Abstract
Nephroblastoma overexpressed gene encodes a matricellular protein (CCN3/NOV) of the CCN family, comprising CCN1 (CYR61), CCN2 (CTGF), CCN4 (WISP-1), CCN5 (WISP-2), and CCN6 (WISP-3). CCN proteins are involved in the regulation of mitosis, adhesion, apoptosis, extracellular matrix production, growth arrest and migration in multiple cell types. Compared to CCN2/CTGF, known as a profibrotic protein, the biological role of CCN3/NOV in liver fibrosis remains obscure. In this study we showed ccn3/nov mRNA to increase dramatically following hepatic stellate cell activation, reaching peak levels in fully transdifferentiated myofibroblasts. In models of experimental hepatic fibrosis, CCN3/NOV increased significantly at the mRNA and protein levels. CCN3/NOV was found mainly in non-parenchymal cells along the areas of tissue damage and repair. In the bile-duct ligation model, CCN3/NOV was localized mainly along portal tracts, while the repeated application of carbon tetrachloride resulted in CCN3/NOV expression mainly in the centrilobular areas. In contrast to CCN2/CTGF, the profibrotic cytokines platelet-derived growth factor-B and -D as well as transforming growth factor-β suppressed CCN3/NOV expression. In vitro, CCN3/NOV siRNA attenuated migration in the cirrhotic fat storing cell line CFSC well in line with in vivo findings that various types of cells expressing CCN3/NOV migrate into the area of tissue damage and regeneration. The suppression of CCN3/NOV enhanced expression of profibrotic marker proteins, such as α-smooth muscle actin, collagen type I, fibronectin, CCN2/CTGF and TIMP-1 in primary rat hepatic stellate cells and in CFSC. We further found that adenoviral overexpression of CCN2/CTGF suppressed CCN3/NOV expression, while the overexpression of CCN3/NOV as well as the suppression of CCN3/NOV by targeting siRNAs both resulted in enhanced CCN2/CTGF expression. These results indicate the complexity of CCN actions that are far beyond the classic Yin/Yang interplay.
Electronic supplementary material
The online version of this article (doi:10.1007/s12079-011-0141-3) contains supplementary material, which is available to authorized users.
Keywords: CCN protein, CCN3/NOV, CCN2/CTGF, Hepatic stellate cells, Myofibroblasts, Hepatocytes, Inflammation, Yin/Yang, Fibrosis, Animal models
Introduction
The CCN protein family consists of six members, CCN1/CYR61, CCN2/CTGF, CCN3/NOV, CCN4/WISP1, CCN5/WISP2, and CCN6/WISP3. They share a modular architecture with up to four distinct modules including an insulin-like growth factor-binding domain, a von Willebrand factor type C domain, a thrombospondin type I homology domain and a C-terminal cystine knot (Bork 1993; Weiskirchen 2011). All six proteins contain an N-terminal leader sequence of 20 to 40 amino acids characterizing them as secreted proteins.
The most familiar member of the CCN protein family, i.e. CCN2/CTGF, was first described in human umbilical vein endothelial cells as a 349-amino acid mitogen with biological activities similar to PDGF and termed connective tissue growth factor (CTGF) (Bradham et al. 1991). In liver, CCN2/CTGF is of particular interest since several studies independently showed that this CCN protein is a key modulator regulating transforming growth factor-β (TGF-β) activity, a cytokine involved in initiation, maintance and progression of hepatic injury by triggering extracellular matrix-synthesis and -remodelling.
CCN3/NOV was originally isolated from myeloblastosis-associated virus 1-induced avian nephroblastoma (Soret et al. 1989; Joliot et al. 1992), representing a well established animal model of pediatric Wilms tumor. Early expression analysis of embryonic and adult livers demonstrated ccn3/nov transcripts to be virtually absent in liver (Joliot et al. 1992). Based on its expression profile, it was first speculated that ccn3/nov is a novel proto-oncogene overexpressed in nephroblastoma while the expression is probably not transforming in all tissues per se.
