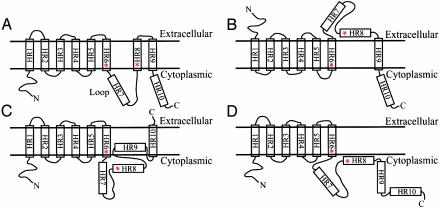
Mutations of presenilin 1 (PS1) account for up to 60% of early-onset familial Alzheimer's disease (AD) (1). Because PS1 is a polytopic membrane protein, deciphering its topology is crucial to understanding its important functions. Hydropathy analysis of the PS1 primary amino acid sequence identified 10 hydrophobic regions (HR) [see figure 1A of Dewji et al. (2) in this issue of PNAS]. An eight transmembrane domain (TM) topology, where only 8 of 10 HRs traverse membranes with amino (N) and carboxy (C) tails facing the cytoplasm, has been proposed and is generally accepted by most investigators in this field (Fig. 1 A) (3, 4). In contrast, Dewji and Singer (5) have suggested an alternative in which PS1 has seven TMs with the N tail facing the extracellular space and the C tail facing the cytoplasm (Fig. 1B). This view is now confirmed in studies employing immunofluorescence microscopy with monoclonal antibodies specific either to the N tail or the loop, and polyclonal antipeptide antibodies to the C tail of PS1. The authors offer a provocative suggestion that PS1 may be a member of the superfamily of seven-TM heterotrimeric G protein-coupled receptors (2).
Fig. 1.
Suggested topologies of PS1. The conserved active site aspartate residues D257 and D385 on HR6 and HR8, respectively, are marked with asterisks. The endoproteolytic sites of PS1 are located in HR7, resulting in N- and C-terminal fragments. According to an eight-TM topology (A), the loop consists of residues between HR6 and HR8.
PS1 is believed to be a catalytic subunit of γ-secretase (see ref. 6 for review). A slightly altered proteolytic activity of this enzyme, in concert with β-secretase, increases production of a more aggregation-prone peptide, Aβ42, rather than Aβ40 from a type I membrane protein, amyloid precursor protein (APP) (6). This increased level of Aβ42 fragments induces formation of plaques in the brains of AD patients (6). Two conserved aspartate residues, D257 and D385, on the sixth and eighth HRs of human PS1, respectively, are suggested to serve as active site residues, defining γ-secretase as an aspartyl protease (Fig. 1) (7). Recent studies suggest that γ-secretase is a multisubunit protease composed of the membrane proteins PS1, APH-1, nicastrin (Nct), and PEN-2 (see literatures in ref. 8). Expression of these four genes in yeast successfully reconstituted the γ-secretase activity, confirming that they are the minimal constituents of this enzyme (8). However, because γ-secretase has not been purified to homogeneity, a stoichiometry of each component in the complex and the catalytic properties of this enzyme remain unclear.
Clearly, the topology of the PS1 subunit of γ-secretase will influence the interpretation of the role of the conserved aspartates in the enzyme complex. Thus, a review of the evidence for the accepted view and possible alternatives is instructive.
The standard model was first proposed based on gene fusion studies with truncated SEL-12, a Caenorhabditis elegans PS1 homologue, fused to β-galactosidase after each HR of SEL-12 and corresponding fusions of truncated human PS1 (3, 4). This approach takes advantage of the observation that β-galactosidase is active within the cytoplasm but not in the extracytosolic compartment. Briefly, the activity of β-galactosidase was detected when this enzyme was fused after HR2, -4, -6, -7, -9, and -10 (Fig. 1 A), suggesting that PS1 has an eight-TM topology. This interpretation was further supported by the analysis of intracellular immunostaining profiles of cells transfected with intact PS1 by using polyclonal antibodies specific for the N tail, the loop, and the C tail (9).
In a similar approach, using fusion to the cytoplasmic portion of Escherichia coli leader peptidase (LP) or to newt growth hormone, Nakai et al. (10) provided independent evidence that PS1 has a seven-TM topology (Fig. 1C), however, quite different from that proposed by Dewji and colleagues (Fig. 1B). Because the cytoplasmic region of LP possesses N-glycosylation acceptor sites, glycosylation of a fusion protein indicates exposure to the lumen of endoplasmic reticulum (ER) and therefore translocation to the ER. Consistent with their seven-TM model, LP fusion constructs after HR1, -3, and -5 of PS1 produced glycoproteins. However, LP fused after HR10 was glycosylated, but fusions after HR7, -8, and -9 were not. Thus, their seven-TM model was altered to suggest that HR9 is membrane embedded but does not span the bilayer and that the C tail is extracellular. In all models other than that of Nakai et al., the C tail is cytoplasmic.
