, we saw a strong reduction in H3K36me3, which was accompanied by either no change or a slight increase in the levels of H3K36me2 in these cells.
FIGURE 3.
RNAPII CTD phosphorylation regulates Set2 protein levels. A, Set2 protein levels as measured by Western blotting of whole cell extract taken from WT and ctk1Δ strains. B, Western blots of extracts taken from PAF complex deletions strains as well as set2Δ cells to measure Set2 and H3K36 methylation. Ser2phos refers to phosphorylation at Ser2 within the heptapeptide repeat of the RNAPII CTD. * denotes a cross-reactive band for the indicated antibody. C, Set2 and H3K36 methylation levels in RNAPII CTD mutants. S2A and S5A refer to mutations at either Ser2 or Ser5 within the CTD heptapeptide repeat, respectively. The number in parentheses refers to the number of tandem repeats in a given CTD sequence. The black line between the third and fourth lanes signifies that two unrelated lanes were removed during the editorial process. In all cases, all of the lanes originate from the same gel.
Overexpression of More Stable Form of Set2(1–261) Is Lethal in Combination with Various Transcription Mutants
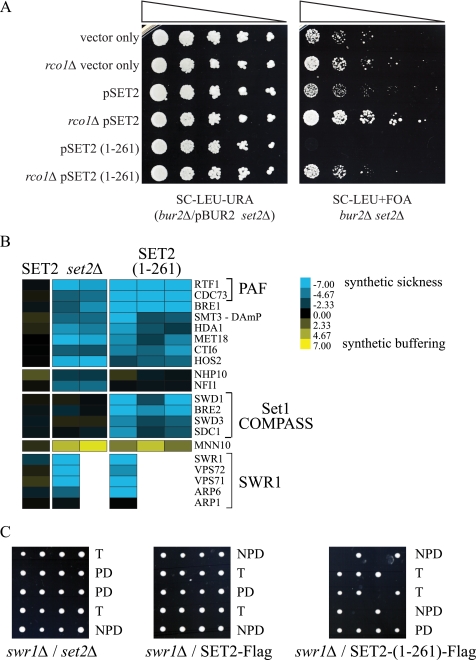
A well studied phenotype for set2Δ is the ability to bypass deletion of essential transcription factors such as members of the Bur1 complex and Spt16 (11, 45, 46). We therefore hypothesized that Set2 degradation might alter the levels of H3K36me2/me3 to facilitate transcription elongation during periods of transcriptional stress, when, for example, chromatin compaction by Rpd3(S) would inhibit rapid gene activation of stress response genes (47, 48). To examine this possibility, by way of using mutant forms of Set2 that have different behaviors in degradation and H3K36me, we expressed the different truncated forms of Set2 (see Fig. 2C) on plasmids in a bur2Δ set2Δ double deletion strain harboring a wild-type copy of BUR2 and then analyzed the growth of these mutants after selection for the loss of the BUR2 plasmid on 5-FOA (Fig. 4A). As expected, deletion of SET2 bypasses the poor growth of bur2Δ strain. However, reintroduction of Set2(1–261) resulted in worse growth than either the SET2 control or vector-only controls. These results suggest that the inability to degrade Set2 and/or reduce H3K36me2 levels results in impaired growth due to inappropriate transcriptional repression through maintained recruitment of Rpd3(S). Accordingly, we next deleted RCO1 (rco1Δ), a unique subunit of the Rpd3(S) complex, to see whether Rpd3(S) activity was necessary for this lethality. In fact, rco1Δ was able to rescue the lethality caused by expression of Set2(1–261) in the bur2 background (Fig. 4A).
FIGURE 4.
Set2(1–261) shows unique genetic interactions with transcription mutants. A, growth of bur2Δ/set2Δ or bur2Δ/set2Δ/rco1Δ strains harboring Set2 truncation plasmids plated on media to select for (−URA) or against (+FOA) a plasmid carrying a wild-type copy of BUR2 (pBUR2). B, genetic interactions between set2 mutants and a subset of the yeast deletion mutant collection. Blue color denotes combined sickness in the double mutant where yellow denotes better than expected growth (40). B, representative spores grown on YPD from the dissection of genetic crosses between swr1Δ and set2 mutant strains (PD, parental ditype; NPD, nonparental ditype; T, tetratype).