In more recent work, it was demonstrated that individual CCN proteins possess a capacity to bind a broad repertoire of different growth factors and cytokines including the transforming growth factor-β (TGF-β), bone morphogenetic protein, and vascular endothelial growth factor families that regulate cell surface localization and interaction with the respective cytokine receptors (Abreu et al. 2002; Minamizato et al. 2007; Rydziel et al., 2007). However, precise formation of the predicted complexes and underlying mechanisms of this potential interaction and their impact on cellular signaling is currently not available. Additionally, several intrinsic activities were reported for some of the CCN proteins. Based on the finding that the binding site of CCN2/CTGF at the cell surface of murine fibroblasts was similar to that of recombinant PDGF-B, it was initially suggested that CCN2/CTGF has similar recognition sites and biological activities as PDGF (Bradham et al. 1991). In liver, the stimulation with recombinant CCN2/CTGF promote phosphorylation of the ets oncogene family member Elk-1 and the extracellular signal-regulated kinases ERK1 and ERK2, thus increasing the expression of c-fos and cellular proliferation in primary hepatic stellate cells (HSC) (Gao et al. 2004). These findings demonstrate that CCN2/CTGF either has intrinsic activities of its own or has the capacity to modulate the activity of special cytokines involved in regulation of afore mentioned processes during ongoing hepatic fibrogenesis.
Similar intrinsic activities were reported for the CCN3/NOV protein. It was found that stimulation of 3T3 cells with recombinant CCN3/NOV resulted in a dose-dependent increase of cellular proliferation and tyrosine phosphorylation of several proteins (Liu et al. 1999). CCN3/NOV expression is also up-regulated in both in vitro activated HSC and in vivo models of experimentally-induced liver fibrosis (Lee et al. 2004). CCN3/NOV protein expression in fibrotic rat and human livers is found predominantly in areas of ductular proliferation and HSC of the fibrous septa (Lee et al. 2004). Stimulation with TGF-β and dexamethasone has been shown to induce expression of CCN3/NOV, CCN2/CTGF and CCN1/CYR61 in human glioma cell line U87 (Liu et al. 1999), a phenomenon also found in culture-activated HSC (Lee et al. 2004). Bile acids including cholic acid, chenodeoxycholic acid and ursodeoxycholic acid are frequent counterparts of liver injury and recognized to modulate ccn3/nov mRNA expression in HSC, underpinning the assumption that this CCN protein has a pathogenic role in liver fibrogenesis. However, analysis of ccn3/nov gene expression in diseased human liver is somewhat conflicting. In one study the expression in resected specimen of patients suffering from small nodular or solitary large hepatocellular carcinoma revealed no significant expression differences (Zeng et al. 2004) while the prevalence of ccn3/nov expression in tumour tissue was higher than in the surrounding para-cancerous tumour tissue in another cohort of patient with hepatocellular carcinoma and metastatic liver tumours (Hirasaki et al. 2001). Hence, the significance of ccn3/nov gene expression in hepatocellular carcinoma is still subject of debate.
Interestingly, exposure of kidney mesangial cells to exogenous CCN3/NOV resulted in a dose-dependent attenuation or blockade of the TGF-β-stimulated increase in CCN2/CTGF transcript and an overall reduction in TGF-β stimulated synthesis, secretion and redistribution of intracellular collagen type I, suggesting that endogenous CCN3/NOV activity plays a role in extracellular matrix metabolism in mesenchymal cells, and that CCN2/CTGF and CCN3/NOV are opposing factors in regulating collagen promoter activity and secretion of this extracellular matrix protein (Riser et al. 2009). Although the underlying mechanisms of this “Yin Yang” concept are not yet fully understood, the results clearly indicate that different combinations of CCN proteins can affect each other and might mediate divergent biological effects (Kawaki et al. 2008).
Based on actual knowledge it is tempting to speculate that in liver most of the biological activities of an individual CCN protein become foremost relevant during hepatic injury and remodelling (Weiskirchen 2011).
To study the biological functions of CCN3/NOV in liver fibrogenesis, we investigated ccn3/nov expression in culture-activated HSC and experimental fibrotic liver models. We demonstrate that the application of CCN3/NOV siRNA enhance expression of profibrogenic genes at both mRNA and protein levels while CCN3/NOV overexpression inhibit profibrogenic gene upregulation. Additionally, we found that CCN3/NOV siRNA enhance CCN2/CTGF expression and inhibit migration of cirrhotic fat storing rat cells (CFSC). Collectively these data indicate that CCN3/NOV participates in liver injury and tissue repair.
Material and methods
Primary liver cells and cell lines HSC were isolated from male Sprague–Dawley rats by density gradient centrifugation technique and cultured as described previously (Schäfer et al. 1987; Fehrenbach et al. 2001). MFB were obtained by sub-cultivation of HSC at day 7 of initial culturing. The immortalized rat hepatic stellate cell line (Greenwel et al. 1991; Greenwel et al. 1993) were cultured in medium containing 10% FCS, 4 mM L-glutamine, 100 IU/ml penicillin, 100 μg/ml streptomycin, and non-essential amino acids.