Somewhat different results were reported by Lehmann et al. (11) (Fig. 1D). They fused a portion of prolactin containing glycosylation sites after each HR of PS1 and monitored glycosylation patterns of the chimeric proteins. Glycoproteins were formed in chimeras with fusion junctions after HR1, -3, and -5 of PS1. Based on these results, they concluded that PS1 traverses the membrane six times with both N and C termini located in the cytoplasm.
One must conclude that the exact topology deduced from fusion studies depends on the nature of the reporter protein. Perhaps, PS1 assumes multiple conformations around HR8, -9, and -10, as contemplated by Nakai et al. (10), and the fate of each of these HRs is directly influenced by the polar sequence that follows. In E. coli, SecG, a subunit of the protein translocation machinery with two TMs, undergoes inversion during the protein translocation cycle (12). SecG also possesses a weakly hydrophobic stretch of 14-aa residues between two TMs that facilitates inversion processes. Perhaps the discrepancy near the C-terminal region of PS1 is related to the weakly hydrophobic nature of HR8 and 10 [see figure 1A of Dewji et al. (2)].
It is noteworthy that the pool of PS1 monitored by Dewji et al. (2) is different from that of PS1 analyzed by other investigators. Dewji et al. (2) probed the topology of intact PS1, both endogenous and transfected, at the cell surface, whereas others have focused on hybrid protein or overexpressed intact PS1 detected in ER and Golgi membranes. However, in agreement with Dewji and Singer, Schwarzman et al. (13) detected an extracellular location of the N tail of PS1 in Jurkat cells. The system as studied by Dewji et al. (2) most closely approaches the native membrane state and physiological amount of PS1.
The topology supported by the work of Dewji et al. (2, 5) inverts the orientation of the first six HRs and positions the presumed active site aspartic residue on HR8 and the loop in the endomembrane lumen or extracellular space as compared to the eight-TM model (Fig. 1B). This seven-TM topology would appear to conflict with other data. For example, this topology cannot explain the observation that a caspase-3 family protease, a cytosolic enzyme, cleaves at the C terminus of D326 and D329 in the loop region of PS1 (14). In addition, the binding sites of other cytosolic armadillo family proteins such as β-catenin, δ-catenin, and p0071 have been mapped to the loop region of PS1 (15–18). Similarly, residue S346 of the loop could not be a substrate for PKC (19), and D385 in HR8 could not contribute directly to a cytosolic proteolytic cleavage of APP and NOTCH (6). Furthermore, considering the fact that aspartyl proteases require two juxtaposed aspartate residues in the active site (20), the two aspartates on HR6 and -8 could not be part of a single catalytic domain. Similar difficulties are encountered with the alternative models depicted in Fig. 1 C and D.
Of course, two pools of PS1, one in an eight-TM topology retained within the cell and the other transported to the cell surface in a seven-TM orientation, would satisfy these seemingly contradictory observations. There are several proteins exhibiting more than one topological orientation (see ref. 21 for review). For example, ductin, which contains four TMs, functions as the major component of a connexon channel of gap junctions as well as subunit c of the vacuolar H+-ATPase. The N and C tails of ductin in the gap junction orient to the cytoplasm, whereas these tails in the vacuolar H+-ATPase face the vacuolar lumen (22).
Protein topology assignment is a risky business. Gene fusion probes designed to report cytoplasmic or extracellular location of a polypeptide domain may themselves influence the fate of a hybrid protein by contributing unanticipated localization signals. Immunostaining reagents are preferable but are only as reliable as they are domain-specific. However, even with perfect reagents, proteins that assume multiple conformations within membranes may yield confusing and conflicting results. Therefore, until more is known about the substrates of PS1 and about the enzymes that act on or assemble with PS1, and ultimately until an atomic resolution structure(s) of the PS1 complex within the membrane is established, we are not likely to see this issue satisfactorily resolved. In the meantime, the results of Dewji et al. (2) lend weight to the seven-TM model of PS1 topology.