High Throughput Genetic Analysis of Set2 Truncation Mutants
The truncated form of Set2(1–261) offered a unique tool to assess the biological consequence of having a stable form of catalytically active Set2, which is not able to catalyze H3K36me3. We therefore performed epistatic miniarray analysis against 1,534 yeast mutant strains (Fig. 4B and supplemental Table S1). Our analysis revealed several classes of interactions. For example Set2(1–261) and set2Δ exhibit similar negative (synthetically sick) interactions with members of the SWR1 and PAF complexes, whereas only Set2(1–261) results in synthetic sickness with members of the COMPASS complex (Fig. 4B). We were able to confirm several interactions by classical genetic analysis, most notably those with SWR1 (Fig. 4C). Five representative tetrads from crosses between set2 mutant constructs and a swr1Δ strain are shown in Fig. 4C. We found that Set2(1–261), but neither Set2-FLAG nor set2Δ was lethal in combination with swr1Δ (Fig. 4C and Table 1). Because the SWR1 complex is primarily responsible for depositing the histone variant HTZ1 into nucleosomes, we also tested genetic interactions between our set2 mutant constructs and HTZ1 (Table 1). In this instance, both deletion of SET2 and expression of the Set2(1–261) construct resulted in synthetic lethality (supplemental Table S2).
TABLE 1.
Tetrad analysis of set2 mutant strains with htz1Δ and swr1Δ
| Mutant | Tetrads | Viable double mutant spores |
|---|---|---|
| % | ||
| swr1Δx | ||
| SET2 | 12 | 100 (12/12) |
| SET2(1–261) | 14 | 0 (0/15) |
| set2Δ | 13 | 100 (15/15) |
| htz1Δx | ||
| SET2 | 20 | 100 (15/15) |
| SET2(1–261) | 14 | 0 (0/13) |
| set2Δ | 18 | 11 (2/19) |
DISCUSSION
Using a combination of genetics and biochemistry, we have uncovered a significant mechanism for the regulation of Set2 methyltransferase activity in yeast. The SRI domain is known to bind to the CTD of RNAPII when it is phosphorylated at both the Ser2 and Ser5 positions within the CTD repeats. It was shown previously that disruption of this interaction, by deleting the CTD kinase Ctk1, resulted in a loss of H3K36 methylation without affecting Set2 mRNA levels (25). Here, we show that loss of H3K36me3 results from changes in the stability of the Set2 enzyme itself, which we demonstrate is mediated via its interaction with the phosphorylated RNAPII CTD.
In support of a role for Set2-RNAPII interaction in governing Set2 stability, we found that mutations in enzymes that either act on the CTD (Ctk1, Kin28, Bur1, Ssu72, Fcp1), or mutations to the CTD sequence itself resulted in lower levels of Set2 protein and H3K36me3 (Fig. 3 and supplemental Fig. S1). Interestingly, we also found that deletion of CTK1 altered the protein levels of other factors that associate with the CTD, suggesting this mechanism may not be restricted to just Set2 (supplemental Fig. S2).
Set2 levels correlated strongly with changes in CTD phosphorylation as wild-type cells pass through log phase and into diauxic shift (t ∼ 8–10 h) (Fig. 1, A and B). Interestingly, we observed H3K36me2 in whole cell extracts at time points where Set2 was barely detectible but H3K36me3 was not observed until log phase where Set2 and CTD Ser(P)2 levels peaked (Fig. 1). These data indicate that the low levels of transcription found in stationary phase are accompanied by low levels of Set2 and CTD phosphorylation, yet H3K36me2 levels are maintained. However, during rapid log phase growth where robust transcription is needed, an increase in CTD phosphorylation accompanied by increased stability of Set2 and H3K36me3 levels is observed. Not only are these results consistent with previous findings that the interaction of Set2 with the CTD is required for H3K36me3 (14), but they provide important insight into the dynamic regulation of H3K36me2 versus H3K36me3 and suggest that these two methyl marks have distinct functions in the cell.