RNA isolation, RT-PCR, and qPCR Total RNA from HSC/MFB and CFSC cells was isolated through QIAzol Lysis Reagent and RNeasy Mini Kits (Qiagen, Hilden, Germany) according to manufacturer’s instructions. Organ RNA was isolated according to guanidine thiocyanate/CsCl method (Weiskirchen and Bister 1993), followed by DNAse digestion and RNeasy clean up. Primers for amplification were selected from sequences deposited in the GenBank database (Table 1) using the Primer Express software (Applied Biosystems Inivtrogen, Darmstadt, Germany). First-strand cDNA was synthesized from 1 to 2 μg RNA in 20 μl volume using SuperScript™ II RNAse H reverse transcriptase and random hexamer primers (Invitrogen, Karlsruhe, Germany). Subsequently, the cDNA was subjected to conventional PCR under following conditions: 95°C for 1 min, annealing for ccn3/nov at 59°C, GAPDH at 56°C for 1 min and 72°C extension for 1 min as per indicated cycles. To allow semi-quantitative analysis of mRNA expression, cDNAs were amplified at low cycle numbers in the range of linear amplification. Real-time quantitative PCR, cDNA derived from 25 ng RNA was amplified in 25 μl volume using qPCR Core Kits (Eurogentec, Cologne, Germany). PCR conditions were 50°C for 2 min, 40 cycles of 95°C for 15 s and 60°C for 1 min. TaqMan primers are given in Table 1. RNA normalization was achieved through TaqMan Ribosomal RNA Control Reagents (Applied Biosystems) designed for 18S ribosomal RNA (rRNA) detection.
Table 1.
Primers used in this study
| Gene | Acc. No. | Primera |
|---|---|---|
| CCN3/NOV | NM_030868 | for: CTGAGATGAGACCCTGCGAC |
| rev: CTCTGAGCCACAGGTCCACT | ||
| GAPDH | NM_017008 | for: TGTGGATCTGACATGCCGCC |
| rev: AACCACCCTGTTGCTGTAGC | ||
| CCN3/NOV | NM_030868 | for: CTACAGAGTGGAGCGCGTGTT |
| rev: GGAAGATTCCTGTTGGTGACCC | ||
| probe: AAGAGCTGTGGAATGGGCTTGTCCAC | ||
| CCN2/CTGF | NM_001010 | for: AGCAGCTGGGAGAACTGTGC |
| rev: GCCGAAGTCGCAGAAGAGAC | ||
| probe: CGTGATCCCTGCGACCCACACAA | ||
| Col type I(α) | NM_053304 | for: GAAGGCAACAGTCGATTCACC |
| rev: GACTGTCTTGCCCCAAGTTCC | ||
| probe: ACAGCACGCTTGTGGATGGCTGC | ||
| α-SMAb | NM_031004 | for: GAGGAGCATCCGACCTTGC |
| rev: ATTTTCTCCCGGTTGGCC |
a the first two primer sets were used in conventional PCR, all others primers were used for quantitative real-time PCR analysis on TaqMan.
bthe quantification of α-SMA was done with the SybrGreen dye.
Inhibition and overexpression of ccn3/nov gene expression in HSC and CFSC Cells were transfected with either the appropriate CCN3/NOV small interfering RNA (siNOV3: SI01920793; siNOV4: SI01920800; siNOV5:SI03084319; siNOV6:SI03119508 according to NM_030868) or a control non-silencing siRNA (Allstar negative control: 1027280) using the HiPerFect transfection reagent (all siRNAs and reagents from Qiagen). Cells were seeded 1 day before transfection in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% FCS, 4 mM L-glutamine, 100 IU/ml penicillin, 100 μg/ml streptomycin, while non-essential amino acids were added for CFSC. Cells were transfected with siRNA according to manufacturer’s manual. For transient overexpression of CCN3/NOV, we used a plasmid (clone IRAKp961P24175Q, in the following denoted as vector rat NOV-1) that was obtained on commercially basis (ImaGENE, Berlin, Germany) containing full-length rat NOV cDNA in vector pExpress-1.
Scratch-induced migration assay For this assay, CFSC were seeded at 200,000 cells per well in 6-well plates in DMEM containing 10% FCS and non-essential amino acid. Cells reached 70–80% confluence after one day and shortly before transfection the medium was exchanged with 2,300 μl medium containing 10% FCS and antibiotics. Subsequently, the cells were transfected with control siRNA and siNOV3 to siNOV6 by diluting 150 ng siRNA in 100 μl DMEM without serum, giving a final siRNA concentration of 5 nM after adding the complexes to the cells. 12 μl of HiPerFect transfection reagent were added to the diluted siRNA and mixed by vortexing, followed by 10 min incubation at room temperature to allow formation of the transfection complexes. The complexes were added to the cells and maintained under normal growth conditions. After 24 h the medium was renewed with medium containing 0.5% FCS and non-essential amino acid followed by wound scratching. Subsequent gap closing was monitored by phase contrast microscope. CCN3/NOV gene silencing was confirmed in ELISA measuring cellular supernatant.
