Abstract
Comparative analysis of genomic sequences is becoming a standard technique for studying gene regulation. However, only a limited number of tools are currently available for the analysis of multiple genomic sequences. An extensive data set for the testing and training of such tools is provided by the SCL gene locus. Here we have expanded the data set to eight vertebrate species by sequencing the dog SCL locus and by annotating the dog and rat SCL loci. To provide a resource for the bioinformatics community, all SCL sequences and functional annotations, comprising a collation of the extensive experimental evidence pertaining to SCL regulation, have been made available via a Web server. A Web interface to new tools specifically designed for the display and analysis of multiple sequence alignments was also implemented. The unique SCL data set and new sequence comparison tools allowed us to perform a rigorous examination of the true benefits of multiple sequence comparisons. We demonstrate that multiple sequence alignments are, overall, superior to pairwise alignments for identification of mammalian regulatory regions. In the search for individual transcription factor binding sites, multiple alignments markedly increase the signal-to-noise ratio compared to pairwise alignments.
Among the vertebrates alone, whole-genome sequences have been determined for human, mouse, rat, and two pufferfish species. Sequencing has started for chimpanzee, chicken, frog (Xenopus tropicalis), and zebrafish, and projects to sequence the genomes of dog and cow are due to commence in the near future. Faced with the task of identifying putative regulatory elements of a gene, it is already a simple matter to obtain homologous human and mouse genomic sequences for the locus of interest, perform an alignment with one of a variety of algorithms, and examine in more detail those regions that are conserved between the two species. Within these regions, phylogenetic footprinting (Tagle et al. 1988; Gumucio et al. 1993; Shelton et al. 1997) can be used to determine potential transcription factor binding sites. Sequence-specific transcription factor binding sites are much more likely to occur within blocks of complete identity than outside such blocks (Wasserman et al. 2000).
Alignments of more than two species (multiple alignments), separated by similar evolutionary distances, may offer advantages over pairwise alignments. Provided that the evolutionary distances between the chosen species are sufficiently small, problems of detecting only a subset of regulatory elements with distant comparisons (Miles et al. 1998; Göttgens et al. 2000, 2002a; Cioffi et al. 2001; Flint et al. 2001; Gilligan et al. 2002) are minimized. Moreover, having more than two species adds substantial resolving power, because each lineage diverges independently after separation from a common ancestor, resulting in evolutionary distances that are effectively additive (Stojanovic et al. 1999). Thus, the likelihood of residual similarities in nonselected regions is markedly reduced. This additive effect was recently demonstrated with identification of gene regulatory elements by multiple sequence comparisons among primates (Boffelli et al. 2003). However, despite the significance of the latter study, multiple alignments involving eutherian mammals other than primates are likely to prove important. If the approach is to be universally applicable, one is limited to sequence comparisons involving those species for which whole-genome sequences are available. Furthermore, murine models are likely to remain the prime in vivo experimental system for studying mammalian gene expression. Finally, statistical modeling has suggested that comparisons across a wide range of mammalian sequences are likely to provide the most useful resolution at the single-nucleotide level (Cooper et al. 2003).
Given the promise of multiple sequence comparisons, there is a relative paucity of tools for the analysis of multiple genomic sequences (Miller 2001). Essential to their design are data sets that can be used to train and evaluate algorithms. One prime model for such a data set is the locus of the basic helix-loop-helix (bHLH) stem cell leukemia (SCL) gene. There are several reasons for this. Firstly, SCL is a pivotal regulator of hematopoiesis and vascular development (for review, see Begley and Green 1999), with a pattern of expression that is highly conserved throughout vertebrate evolution, from mammals to teleost fish. Second, the SCL locus is not too large—there is evidence that all elements necessary for its transcriptional pattern are contained in a region spanning approximately 54 kb (Barton et al. 2001; Sinclair et al. 2002). Third, there is a wealth of experimental data on the transcriptional regulation of human and mouse SCL loci (Begley and Green 1999, and references therein; Fordham et al. 1999; Sanchez et al. 1999, 2001; Sinclair et al. 1999, 2002; Göttgens et al. 2000, 2001, 2002a,b; Barton et al. 2001). Fourth, SCL sequences are currently available for six species: zebrafish, pufferfish (Fugu and Tetraodon), chicken, mouse, and human (Göttgens et al. 2000, 2002a; Barton et al. 2001). Finally, SCL is not flanked by paralogous genes, unlike, for example, the hox, interleukin, and globin loci. Therefore, characterized functional elements do not regulate multiple genes. Thus, inferring the functional significance of conserved elements may be more straightforward.