Consistent with SRI regulating H3K36me3, several other transcription complexes or kinases have been shown to regulate H3K36 trimethylation, notably the PAF and BUR kinase complexes (46, 49). Prelich and co-workers demonstrated several years ago that deletion of several PAF complex members, cdc73Δ, ctr9Δ, or paf1Δ, resulted in a decrease in H3K36me3 (49). Similarly, Jaehning and co-workers showed that CTD Ser2 phosphorylation is reduced in these same three PAF mutants (44). Our data show that Set2 protein levels are also decreased in cdc73Δ, ctr9Δ, and paf1Δ strains (Fig. 3B). We take these data to conclude that decreased CTD phosphorylation in PAF mutants results in reduced Set2 protein levels which ultimately changes the balance of H3K36 methylation in the cell. Consistent with PAF effects of Ser2 phosphorylation, we also see a decrease of Set2 levels in BUR2 deletions (data not shown). Given that Bur1/Bur2 has recently been demonstrated to function on both Spt5 (DSIF) and the CTD (50), it is plausible that Bur1 also affects H3K36me3/Set2 levels by directly influencing Ser2 phosphorylation and/or through DSIF recruitment of the PAF complex.
In our analysis of Set2 regulation, we noted that Set2 protein turnover was reduced in a temperature-sensitive allele of a proteasome subunit. We take these data to suggest that Set2 is degraded via the proteasome (Fig. 2). Consistent with this notion, our data are in agreement with a high throughput study of protein stability where it was predicted that Set2 is an unstable protein (51). However, perhaps due to the rapid turnover of Set2 or the relatively small amounts of enzyme normally found in cells, we have been unable to identify a ubiquitin ligase, ubiquitin protease, or a site of ubiquitin attachment to Set2 (data not shown).
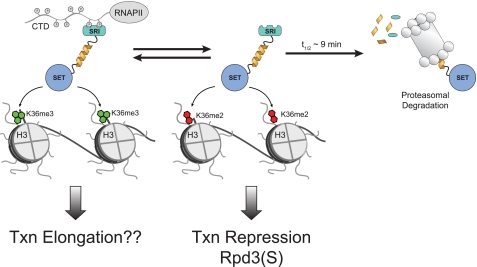
A significant question to then ask is why would cells need to dynamically regulate Set2 levels? In addition to providing a rapid means of regulating H3K36 di- and trimethylation during logarithmic cell growth, the control of Set2 levels might have a role in the response to cellular stress as mentioned above, where the transient changes in H3K36 methylation may alter recruitment of the repressor Rpd3(S) to sites of transcription to alleviate chromatin repression during transcription as proposed in Fig. 5. This would explain why deletion of SET2 is able to bypass a number of transcription elongation mutations including mutations in Spt6, Spt16, and Bur1/2 (11, 14, 45). In this scenario, removal of Set2 and hence both H3K36me2/me3, would result in more open chromatin structure and improved transcription of stress-response genes. In support of this idea, studies have shown that a wide number of stress-response genes are up-regulated in a SET2 deletion (47).
FIGURE 5.
Proposed model of Set2 regulation. Set2 is in equilibrium between a free state and association with the hyperphosphorylated form of RNAPII. The free form of Set2 is degraded in a proteasome-dependent manner with a half-life in yeast of ∼9 min. In its free form, Set2 is capable of dimethylation of H3K36, which is sufficient for the recruitment of the repressive Rpd3(S) complex. Binding to RNAPII is required for H3K36me3, which may perform a positive function in transcription elongation.
As mentioned above, the ability to rapidly alter Set2 levels to control the balance of H3K36me2 and H3K36me3 supports the idea that H3K36me2 and H3K36me3 have different biological functions. To explore this possibility further, we examined the genetic interactions of a Set2 fragment that shows an inability to catalyze H3K36me3 (Set2(1–261)). Importantly, this Set2 fragment is properly recruited to gene coding regions in a transcription-dependent manner (14). Our study compared the Set2(1–261) fragment against wild-type Set2, where all three methylation states at H3K36 are present, and a SET2 deletion, where no H3K36 methylation is observed. We found that Set2(1–261) shows distinct genetics from either deletion of SET2 or the wild-type case when crossed with mutations in other factors important for chromatin structure and/or transcription elongation. This is particularly significant because Set2(1–261) exhibits partial function. This could be attributed to an imbalance in the levels of H3K36 methylation and supports that Set2 has both positive and negative roles in regulating transcription. For example, a lack of H3K36me3 could lead to increased recruitment of repressive factors such as Rpd3(S) to H3K36me2, or it may result in the inability to recruit effector proteins that recognize H3K36me3. Interestingly, Eaf3, the factor that recognizes H3K36me in the Rpd3(S) complex, is also an integral member of the NuA4 histone acetyltransferase complex. Furthermore, evidence shows that a member of the NuA3 histone acetyltransferase complex also interacts with H3K36me3 (and this complex is known to associate with chromatin in a Set2-dependent manner) (19, 20). Perhaps then, H3K36me3 recruits activating complexes such as NuA3 and NuA4 to certain genes to facilitate transcription, and methylation at H3K36 can act as both an activating and repressive mark, as is seen with lysine methylation at H3K4 (52, 53). If this were the case, it is possible that, in response to changes in CTD phosphorylation caused by an environmental cue or transcriptional stress, degradation of Set2, rather than just disruption of the interaction between Set2 and the CTD is necessary to maintain some proper balance of H3K36me2/me3 in the cell. As Keogh et al. proposed several years ago, such a mechanism might also be used to attenuate regulation of certain genes or control transcription from normally silent cryptic promoters (11).