CFSC BrdU incorporation assay Cells were seeded at 80,000 cells per well in 12-well plates and transduced the next days with indicated siRNAs or adenoviral constructs. Following a 16 h starvation period, cells were incubated with PDGF-B (20 ng/ml) in the presence of indicated concentrations of recombinant mouse CCN3/NOV (1976NV, R&D Systems, Wiesbaden, Germany) and labelled with 10 mM 5-bromo-20-deoxyuridine (BrdU) for 24 h, using the Cell ELISA BrdU colorimetric assay kit (Roche Diagnostics, Mannheim, Germany). Upon culture medium removal, cells were fixed and DNA denatured in one step by adding FixDenat solution (Roche) for 1 h at room temperature, followed by incubation with an anti-BrdU antibody conjugated to peroxidase for 2 h that binds to BrdU incorporated into the newly synthesized DNA. After threefold washing, the bound anti-BrdU-peroxidase conjugate was detected with the included substrate reaction mixture for 10–30 min. The reaction was stopped by addition of 1 M H2SO4 and quantified in a standard ELISA plate reader at 450 nm. For cell count the cells were treated identically but without BrdU incorporation. CFSC were trypsinized and re-suspended in PBS-based enzyme-free cell dissociation buffer (Gibco, Invitrogen) in 1:2 to 1:5 dilutions and cells were then counted using the CasyR cell counter model DT (Schärfe System GmbH Reutlingen, Germany). Primary HSC were removed by incubation with Accutase (PAA Laboratories, Pasching, Austria) and counted in a Neubauer hemocytometer.
SDS-PAGE and Western blot analysis Cell and tissue lysates were prepared using RIPA buffer containing 20 mM Tris–HCl (pH 7.2), 150 mM NaCl, 2% (w/v) NP-40, 0.1% (w/v) SDS, 0.5% (w/v) sodium deoxycholate and the Complete™-mixture of proteinase inhibitors (Roche). Equal amounts of cellular protein extracts or supernatants were diluted with Nu-PAGE™ LDS electrophoresis sample buffer with DTT as reducing agent, heated at 95°C for 10 min, and separated in 4–12% Bis-Tris gradient gels, using MOPS or MES running buffer (Invitrogen). Proteins were electroblotted onto nitrocellulose membranes, and equal loading was shown in Ponceau S stain. Subsequently, non-specific binding sites were blocked in TBS containing 5% (w/v) non-fat milk powder. All antibodies (see Table 2) were diluted in 2.5% (w/v) non-fat milk powder in Tris-buffered saline. Primary antibodies were visualized using horseradish peroxidase conjugated anti-mouse-, anti-rabbit- or anti-goat IgG (Santa Cruz Biotech, Santa Cruz, CA) and the SuperSignal chemiluminescent substrate (Pierce, Bonn, Germany).
Table 2.
Antibodies used in this study
| Antibody | Cat no. | Supplier | Speciesa | Dilution |
|---|---|---|---|---|
| mCCN3/NOV | AF1976 | R&D Systems | m, r | 1:1000, 1:50 (IHC) |
| mCCN3/NOV | BF1976 | R&D Systems | m, r | 1:500 (ELISA) |
| CCN2/CTGF | 14939 | Santa Cruz | m, r | 1:1000 |
| rCollagen type I | PS065 | Monosan, Beutelsbach, Germany | m, r | 1:1000 |
| Fibronectin | AB1954 | Millipore, Schwalbach, Germany | m, r | 1:2000 |
| α-SMA | CBL1 | Cymbus Biotechn., Hampshire, UK | h, m, r | 1:2000 |
| β-Actin | A5441 | Sigma, Taufkirchen, Germany | h, m, r | 1:10000 |
| MMP9 | 2270 | Cell Signaling Technology, Frankfurt, Germany | h, m, r | 1:1000 |
| TIMP-1 | AF580 | R&D Systems | r | 1:1000 |
| Vimentin | ab92547 | Abcam | h, m, r | 1:2500 |
| Ribosomal rS6 | 2317 | Cell Signaling Technology | h, m, r | 1:1000 |
| ED2 (CD163) | MCA342GA | AbD Serotec, Düsseldorf, Germany | r | 1:50 (IHC) |
| phosphoTyr clone 4 G10 | 05-312 | Millipore | h, m, r | 1:1000 |
| PDGFR-α | 3164 | Cell Signalling Technology | h, m, r | 1:1000 |
| PDGFR-β | 3169 | Cell Signalling Technology | h, m, r | 1:1000 |
| Phospho AKT | 4051 | Cell Signalling Technology | h, m, r | 1:1000 |
| Total AKT | 2920 | Cell Signalling Technology | h, m, r | 1:1000 |
| Phospho ERK1/2 | 9101 | Cell Signalling Technology | h, m, r | 1:1000 |
| Total ERK1/2 | 9102 | Cell Signalling Technology | h, m, r | 1:1000 |