This article describes the addition of dog and rat sequences to the SCL data set and the development of a Web server that provides a highly annotated testing and training set as a resource for the bioinformatics community. To examine the practical utility of multiple alignments, all combinations of mammalian SCL alignment—six pairwise, four three-way, and one four-way alignment—were generated. These alignments were compared using the annotations and with new tools designed to analyze multiple alignments, made available to run via the server. Multiple alignments, and particularly the four-way alignment, markedly narrowed the search for putative transcription factor binding sites in the locus, with minimal loss of sensitivity.
RESULTS
Development of the SCL Data Set as a Genomics Research Resource
The first goal was to extend the SCL data set. A single clone of 169,460 bp in length, containing the entire dog SIL, SCL, and MAP17 genes, was therefore isolated and fully sequenced (see Methods). The locations of noncoding exons were defined by RT-PCR using RNA from two dog cell lines. Exons 1a and 1b were spliced directly to exon 3, suggesting that there are no dog homologs to human exons 2a and 2b. The rat SCL sequence was obtained from the Berkeley Genome Pipeline (http://pipeline.lbl.gov/rat) and fully annotated. The extracted region extended from within the SIL gene to approximately 2 kb downstream of MAP17. The choice of upstream and downstream boundaries for the contig was based on the minimum region of conserved synteny previously reported (Barton et al. 2001) and the observation that a human YAC containing this region could rescue SCL -/- mouse embryos from the normally lethal phenotype (Sinclair et al. 2002).
Algorithms for predicting regulatory regions benefit considerably from training and testing data sets with experimentally confirmed functional annotations (Miller 2001; Elnitski et al. 2003). The SCL data set now includes eight vertebrate species with a wealth of experimental information on regulatory elements within the locus. A Web server that provides access to the SCL sequences together with annotations of functional noncoding regions has therefore been implemented (http://hscl.cimr.cam.ac.uk/genomic_datasets.html). From here, there are three main links. One is to the SCL sequences themselves. A second is to annotations of these sequences in General Feature Format (GFF) files (see http://www.sanger.ac.uk/Software/formats/GFF/). Each GFF file details the locations of coding and noncoding exons, which have been determined experimentally in all species except rat, and repeat regions. The known SCL regulatory regions have been defined experimentally in the murine locus. These have therefore been annotated in the mouse GFF file, and the equivalent regions are defined in the GFF files for the other mammals. The final link is to a table (Table 1), which serves as an index to detailed descriptions of these known functional regions. The nomenclature of the regions refers to their distance from promoter 1a in the murine SCL locus. The extent of all regions except the +23 kb enhancer has been experimentally confirmed to be in an open chromatin configuration. The core region with full activity is described, together with the positions of transcription factor binding sites where these have been experimentally determined.
Table 1.
Functional Annotation of the Mouse SCL Locus
| Region | Open chromatin (approx. location) | Minimally defined active element | Transcription factor binding site |
|---|---|---|---|
| –8/–9 kb | 20265–20665 | 18884–22017 | Unknown |
| 20765–21165 | |||
| –4 kb | 24965–25445 | 25095–25488 | Ets family 25213 |
| Ets family 25262 | |||
| Ets family 25361 | |||
| Ets family 25369 | |||
| Promoter 1a | 28965–29265 | 29044–29257 | C/EBP 29119 |
| Ap1 29130 | |||
| Gata1 29162 | |||
| Sp1 29168 | |||
| Gata1 29194 | |||
| Promoter 1b | 29425–29585 | 29440–29558 | Sp1/3 29472 |
| Ets family 29494 | |||
| Ets family 29498 | |||
| Ets family 29502 | |||
| C/EBP 29520 | |||
| +1 kb | 29865–30165 | 29558–30376 | Unknown |
| +3 kb | 31865–32065 | 30377–36071 | Unknown |
| +7 kb | 35625–36065 | 34510–37251 | Unknown |
| +18 kb | 47265–47595 | 47299–47728 | Unknown |
| +19 kb | 48045–48345 | 47728–48369 | AATAA 48210 |
| Ets family 48217 | |||
| Ets family 48237 | |||
| Gata2 48262 | |||
| +23 kb | Unknown | 50934–52609 | Unknown |
The names of the regulatory regions, approximate regions of open chromatin, the minimally defined active element, and known transcription factor binding sites are indicated. On the Web server, entries in the first column serve as hypertext links to detailed information on the individual regions.