Supplementary Material
Acknowledgments
We thank members of the Strahl laboratory for critical reading of this manuscript and Michael Keogh, Mike Hampsey, Bill Tansey, David Stillman, Danny Reinberg, Dirk Eick, Steve Hanes, and David Atencio for valuable technical suggestions, plasmids, yeast strains, and antibodies.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM68088 (to B. D. S.). This work was also supported by National Research Service Award Postdoctoral Fellowship F32 GM80896 (to S. M. F.).

This article contains supplemental Figs. S1 and S2, Tables S1 and S2, and additional references.
- H3K36
- lysine 36 on histone H3
- CTD
- C-terminal domain
- 5-FOA
- 5-fluoroorotic acid
- H3K36me2
- H3K36 dimethylation
- H3K36me3
- H3K36 trimethylation
- RNAPII
- RNA polymerase II
- SRI
- Set2-Rpb1 interacting
- PAF
- RNA polymerase-associated factor.
REFERENCES
- 1. Yun M., Wu J., Workman J. L., Li B. (2011) Readers of histone modifications. Cell Res. 21, 564–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gardner K. E., Allis C. D., Strahl B. D. (2011) Operating on chromatin, a colorful language where context matters. J. Mol. Biol. 409, 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ruthenburg A. J., Li H., Patel D. J., Allis C. D. (2007) Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8, 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., Patel D. J. (2007) How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14, 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Strahl B. D., Grant P. A., Briggs S. D., Sun Z. W., Bone J. R., Caldwell J. A., Mollah S., Cook R. G., Shabanowitz J., Hunt D. F., Allis C. D. (2002) Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 22, 1298–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pokholok D. K., Harbison C. T., Levine S., Cole M., Hannett N. M., Lee T. I., Bell G. W., Walker K., Rolfe P. A., Herbolsheimer E., Zeitlinger J., Lewitter F., Gifford D. K., Young R. A. (2005) Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122, 517–527 [DOI] [PubMed] [Google Scholar]
- 7. Rando O. J. (2007) Global patterns of histone modifications. Curr. Opin. Genet. Dev. 17, 94–99 [DOI] [PubMed] [Google Scholar]
- 8. Rao B., Shibata Y., Strahl B. D., Lieb J. D. (2005) Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol. Cell. Biol. 25, 9447–9459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carrozza M. J., Li B., Florens L., Suganuma T., Swanson S. K., Lee K. K., Shia W. J., Anderson S., Yates J., Washburn M. P., Workman J. L. (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123, 581–592 [DOI] [PubMed] [Google Scholar]
- 10. Joshi A. A., Struhl K. (2005) Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to pol II elongation. Mol. Cell 20, 971–978 [DOI] [PubMed] [Google Scholar]
- 11. Keogh M. C., Kurdistani S. K., Morris S. A., Ahn S. H., Podolny V., Collins S. R., Schuldiner M., Chin K., Punna T., Thompson N. J., Boone C., Emili A., Weissman J. S., Hughes T. R., Strahl B. D., Grunstein M., Greenblatt J. F., Buratowski S., Krogan N. J. (2005) Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123, 593–605 [DOI] [PubMed] [Google Scholar]
- 12. Li B., Gogol M., Carey M., Lee D., Seidel C., Workman J. L. (2007) Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science 316, 1050–1054 [DOI] [PubMed] [Google Scholar]
- 13. Li B., Jackson J., Simon M. D., Fleharty B., Gogol M., Seidel C., Workman J. L., Shilatifard A. (2009) Histone H3 lysine 36 dimethylation (H3K36me2) is sufficient to recruit the Rpd3s histone deacetylase complex and to repress spurious transcription. J. Biol. Chem. 284, 7970–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Youdell M. L., Kizer K. O., Kisseleva-Romanova E., Fuchs S. M., Duro E., Strahl B. D., Mellor J. (2008) Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol. Cell. Biol. 28, 4915–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cheung V., Chua G., Batada N. N., Landry C. R., Michnick S. W., Hughes T. R., Winston F. (2008) Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 6, e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li B., Gogol M., Carey M., Pattenden S. G., Seidel C., Workman J. L. (2007) Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev. 21, 1422–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lickwar C. R., Rao B., Shabalin A. A., Nobel A. B., Strahl B. D., Lieb J. D. (2009) The Set2/Rpd3S pathway suppresses cryptic transcription without regard to gene length or transcription frequency. PLoS One 4, e4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaplan C. D., Laprade L., Winston F. (2003) Transcription elongation factors repress transcription initiation from cryptic sites. Science 301, 1096–1099 [DOI] [PubMed] [Google Scholar]