| Phospho p38 | 612288 | BD Biosciences, Heidelberg, Germany | h, m, r | 1:2500 |
| Total p38 | 612168 | BD Biosciences | h, m, r | 1:2500 |
aabbreviations used are: m = mouse, r = rat, h = human
CCN3/NOV ELISA Culture media were analyzed by sandwich ELISA using an affinity purified polyclonal anti-mouse CCN3/NOV antibody (AF1976; R&D Systems) as capture antibody and a biotinylated anti-mouse CCN3/NOV antibody (BAF1976; R&D Systems) as detection antibody. Formed immunocomplexes were visualized with a streptavidin-horseradish-peroxidase conjugate (DAKO, Hamburg, Germany) and the 1-StepTM Ultra TMB-ELISA reagent (Thermo Scientific, Rockford, IL). After addition of stop solution, the optical density of each well was determined immediately using a micro-plate reader set to 450 nm. CCN3/NOV concentrations were calculated against a mouse CCN3/NOV (1976NV, R&D System) standard curve.
Animal experiments and specimen collection All animal protocols were in compliance with the guidelines for animal care approved by the German Animal Care Committee. Sprague–Dawley rats were subjected to bile duct ligation (BDL) as described previously (Arias et al. 2003), and sacrificed at indicated time intervals. Liver specimen were snap frozen in liquid nitrogen for protein and RNA isolation or fixed in 4% buffered paraformaldehyde for histology. For the CCl4 treatment, male Sprague–Dawley rats weighing 180–200 g received intraperitoneal injections of 1 ml/kg of CCl4 in an equal volume of mineral oil twice weekly for up to 12 weeks following established protocols (Borkham-Kamphorst et al. 2008).
Immunohistochemistry Liver tissue sections were treated as described before (Borkham-Kamphorst et al. 2008). For CCN3/NOV detection we applied goat anti-mouse CCN3/NOV (AF1976) overnight. Liver slides were incubated with biotinylated secondary antibodies (BA-9200, Vector Laboratories, Eching, Germany) and developed with the Vectastain ABC-Elite reagent (Vector Laboratories) and substrates from the Vector NovaRED Peroxidase substrate kit (SK-4800; Vector Laboratories) or the 3,3’-diaminobenzidine substrate (DAKO).
Generation of adenoviral expression vectors For construction of adenoviral expression vector Ad-CMV-CTGF for expression of human CCN2/CTGF, the ~1.5 kbp KpnI fragment from clone pCEP[hCTGF] (kind gift of Walter Sebald, Department of Physiological Chemistry II, Biocenter, University of Wuerzburg Germany) was cloned into the KpnI site of shuttle vector pShuttle-CMV (Stratagene, Agilent Technologies, Waldbronn, Germany). Integrity of resulting plasmid pShuttle-CMV-CTGF was verified by restriction mapping and sequencing. Subsequently, 1 μg of respective plasmid was linearized with PmeI and co-transformed with 100 ng pAdEasy-1 vector (Agilent Technologies) into BJ5183 electroporation competent cells (Stratagene) following standard protocols. Recombinant adenoviral plasmids were selected with kanamycin and a recombinant vector pAdEasy-1-CMV-CTGF was isolated, characterized by restriction digest and sequencing. Respective DNA was cut with PacI and transfected into 293A cells to prepare adenoviral particles. The cloning of Ad-CMV-NOV driving expression of full-length rat NOV protein fused to a short C-terminal 22-amino acid peptide was recently described (Bohr et al. 2010). Detailed protocols for amplification and purification of recombinant viral particles using CsCl gradient centrifugation and the BD Adeno-XTM Purification Filter system (BD Biosciences, Clontech, Palo Alto, CA) were described before (Weiskirchen et al. 2000; Nevzorova et al. 2009).
Statistics Statistical analyses was performed using the Kruskal–Wallis test for nonparametric multiple comparison with a statistical software program (STATGRAPHICS Plus, version 5.1). Results are expressed by the median values followed by range. Probability values of less than 0.05 or 0.01 were considered as statistically significant and marked with or “*” “**”. To establish the differences within individual groups, we used the Mann–Whitney test.