Within the Web site, Table 1 contains hyperlinks for each regulatory region, each of which leads to a more detailed review of underlying experimental evidence. These functional data include (where determined) methylation status, activity in transient and stable transfection assays, activity in transgenic mice, and the effect of mutating known transcription factor binding sites in the various experimental systems. All transcription factor binding sites known to be involved in the regulation of SCL are preserved in alignments of the four mammalian species (see below). These binding sites thus represent rigorous sensitivity criteria for training or testing algorithms concerned with genomic sequence alignment and binding site prediction. Therefore, for each region, known binding sites are indicated on an alignment of the mammalian sequences in that region. Finally, references to the original publications are given. The Web site will continue to be updated as more data on SCL regulation emerge.
Use of Multiple Mammalian Alignments to Identify SCL Regulatory Regions
It has been proposed that multiple mammalian sequences represent the best comparison for detecting functional regulatory regions (Frazer et al. 2003). Statistical modeling has suggested that they also provide a more useful resolution at the nucleotide level than pairwise alignments (Cooper et al. 2003). The new data set provided the opportunity to address these issues by comparing the abilities of multiple and pairwise alignments of mammalian SCL sequences to detect regulatory elements. To this end, the SynPlot module (Göttgens et al. 2000, 2001) was modified to process multiple alignments and to run via the Web server (http://hscl.cimr.cam.ac.uk/syn_plot.html). SynPlot displays a plot of alignment score within a sliding window, which moves across the alignment. To accommodate multiple alignments, the alignment score is generated by the sum of pairs method (see Methods) and expressed as the proportion of the maximum attainable score in the window. All possible two-, three-, and four-way alignments of human, dog, mouse, and rat SCL were generated and a SynPlot produced. In addition to exons and repeats, all known regions of open chromatin were shaded. All such sites in the SCL locus have transcriptional regulatory activity, or map to larger regions with such activity, in transfection and/or transgenic assays. In addition, the +23 kb enhancer (outside mapped regions of open chromatin) was shaded.
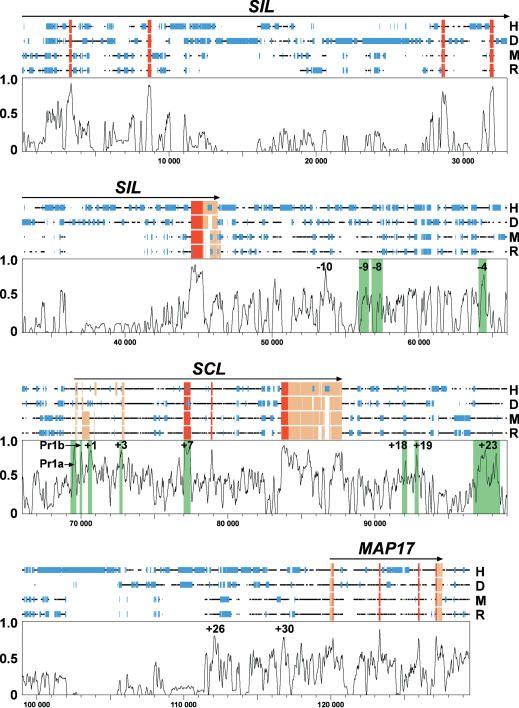
Figure 1 is the SynPlot for the four-way alignment (SynPlots for all the other alignments are available as Supplemental data at www.genome.org and hscl.cimr.cam.ac.uk/supplementary_data.html). As expected, coding exons are highly conserved, each represented by a peak with 77% of the maximum possible alignment score or greater. The regions of open chromatin and the +23 kb region—known functional regions, as depicted in Table 1—are shaded in green. They are also highly conserved between the four species, the only exceptions being the -9 kb and -8 kb regions (71% and 64% of maximum score, respectively). In addition, there are a few other noncoding regions with a high degree of sequence conservation, most notably the -10 kb, +26 kb, and +30 kb regions indicated (92%, 80%, and 83% of maximum score, respectively). The pattern of peak distribution was similar in the SynPlots of the other mammalian SCL alignments (Supplemental data).
Figure 1.
A long-range four-way mammalian alignment is capable of detecting all known SCL regulatory regions. A SynPlot representation of a human/dog/mouse/rat alignment of the SCL locus. Numbers on the horizontal axis do not refer to any one of the sequences in the alignment, but instead represent distance in nucleotides from the beginning of the aligned file (i.e., gaps in the alignment are counted). Numbers on the vertical axis represent an alignment score in a 100-bp window moved by 25-bp increments across the entire alignment. The horizontal black lines above the profile represent the human (H), dog (D), mouse (M), and rat (R) sequences and illustrate the position of gaps introduced to permit optimum alignment. Coding and noncoding exons are demonstrated by red and brown boxes, respectively, and repeats are illustrated by the smaller blue boxes. Regions of open chromatin and the +23 kb enhancer are shaded in green over the plot. Peaks of conserved sequence can be seen within each known regulatory region. Three well conserved peaks in regions of unknown function (-10 kb, +26 kb, and +30 kb) are also indicated. Pr1a, promoter 1a; Pr1b, promoter 1b.