- 19. Bua D. J., Kuo A. J., Cheung P., Liu C. L., Migliori V., Espejo A., Casadio F., Bassi C., Amati B., Bedford M. T., Guccione E., Gozani O. (2009) Epigenome microarray platform for proteome-wide dissection of chromatin-signaling networks. PLoS One 4, e6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martin D. G., Grimes D. E., Baetz K., Howe L. (2006) Methylation of histone H3 mediates the association of the NuA3 histone acetyltransferase with chromatin. Mol. Cell. Biol. 26, 3018–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kizer K. O., Phatnani H. P., Shibata Y., Hall H., Greenleaf A. L., Strahl B. D. (2005) A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 25, 3305–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krogan N. J., Kim M., Tong A., Golshani A., Cagney G., Canadien V., Richards D. P., Beattie B. K., Emili A., Boone C., Shilatifard A., Buratowski S., Greenblatt J. (2003) Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23, 4207–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li B., Howe L., Anderson S., Yates J. R., 3rd, Workman J. L. (2003) The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 278, 8897–8903 [DOI] [PubMed] [Google Scholar]
- 24. Schaft D., Roguev A., Kotovic K. M., Shevchenko A., Sarov M., Shevchenko A., Neugebauer K. M., Stewart A. F. (2003) The histone 3 lysine 36 methyltransferase, SET2, is involved in transcriptional elongation. Nucleic Acids Res. 31, 2475–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xiao T., Hall H., Kizer K. O., Shibata Y., Hall M. C., Borchers C. H., Strahl B. D. (2003) Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 17, 654–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chapman R. D., Heidemann M., Albert T. K., Mailhammer R., Flatley A., Meisterernst M., Kremmer E., Eick D. (2007) Transcribing RNA polymerase II is phosphorylated at CTD residue serine 7. Science 318, 1780–1782 [DOI] [PubMed] [Google Scholar]
- 27. Egloff S., O'Reilly D., Chapman R. D., Taylor A., Tanzhaus K., Pitts L., Eick D., Murphy S. (2007) Serine 7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 318, 1777–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fuchs S. M., Laribee R. N., Strahl B. D. (2009) Protein modifications in transcription elongation. Biochim. Biophys. Acta 1789, 26–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vojnic E., Simon B., Strahl B. D., Sattler M., Cramer P. (2006) Structure and carboxyl-terminal domain (CTD) binding of the Set2 SRI domain that couples histone H3 Lys-36 methylation to transcription. J. Biol. Chem. 281, 13–15 [DOI] [PubMed] [Google Scholar]
- 30. Buratowski S. (2003) The CTD code. Nat. Struct. Biol. 10, 679–680 [DOI] [PubMed] [Google Scholar]
- 31. Phatnani H. P., Greenleaf A. L. (2006) Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 20, 2922–2936 [DOI] [PubMed] [Google Scholar]
- 32. Tietjen J. R., Zhang D. W., Rodríguez-Molina J. B., White B. E., Akhtar M. S., Heidemann M., Li X., Chapman R. D., Shokat K., Keles S., Eick D., Ansari A. Z. (2010) Chemical-genomic dissection of the CTD code. Nat. Struct. Mol. Biol. 17, 1154–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gelbart M. E., Rechsteiner T., Richmond T. J., Tsukiyama T. (2001) Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol. Cell. Biol. 21, 2098–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. (2004) A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 [DOI] [PubMed] [Google Scholar]
- 35. Tong A. H. Y., Boone C. (2007) High-throughput strain construction and systematic synthetic lethal screening in Saccharomyces cerevisiae. Method Microbiol. 36, 369–386 [Google Scholar]