Results and discussion
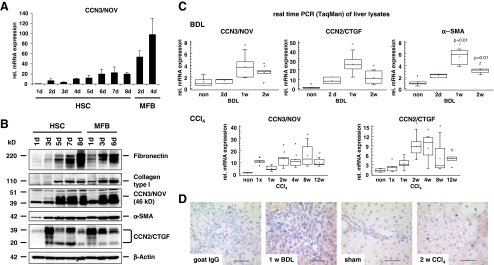
CCN3/NOV expression in isolated primary HSC and hepatocytes Although different types of liver cells possess capacity to synthesize extracellular matrix components, HSC are considered the main connective tissue-producing cells in fibrotic liver. HSC transdifferentiation from a quiescent, vitamin A-storing cell phenotype into a proliferative, contractile myofibroblast (MFB) remains the most informative discovery to date in unlocking the basics of hepatic fibrogenesis. To get a first impression of ccn3/nov expression in culture-activated HSC, we performed qRT-PCR and found that ccn3/nov mRNA is virtually undetectable in freshly isolated HSC and dramatically increases following HSC activation and reaching peak levels in fully transdifferentiated MFB (Fig. 1a). The expression kinetics observed for ccn3/nov mRNA correlated well with the expression of ccn2/ctgf and α-SMA that are known to be closely linked to the transdifferentiation process (Suppl. Fig. 1A). Surprisingly, no ccn3/nov mRNA expression was detected in cultured hepatocytes isolated from naive and early CCl4 treated animals (not shown) confirming previous studies showing that ccn3/nov expression is absent in human liver carcinoma cell line HepG2 (Liu et al. 1999). In contrast, ccn2/ctgf mRNA expression increased markedly during culturing in primary hepatocytes (Suppl. Fig. 1B).Additionally, we found expression of ccn3/nov in cultured Kupffer cells (KC) and primary isolated liver sinusoidal endothelial cells (LSEC) lower than the expression observed in HSC and subcultures of periportal myofibroblasts (pMF) (Suppl. Fig. 1C). Western blot analysis of extracts prepared from primary HSC/MFB cultured for different time intervals confirmed that CCN3/NOV expression is markedly increased during culture-activation and transdifferentiation in HSC/MFB (Fig. 1b). The increase of CCN3/NOV protein showed correlation with fibronectin and collagen type I and the degree of cellular confluence upon prolonged culturing of both HSC and sub-cultivated MFB in a time dependent manner. CCN2/CTGF by contrast showed down regulation when the cells reached their highest density. This finding is in line with a previous report showing that the amount of CCN3/NOV production per cell depends on cell density (Bleau et al. 2007). We also found that the amount of CCN3/NOV from an equal protein quantity taken form HSC that were cultured for 4 days was higher in cultures with higher plating efficiency (data not shown).
Fig. 1.
CCN3/NOV expression in cultured HSC and in experimental hepatic fibrosis in vivo. a Quantitative RT-PCR (TaqMan) from culture-activated HSC showed significant upregulation of CCN3/NOV, reaching peak levels in fully transdifferentiated MFB. b Western blot analysis of HSC protein lysate, showing increased CCN3/NOV protein expression in culture-activated HSC and MFB that correlated with increased expression of fibronectin, collagen type I, α-SMA and CCN2/CTGF. In this analysis, β-Actin was used as a loading control. c Relative mRNA expression of CCN3/NOV in BDL and chronic CCl4-treated rat livers. Quantitative RT-PCR (TaqMan) showed the relative mRNA levels of CCN3/NOV to increase significantly upon BDL and CCl4 administration. Expression levels were comparable to CCN2/CTGF and α-SMA expression throughout the period of liver fibrogenesis. d CCN3/NOV immunohistochemistry of BDL and CCl4 rat livers reflecting positive staining in mesenchymal cells and proliferative bile duct epithelia along the fibrotic septa of BDL rat livers. The CCl4 model showed positive staining in perisinusoidal areas peripheral to centrilobular hepatic necrosteatosis
CCN3/NOV expression in experimental fibrotic models We next investigated CCN3/NOV expression in in vivo models of liver fibrogenesis. Quantitative RT-PCR of RNA liver samples isolated from rats subjected to BDL and CCl4 again revealed that CCN3/NOV during ongoing fibrogenesis that was comparable to the increase that was observed for CCN2/CTGF and α-SMA representing well established markers of hepatic fibrogenesis (Fig. 1c). Furthermore, CCN3/NOV immunohistochemistry of specimen taken from rat livers of animals subjected to BDL and CCl4 showed positive staining in mesenchymal cells and proliferative bile duct epithelia along the fibrotic septa, and perisinusoidal areas around the centrilobular hepatic necrosteatosis (Fig. 1d). Double staining of CCN3/NOV and a KC marker (i.e. ED2) reflected no co-localization (Suppl. Fig. 1D) demonstrating that KC are not the primary mesenchymal hepatic cell fraction responsible for expression of CCN3/NOV in injured livers. In our view, the finding that the expression of the matricellular protein CCN3/NOV is markedly increased in response to liver injury points to an important role of CCN3/NOV in hepatic insult and repair. A similar CCN protein expression profile is found in granulation tissue during cutaneous wound healing following skin injury (Lin et al. 2005) suggesting that CCN3/NOV is a general marker or mediator that is linked to inflammation, wound healing and associated tissue remodelling processes.