Noncoding peaks were divided into two classes: those corresponding to regions of open chromatin and the +23 kb region (known regulatory regions), and those representing all other noncoding sequence conservation. A number of analyses were then performed to compare the abilities of the various two-, three-, and four-way alignments to selectively identify SCL regulatory elements and transcription factor binding sites. The methods and a detailed discussion of the results are available from http://hscl.cimr.cam.ac.uk/additional_info.html. The tools used in the analysis were designed to run via a user-friendly Web interface (http://hscl.cimr.cam.ac.uk/genomic_tools.html), and form an additional resource for biologists working in the field of gene regulation. The results of the analyses are summarized below.
In terms of identifying regulatory regions of the order of a few hundred base pairs in length, multiple alignments were, on the whole, more selective than pairwise alignments, with no loss of sensitivity. The four-way alignment was the most specific. Some pairwise alignments performed well, but this could not be predicted from the evolutionary distances separating the species. In terms of individual transcription factor binding sites known to regulate SCL, all were conserved in all alignments. When the alignments are searched for other putative conserved transcription factor binding sites, fewer are found in three-way than pairwise alignments, and the fewest are found in the four-way alignment, as would be expected. However, putative sites become concentrated within the known regulatory regions as the number of species is increased, suggesting a genuine increase in the signal-to-noise ratio of multiple over pairwise mammalian alignments in the search for transcription factor binding sites.
DISCUSSION
Visual Representation of Multiple Alignments
Visual interfaces to alignments are required to gain an appreciation of the quality of the alignment and to identify regions of sequence conservation, which may represent functional elements. Effective representation of pairwise alignments can be achieved by using a sliding window to calculate the percentage identities across the alignment, which can then be plotted, for example, PipMaker (http://bio.cse.psu.edu/pipmaker; Schwartz et al. 2000), Vista (http://www-gsd.lbl.gov/vista/vista_cite.html; Loots et al. 2000), and the original version of SynPlot (Göttgens et al. 2000). However, percentage identity cannot be calculated for more than two species simultaneously. Therefore, to deal with multiple alignments, Vista and MultiPipMaker (Schwartz et al. 2003) construct a series of successive pairwise plots comparing various species in an alignment with a reference sequence. This may be the most efficient way of visualizing very large-scale alignments, for example, whole genomes. However, there is necessarily a loss of information: Differences in genomic organisation cannot be seen; misalignments are often not clear; and prioritizing peaks of sequence conservation can be difficult. Thus, we felt that for examining multiple alignments of single gene loci, an alternative approach could be useful.
SynPlot was modified to process multiple alignments, generating an alignment score, to replace percentage identity, via a sliding window. The alignment score is calculated by the sum of pairs method (see Methods). This allows all the information from a multiple alignment to be considered simultaneously, rather than requiring a reference sequence. It may therefore better delineate the very highest peaks of sequence conservation. It does not yet take into account the evolutionary relationships between the species involved, but this approach may be implemented in future versions. Another advantage of the SynPlot approach is that all features of all the sequences can be visualized above the plot, which is important when the genomic organization is not identical.
SCL Data Set
One of the many uses of comparative genomic sequence analysis is the identification of gene regulatory elements. Central to its success are algorithms that are capable of selectively identifying biologically meaningful conserved regions. In particular, with the increasing availability of whole-genome sequences, tools for analyzing multiple sequences are likely to gain prominence. However, such algorithms are in their infancy. Their success will depend on robust data sets (Miller 2001).
The SCL locus is a prime candidate for such a testing or training data set, and its use has precedents. It was used in the training of the LAGAN algorithm and in the testing of the Chaos-Dialign combination (Brudno et al. 2003, Brudno et al. 2004). The data set has now been extended to eight species by the complete sequencing and annotation of dog SCL and annotation of rat SCL. By implementing a Web server, convenient access to the full data set is now provided. The sequences can be easily obtained from a single source. Also supplied are complete functional annotations. There are other multiple mammalian genomic sequence data sets available (e.g., Stojanovic et al. 1999; Thomas et al. 2003). However, these loci have not been subject to the same degree of in-depth analysis (including extensive transgenic experiments) as the SCL locus. In the case of the CFTR data set, the length of the locus means that there are large stretches of sequence that have not been subjected to any functional studies. As a result, at 63% sensitivity, 49 times as many conserved noncoding regions without known function than those with known function are identified in the CFTR locus (Thomas et al. 2003). In the SCL data set, at 63% sensitivity, three times fewer conserved noncoding regions without known function than those with known function are identified. The β-globin data set (Stojanovic et al. 1999) is weakened by incomplete genomic coverage and the complexities of interpreting comparative sequence analyses in a locus containing clusters of paralogous genes. Neither the β-globin data set nor the CFTR data set offers a single resource with a comprehensive review of the underlying experimental data to support the definition of minimally defined regulatory elements and the transcription factor binding sites contained therein.