- 36. Gietz R. D., Schiestl R. H. (2007) Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 35–37 [DOI] [PubMed] [Google Scholar]
- 37. Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. (1987) 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154, 164–175 [DOI] [PubMed] [Google Scholar]
- 38. Burke D., Dawson D., Stearns T. (2000) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, p. 174, Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- 39. Pal B., Chan N. C., Helfenbaum L., Tan K., Tansey W. P., Gething M. J. (2007) SCFCdc4-mediated degradation of the Hac1p transcription factor regulates the unfolded protein response in Saccharomyces cerevisiae. Mol. Biol. Cell 18, 426–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schuldiner M., Collins S. R., Weissman J. S., Krogan N. J. (2006) Quantitative genetic analysis in Saccharomyces cerevisiae using epistatic miniarray profiles (E-MAPs) and its application to chromatin functions. Methods 40, 344–352 [DOI] [PubMed] [Google Scholar]
- 41. Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D., Pagé N., Robinson M., Raghibizadeh S., Hogue C. W., Bussey H., Andrews B., Tyers M., Boone C. (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294, 2364–2368 [DOI] [PubMed] [Google Scholar]
- 42. Collins S. R., Miller K. M., Maas N. L., Roguev A., Fillingham J., Chu C. S., Schuldiner M., Gebbia M., Recht J., Shales M., Ding H., Xu H., Han J., Ingvarsdottir K., Cheng B., Andrews B., Boone C., Berger S. L., Hieter P., Zhang Z., Brown G. W., Ingles C. J., Emili A., Allis C. D., Toczyski D. P., Weissman J. S., Greenblatt J. F., Krogan N. J. (2007) Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446, 806–810 [DOI] [PubMed] [Google Scholar]
- 43. West M. L., Corden J. L. (1995) Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics 140, 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nordick K., Hoffman M. G., Betz J. L., Jaehning J. A. (2008) Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryot. Cell 7, 1158–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Biswas D., Dutta-Biswas R., Mitra D., Shibata Y., Strahl B. D., Formosa T., Stillman D. J. (2006) Opposing roles for Set2 and yFACT in regulating TBP binding at promoters. EMBO J. 25, 4479–4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chu Y., Sutton A., Sternglanz R., Prelich G. (2006) The BUR1 cyclin-dependent protein kinase is required for the normal pattern of histone methylation by SET2. Mol. Cell. Biol. 26, 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lenstra T. L., Benschop J. J., Kim T., Schulze J. M., Brabers N. A., Margaritis T., van de Pasch L. A., van Heesch S. A., Brok M. O., Groot Koerkamp M. J., Ko C. W., van Leenen D., Sameith K., van Hooff S. R., Lijnzaad P., Kemmeren P., Hentrich T., Kobor M. S., Buratowski S., Holstege F. C. (2011) The specificity and topology of chromatin interaction pathways in yeast. Mol. Cell 42, 536–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shivaswamy S., Iyer V. R. (2008) Stress-dependent dynamics of global chromatin remodeling in yeast: dual role for SWI/SNF in the heat shock stress response. Mol. Cell. Biol. 28, 2221–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chu Y., Simic R., Warner M. H., Arndt K. M., Prelich G. (2007) Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. EMBO J. 26, 4646–4656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhou K., Kuo W. H., Fillingham J., Greenblatt J. F. (2009) Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc. Natl. Acad. Sci. U.S.A. 106, 6956–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Belle A., Tanay A., Bitincka L., Shamir R., O'Shea E. K. (2006) Quantification of protein half-lives in the budding yeast proteome. Proc. Natl. Acad. Sci. U.S.A. 103, 13004–13009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim T., Buratowski S. (2009) Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell 137, 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Taverna S. D., Ilin S., Rogers R. S., Tanny J. C., Lavender H., Li H., Baker L., Boyle J., Blair L. P., Chait B. T., Patel D. J., Aitchison J. D., Tackett A. J., Allis C. D. (2006) Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol. Cell 24, 785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.