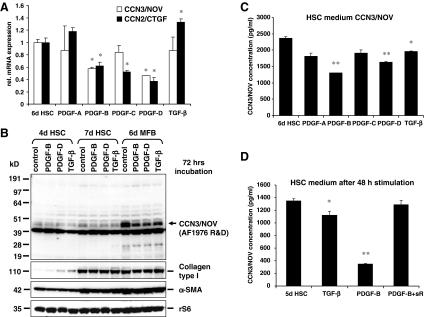
Down regulation of CCN3/NOV expression in HSC by PDGF and TGF-β Extensive HSC proliferation, migration and formation of extracellular matrix occur in response to polypeptide growth factors and cytokines locally released as part of the inflammatory response in the tissue repair process. PDGF-B and -D are the most potent stimulators of HSC proliferation and migration through PDGF receptor type β (PDGFRβ) signalling (Pinzani et al. 1989; Borkham-Kamphorst et al. 2007). Also TGF-β plays a definite pro-fibrogenic role through increased synthesis, deposition and capability of retarding degradation of ECM components.To test the impact of these profibrogenic cytokines on CC3/NOV expression, HSC were incubated with different PDGF isoforms (PDGF-A, PDGF-B, PDGF-C, PDGF-D) or TGF-β in medium containing 0.5% serum at indicated doses for 24 h. Following stimulation RNA, protein extracts, and conditioned media were taken and subjected to qRT-PCR (Fig. 2a), Western blot analysis (Fig. 2b), and ELISA testing (Fig. 2c) for analysis of CCN3/NOV expression and secretion. This analysis revealed that PDGF-B and –D as well as TGF-β were able to significantly down regulate CCN3/NOV expression at both mRNA and protein levels. Moreover, PDGF-B and –D that exert high mitogenic and fibrogenic effects on HSC/MFB (Borkham-Kamphorst et al. 2007) suppressed both CCN2/CTGF and CCN3/NOV protein production. This effect was rescued by the application of a soluble PDGFRβ (Fig. 2d) that specifically antagonize PDGF-B and -D activity in vitro and in vivo (Borkham-Kamphorst et al. 2004a, b). Contrastingly, TGF-β induced CCN2/CTGF but suppressed CCN3/NOV. Additionally, we found a decline of CCN3/NOV transcripts in HSC that were cultured in the presence of 10% foetal calf serum (data not shown) confirming earlier reports demonstrating that TGF-β strongly induces CCN1 and CCN2, whereas it represses CCN3 (Brunner et al. 1991; Lafont et al. 2002). TGF-β inhibition of CCN3/NOV is Smad independent, but is moderated through activation of c-Jun, AP-1 and MEKK1 (Lafont et al. 2002), supporting our findings of PDGF- and serum-enhanced down regulation of CCN3/NOV in cultured HSC. A similar phenomenon was noticed during mesangioproliferative glomerulonephritis, showing that glomerular CCN3/NOV acts as an endogenous inhibitor of mesangial cell growth and a modulator of PDGF-induced mitogenesis (van Roeyen et al. 2008). CCN2/CTGF overexpression in activated HSC was induced by TGF-β and TGF-β receptor (ALK4/5/7) in Smad-dependent manner (Leask et al. 2008). Thus, CCN3/NOV is regulated in an antithetical manner compared to CCN2/CTGF, suggesting that CCN2/CTGF and CCN3/NOV may serve opposing functions in HSC.
Fig. 2.