The comprehensive functional annotation of the SCL data set and the successful alignment of all known functional elements and binding sites allow strict performance criteria to be defined for multiple species analysis tools. These criteria can be used to set parameters during algorithm training or as a gold standard for sensitivity in evaluation. The detailed experimental work underlying functional annotation of the SCL locus allowed us to test the performance of pairwise and multiple alignments in their identification of regulatory elements with a degree of rigor that was probably hitherto impossible. Multiple alignments were superior to pairwise alignments in the selective identification of both regulatory elements a few hundred base pairs in length and individual transcription factor binding sites. The four-way alignment performed better than any of the three-way alignments. This suggests that increasing the number of species progressively increases the signal-to-noise ratio of the alignment. It is important to note that our approach in these analyses has been empirical. Thus, although it would be interesting to predict how much more useful multiple alignments would become as the number of mammalian species is increased (above the four presented herein), the lack of available sequence data for the SCL locus in other mammalian species precludes this. We did not use the chicken or fish SCL loci in our analysis, as most of the mammalian regulatory elements cannot be found in these sequences (Göttgens et al. 2000, 2002a). However, they are included in the data set, as comparisons of more divergent sequences may be useful in addressing other important questions, such as how similar patterns of gene expression can be achieved with a different organization of regulatory elements.
Functional annotation of the noncoding portion of the human genome represents the next major milestone of the Human Genome Project (Collins et al. 2003). The ENCODE program, to be launched later this year, has been set up with the objective of developing the necessary functional and computational resources. It is anticipated that the wealth of functional information, combined with the comparative sequence data, will make the SCL locus a valuable model for this next phase of genomics research.
METHODS
Production of Probes for Library Screening
The human SCL bHLH probe was amplified by PCR from genomic DNA, using standard procedures. Primers were CCATTCTCCTAACTCTTGTCCTC and CCAGCAGCCTAAGAAC GCC, and the annealing temperature was 55°C. SIL and MAP17 whole cDNA (available from the authors on request) were used as probes for these genes.
Sequencing, Annotation, and Alignments of SCL Loci
To isolate a clone covering the SCL locus, the RPCI canine BAC library 81 (Li et al. 1999) was screened using the SCL bHLH probe as described (Chapman et al. 2003). DNA was sequenced from a single dog clone, RP81-70I6, containing SIL, SCL, and MAP17 as described elsewhere (Lander et al. 2001). Briefly, 2–4 kb of sheared fragments were subcloned into pUC19. The sequences were assembled with 1500 sequence reads. During the finishing process, difficult gaps were subjected to transposon libraries. The locations of coding exons were predicted with BLAST searches (http://www.ncbi.nlm.nih.gov/BLAST/; Altschul et al. 1990). The coding frame was maintained in each, and there was no ambiguity in the start or stop site of the various exons. RT-PCR from two dog cell lines, D064 (bone marrow) and IIIG7 (peripheral blood), was used to locate noncoding exons. Rat SCL sequence was extracted from the Berkeley Genome Pipeline (http://pipeline.lbl.gov/rat/). Pairwise BLAST analysis with mouse sequence was used to predict both noncoding and coding exon locations. Human, dog, mouse, and rat SCL sequences were trimmed from about 2 kb upstream of the fifth last exon of SIL to about 2 kb downstream of the last exon of MAP17. Repeats were masked using RepeatMasker (http://repeatmasker.genome.washington.edu/cgi-bin/RepeatMasker) on slow setting. The LAGAN server (http://lagan.stanford.edu) was used to create all possible two-, three-, and four-way alignments using these sequences.