See companion article on page 1057.
References
- 1.Schellenberg, G. D. (1995) Proc. Natl. Acad. Sci. USA 92, 8552–8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dewji, N. N., Valdez, D. & Singer, S. J. (2004) Proc. Natl. Acad. Sci. USA 101, 1057–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li, X. & Greenwald, I. (1996) Neuron 17, 1015–1021. [DOI] [PubMed] [Google Scholar]
- 4.Li, X. & Greenwald, I. (1998) Proc. Natl. Acad. Sci. USA 95, 7109–7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dewji, N. N. & Singer, S. J. (1997) Proc. Natl. Acad. Sci. USA 94, 14025–14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haass, C. & Steiner, H. (2002) Trends Cell Biol. 12, 556–562. [DOI] [PubMed] [Google Scholar]
- 7.Wolfe, M. S., Xia, W., Ostaszewski, B. L., Diehl, T. S., Kimberly, W. T. & Selkoe, D. J. (1999) Science 398, 513–517. [DOI] [PubMed] [Google Scholar]
- 8.Edbauer, D., Winkler, E., Regula, J. T., Pesold, B., Steiner, H. & Haass, C. (2003) Nat. Cell Biol. 5, 486–488. [DOI] [PubMed] [Google Scholar]
- 9.Doan, A., Thinakaran, G., Borchelt, D. R., Slunt, H. H., Ratovitsky, T., Podlisny, M., Selkoe, D. J., Seeger, M., Gandy, S. E., Price, D. L. & Sisodia, S. S. (1996) Neuron 17, 1023–1030. [DOI] [PubMed] [Google Scholar]
- 10.Nakai, T., Yamasaki, A., Sakaguchi, M., Kosaka, K., Mihara, K., Amaya, Y. & Miura, S. (1999) J. Biol. Chem. 274, 23647–23658. [DOI] [PubMed] [Google Scholar]
- 11.Lehmann, S., Chiesa, R. & Harris, D. A. (1997) J. Biol. Chem. 272, 12047–12051. [DOI] [PubMed] [Google Scholar]
- 12.Nishiyama, K.-I., Suzuki, T. & Tokuda, H. (1996) Cell 85, 71–81. [DOI] [PubMed] [Google Scholar]
- 13.Schwarzman, A. L., Singh, N., Tsiper, M., Gregori, L., Dranovsky, A., Vitek, M. P., Glabe, C. G., St. George-Hyslop, P. H. & Goldgaber, D. (1999) Proc. Natl. Acad. Sci. USA 96, 7932–7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, T.-W., Pettingell, W. H., Jung, Y.-K., Kovacs, D. M. & Tanzi, R. E. (1997) Science 277, 373–376. [DOI] [PubMed] [Google Scholar]
- 15.Zhou, J., Liyanage, U., Medina, M., Ho, C., Simmons, A. D., Lovett, M. & Kosik, K. S. (1997) NeuroReport 8, 2085–2090. [DOI] [PubMed] [Google Scholar]
- 16.Stahl, B., Diehlmann, A. & Südhof, T. C. (1999) J. Biol. Chem. 274, 9141–9148. [DOI] [PubMed] [Google Scholar]
- 17.Saura, C. A., Tomita, T., Soriano, S., Takahashi, M., Leem, J.-Y., Honda, T., Koo, E. H., Iwatsubo, T. & Thinakaran, G. (2000) J. Biol. Chem. 275, 17136–17142. [DOI] [PubMed] [Google Scholar]
- 18.Soriano, S., Kang, D. E., Fu, M., Pestell, R., Chevallier, N., Zheng, H. & Koo, E. H. (2001) J. Cell Biol. 152, 785–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fluhrer, R., Friedlein, A., Haass, C. & Walter, J. (2003) J. Biol. Chem., in press. [DOI] [PubMed]
- 20.Navia, M. A., Fitzgerald, P. M., McKeever, B. M., Leu, C. T., Heimbach, J. C., Herber, W. K., Sigal, I. S., Darke, P. L. & Springer, J. P. (1989) Nature 337, 615–620. [DOI] [PubMed] [Google Scholar]
- 21.Levy, D. (1996) Essays Biochem. 31, 49–60. [PubMed] [Google Scholar]
- 22.Dunlop, J., Jones, P. C. & Finbow, M. E. (1995) EMBO J. 14, 3609–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]