SynPlot
SynPlot is a modification of the SynPlot Perl module (Göttgens et al. 2000), and is now implemented on a worldwide Web server (http://hscl.cimr.cam.ac.uk/syn_plot.html). It runs as described previously, with the exception that more than two sequences can be compared simultaneously to generate a single identity plot. To enable this, the scoring mechanism employs the sum of pairs method. Each base position within a user-defined window is examined sequentially, and the base in each species is compared with those with which it is aligned in the other species. For every matching base, 1 is scored. Therefore, the maximum score at any base position is 1 in a two-way alignment, 3 in a three-way alignment, 6 in a four-way alignment, and so on. The scores are summed for the entire window, and the alignment score is given as a fraction of maximum total possible score (window length multiplied by maximum possible score at each base position). Scores will thus range from 0 to 1.0 for each window. The window is then moved by user-defined increments along the alignment. Gaps introduced into individual sequences by the alignment are considered in the scoring; that is, they contribute to the length of the window, but not to the score. Features can be designed by the user, described in a configuration file, and annotated in GFF files. These are then drawn on an individual line for each sequence above the plot. Gaps introduced into individual sequences by the alignment will also be shown above the plot. SynPlot was designed to process global rather than local alignments; that is, it assumes that all the bases of each sequence have been used in the alignment. For this reason, gaps in the alignment will mean that the numbering on the x-axis refers to the position within the global alignment rather than in any one of the sequences. PeakExtractor is therefore provided on the Web server to enable the alignment in regions of interest to be examined (http://hscl.cimr.cam.ac.uk/peak_extractor.html; see supplementary online material at http://hscl.cimr.cam.ac.uk/additional_info.html for details). The output of the stand-alone SynPlot module is a Postscript file. The Web server uses the ps2pdf package to perform a conversion to PDFif required. For the purposes of this paper, SynPlots were generated for all the two-, three-, and four-way mammalian alignments using a 100- bp window moving at 25-bp increments. Three features in the configuration files were enabled: exons, coding sequence, and repeat regions.
Acknowledgments
This work was supported by the Wellcome Trust, the Sackler Studentship, the Leukaemia Research Fund, and the Cambridge MIT Institute. We thank Dr. R. Huss for providing two dog cell lines, Dr. E. Wingender and colleagues for use of the TRANSFAC database, R. Li and P. de Jong for the canine BAC library, and the Human Genome Mapping Project for supplying BAC library filters and positive clones. We are particularly indebted to sequencing teams 40 and 47 at the Wellcome Trust Sanger Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1759004. Article published online before print in January 2004.
Footnotes
[Supplemental data is available at www.genome.org and http://hscl.cimr.cam.ac.uk/supplementary_data.html. DNA sequence as described in the paper has been deposited in the GenBank database under accession no. AL731652. The following individuals kindly supplied reagents, samples, or unpublished information as indicated in the paper: R. Li, P. de Jong, and R. Huss.]
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. [DOI] [PubMed] [Google Scholar]
- Barton, L.M., Göttgens, B., Gering, M., Gilbert, J.G., Grafham, D., Rogers, J., Bentley, D., Patient, R., and Green, A.R. 2001. Regulation of the stem cell leukemia (SCL) gene: A tale of two fishes. Proc. Natl. Acad. Sci. 98: 6747-6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley, C.G. and Green, A.R. 1999. The SCL gene: From case report to critical hematopoietic regulator. Blood 93: 2760-2770. [PubMed] [Google Scholar]
- Boffelli, D., McAuliffe, J., Ovcharenko, D., Lewis, K.D., Ovcharenko, I., Pachter, L., and Rubin, E.M. 2003. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299: 1391-1394. [DOI] [PubMed] [Google Scholar]
- Brudno, M., Do, C.B., Cooper, G.M., Kim, M.F., Davydov, E., Green, E.D., Sidow, A., and Batzoglou, S. 2003. LAGAN and Multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13: 721-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno, M., Chapman, M., Göttgens, B., Batzoglou, S., and Morgenstern, B. 2004. Fast and sensitive multiple alignment of large genomic sequences. BMC Bioinformatics (in press). [DOI] [PMC free article] [PubMed]
- Chapman, M.A., Charchar, F.J., Kinston, S., Bird, C.P., Grafham, D., Rogers, J., Grutzner, F., Marshall Graves, J.A., Green, A.R., and Gottgens, B. 2003. Comparative and functional analyses of LYL1 loci establish marsupial sequences as a model for phylogenetic footprinting. Genomics 81: 249-259. [DOI] [PubMed] [Google Scholar]
- Cioffi, C.C., Middleton, D.L., Wilson, M.R., Miller, N.W., Clem, L.W., and Warr, G.W. 2001. An IgH enhancer that drives transcription through basic helix-loop-helix and Oct transcription factor binding motifs. Functional analysis of the E(mu)3′ enhancer of the catfish. J. Biol. Chem. 276: 27825-27830. [DOI] [PubMed] [Google Scholar]
- Collins, F.S., Green, E.D., Guttmacher, A.E., and Guyer, M.S. 2003. A vision for the future of genomics research. Nature 422: 835-847. [DOI] [PubMed] [Google Scholar]
- Cooper, G.M., Brudno, M., Green, E.D., Batzoglou, S., and Sidow, A. 2003. Quantitative estimates of sequence divergence for comparative analyses of mammalian genomes. Genome Res. 13: 813-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnitski, L., Hardison, R.C., Li, J., Yang, S., Kolbe, D., Eswara, P., O'Connor, M.J., Schwartz, S., Miller, W., and Chiaromonte, F. 2003. Distinguishing regulatory DNA from neutral sites. Genome Res. 13: 64-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint, J., Tufarelli, C., Peden, J., Clark, K., Daniels, R.J., Hardison, R., Miller, W., Philipsen, S., Tan-Un, K.C., McMorrow, T., et al. 2001. Comparative genome analysis delimits a chromosomal domain and identifies key regulatory elements in the α lobin cluster. Hum. Mol. Genet. 10: 371-382. [DOI] [PubMed] [Google Scholar]
- Fordham, J.L., Göttgens, B., McLaughlin, F., and Green, A.R. 1999. Chromatin structure and transcriptional regulation of the stem cell leukaemia (SCL) gene in mast cells. Leukemia 13: 750-759. [DOI] [PubMed] [Google Scholar]
- Frazer, K.A., Elnitski, L., Church, D.M., Dubchak, I., and Hardison, R.C. 2003. Cross-species sequence comparisons: A review of methods and available resources. Genome Res. 13: 1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilligan, P., Brenner, S., and Venkatesh, B. 2002. Fugu and human sequence comparison identifies novel human genes and conserved non-coding sequences. Gene 294: 35. [DOI] [PubMed] [Google Scholar]
- Göttgens, B., Barton, L.M., Gilbert, J.G., Bench, A.J., Sanchez, M.J., Bahn, S., Mistry, S., Grafham, D., McMurray, A., Vaudin, M., et al. 2000. Analysis of vertebrate SCL loci identifies conserved enhancers. Nat. Biotechnol. 18: 181-186. [DOI] [PubMed] [Google Scholar]
- Göttgens, B., Gilbert, J.G., Barton, L.M., Grafham, D., Rogers, J., Bentley, D.R., and Green, A.R. 2001. Long-range comparison of human and mouse SCL loci: Localized regions of sensitivity to restriction endonucleases correspond precisely with peaks of conserved noncoding sequences. Genome Res. 11: 87-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttgens, B., Barton, L.M., Chapman, M.A., Sinclair, A.M., Knudsen, B., Grafham, D., Gilbert, J.G., Rogers, J., Bentley, D.R., and Green, A.R. 2002a. Transcriptional regulation of the stem cell leukemia gene (SCL)—Comparative analysis of five vertebrate SCL loci. Genome Res. 12: 749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttgens, B., Nastos, A., Kinston, S., Piltz, S., Delabesse, E.C., Stanley, M., Sanchez, M.J., Ciau-Uitz, A., Patient, R., and Green, A.R. 2002b. Establishing the transcriptional programme for blood: The SCL stem cell enhancer is regulated by a multiprotein complex containing Ets and GATA factors. EMBO J. 21: 3039-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumucio, D.L., Shelton, D.A., Bailey, W.J., Slightom, J.L., and Goodman, M. 1993. Phylogenetic footprinting reveals unexpected complexity in trans factor binding upstream from the ε-globin gene. Proc. Natl. Acad. Sci. 90: 6018-6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Li, R., Mignot, E., Faraco, J., Kadotani, H., Cantanese, J., Zhao, B., Lin, X., Hinton, L., Ostrander, E.A., Patterson, D.F., et al. 1999. Construction and characterization of an eightfold redundant dog genomic bacterial artificial chromosome library. Genomics 58: 9-17. [DOI] [PubMed] [Google Scholar]
- Loots, G.G., Locksley, R.M., Blankespoor, C.M., Wang, Z.E., Miller, W., Rubin, E.M., and Frazer, K.A. 2000. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288: 136-140. [DOI] [PubMed] [Google Scholar]
- Miles, C., Elgar, G., Coles, E., Kleinjan, D.J., van Heyningen, V., and Hastie, N. 1998. Complete sequencing of the Fugu WAGR region from WT1 to PAX6: Dramatic compaction and conservation of synteny with human chromosome 11p13. Proc. Natl. Acad. Sci. 95: 13068-13072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, W. 2001. Comparison of genomic DNA sequences: Solved and unsolved problems. Bioinformatics 17: 391-397. [DOI] [PubMed] [Google Scholar]
- Sanchez, M., Göttgens, B., Sinclair, A.M., Stanley, M., Begley, C.G., Hunter, S., and Green, A.R. 1999. An SCL 3′ enhancer targets developing endothelium together with embryonic and adult haematopoietic progenitors. Development 126: 3891-3904. [DOI] [PubMed] [Google Scholar]
- Sanchez, M.J., Bockamp, E.O., Miller, J., Gambardella, L., and Green, A.R. 2001. Selective rescue of early haematopoietic progenitors in Scl(-/-) mice by expressing Scl under the control of a stem cell enhancer. Development 128: 4815-4827. [DOI] [PubMed] [Google Scholar]
- Schwartz, S., Zhang, Z., Frazer, K.A., Smit, A., Riemer, C., Bouck, J., Gibbs, R., Hardison, R., and Miller, W. 2000. PipMaker—A web server for aligning two genomic DNA sequences. Genome Res. 10: 577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S., Elnitski, L., Li, M., Weirauch, M., Riemer, C., Smit, A., Green, E.D., Hardison, R.C., and Miller, W. 2003. MultiPipMaker and supporting tools: Alignments and analysis of multiple genomic DNA sequences. Nucleic Acids Res. 31: 3518-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton, D.A., Stegman, L., Hardison, R., Miller, W., Bock, J.H., Slightom, J.L., Goodman, M., and Gumucio, D.L. 1997. Phylogenetic footprinting of hypersensitive site 3 of the β-globin locus control region. Blood 89: 3457-3469. [PubMed] [Google Scholar]
- Sinclair, A.M., Göttgens, B., Barton, L.M., Stanley, M.L., Pardanaud, L., Klaine, M., Gering, M., Bahn, S., Sanchez, M., Bench, A.J., et al. 1999. Distinct 5′ SCL enhancers direct transcription to developing brain, spinal cord, and endothelium: Neural expression is mediated by GATA factor binding sites. Dev. Biol. 209: 128-142. [DOI] [PubMed] [Google Scholar]
- Sinclair, A.M., Bench, A.J., Bloor, A.J., Li, J., Göttgens, B., Stanley, M.L., Miller, J., Piltz, S., Hunter, S., Nacheva, E.P., et al. 2002. Rescue of the lethal scl(-/-) phenotype by the human SCL locus. Blood 99: 3931-3938. [DOI] [PubMed] [Google Scholar]
- Stojanovic, N., Florea, L., Riemer, C., Gumucio, D., Slightom, J., Goodman, M., Miller, W., and Hardison, R. 1999. Comparison of five methods for finding conserved sequences in multiple alignments of gene regulatory regions. Nucleic Acids Res. 27: 3899-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagle, D.A., Koop, B.F., Goodman, M., Slightom, J.L., Hess, D.L., and Jones, R.T. 1988. Embryonic ε and γ globin genes of a prosimian primate (Galago crassicaudatus). Nucleotide and amino acid sequences, developmental regulation and phylogenetic footprints. J. Mol. Biol. 203: 439-455. [DOI] [PubMed] [Google Scholar]
- Thomas, J.W., Touchman, J.W., Blakesley, R.W., Bouffard, G.G., Beckstrom-Sternberg, S.M., Margulies, E.H., Blanchette, M., Siepel, A.C., Thomas, P.J., McDowell, J.C., et al. 2003. Comparative analyses of multi-species sequences from targeted genomic regions. Nature 424: 788-793. [DOI] [PubMed] [Google Scholar]
- Wasserman, W.W., Palumbo, M., Thompson, W., Fickett, J.W., and Lawrence, C.E. 2000. Human-mouse genome comparisons to locate regulatory sites. Nat. Genet. 26: 225-228. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://bio.cse.psu.edu/pipmaker; PipMaker home page.
- http://lagan.stanford.edu; LAGAN home page.
- http://pipeline.lbl.gov/rat/; Berkeley Genome Pipeline.
- http://repeatmasker.genome.washington.edu/cgi-bin/RepeatMasker; RepeatMasker server.
- http://www-gsd.lbl.gov/vista/; Vista home page.
- http://www.ncbi.nlm.nih.gov/BLAST/; BLAST home page
- http://www.sanger.ac.uk/Software/formats/GFF/; GFF format.
- http://hscl.cimr.cam.ac.uk/supplementary_data.html; Supplemental data for this article.
- http://hscl.cimr.cam.ac.uk/genomic_datasets.html; Data sets are described in this article.