Down regulation of CCN3/NOV in HSC by PDGF and TGF-β. a qRT-PCR, b Western blot and c ELISA of conditioned media showed that PDGF-B and -D as well as TGF-β significantly decrease expression of CCN3/NOV at both mRNA and protein levels in culture-activated HSC. d The inhibitory effect of PDGF was neutralized by a soluble PDGF-β receptor (soluble PDGFRβ, sR) that acts as a specific antagonist of PDGF-B and PDGF-D
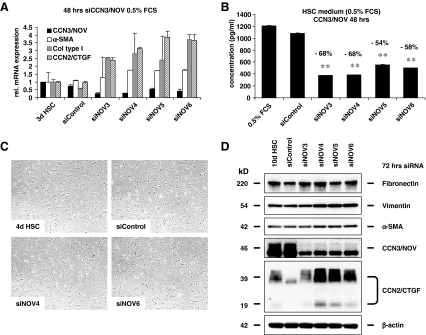
CCN3/NOV siRNA-induced profibrotic gene expression in culture-activated HSC Since CCN3/NOV expression increases in HSC during activation and transdifferentiation to MFB, we studied CCN3/NOV functions after ccn3/nov knock down using siRNA technology in cultured HSC and evaluate expression of relevant fibrotic gene. Therefore, HSC were transfected one-day after initial plating with control siRNA and 4 different siRNAs (i.e. siNOV3, siNOV4, siNOV5, siNOV6) targeting ccn3/nov gene expression. After 48 h, CCN3/NOV showed significant downregulation, evidenced by qRT-PCR (Fig. 3a) and further confirmed in CCN3/NOV ELISA (Fig. 3b) analysing culture supernatants of respective cells. HSC cell morphology 72 h after transfection with CCN3/NOV siRNA showed slightly more advanced transdifferentiation toward the MFB phenotype (Fig. 3c). Interestingly, the decrease of CCN3/NOV correlated with an increase of α-SMA, collagen type I(α) and CCN2/CTGF mRNA expression (cf Fig. 3a). Western blot analysis of HSC confirmed that siRNA targeting CCN3/NOV expression increased expression of α-SMA, vimentin and CCN2/CTGF (Fig. 3d).
Fig. 3.
CCN3/NOV siRNA induced profibrotic gene expression in culture-activated HSC. Primary HSC cultured for one day were the transfected for 48 h with control siRNA and four different CCN3/NOV siRNAs. Successful down regulation of CCN3/NOV was demonstrated in a qRT-PCR analysis and b ELISA using culture supernatants from identical experiments. Within 48 h all CCN3/NOV siRNA increased α-SMA, collagen type I and CCN2/CTGF mRNA expression. c Phase contrast microscopy depicting HSC cell morphology upon 72 h siRNA transfection and showing that CCN2/NOV siRNA transfected cells contain lesser fat droplets and more advanced transdifferentiation toward the MFB phenotype. d Western blot of 72 h siRNA transfected confirmed that CCN3/NOV gene knock down increase expression of α-SMA, fibronectin, vimentin and CCN2/CTGF. At the point of analysis HSC were kept in total 10 days after initial plating in primary culture
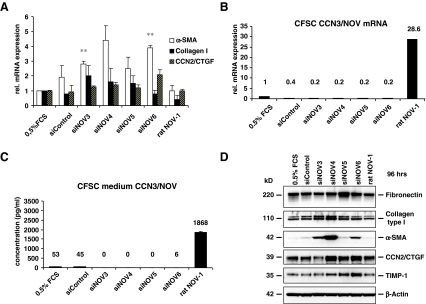
Inhibitory effects of CCN3/NOV on fibrogenic gene expression in CSFC Besides in HSC, we found CCN3/NOV expressed in other non-parenchymal liver cells like portal myofibroblast (pMF) and several cell lines derived from HSC i.e. rat CFSC and human hepatic stellate cell line LX-2 (not shown). Using diverse chemically-enhanced transfection technologies we found that only approximately 6% of primary HSC and even a smaller fraction (< 1%) of fully transdifferentiated MFB are susceptible to gene transfer (Weiskirchen et al. 2000). Therefore, we used in the following the immortalized rat cirrhotic fat storing cell line CFSC to test the impact of CCN3/NOV overexpression. CFSC showed enough CCN3/NOV expression to allow for real time PCR quantification (Fig. 4a and b) and culture medium ELISA, but the quantity of CCN3/NOV protein is low compared to CCN3/NOV production in HSC and MFB. CFSC were transfected with an unspecific negative control siRNA (siControl), different siRNAs targeting CCN3/NOV (siNOV3, siNOV4, siNOV5, siNOV6) and an expression vector (rat NOV-1) that effectively drives expression of rat CCN3/NOV. Cells were maintained in medium containing 0.5% serum and harvested 48 or 96 h later for RNA and protein isolation. Quantitative RT-PCR (Fig. 4a and b) and ELISA measurement (Fig. 4c) of cell supernatants confirmed the downregulation of CCN3/NOV by the four individual siRNAs targeting CCN3/NOV and the overexpression of CCN3/NOV by the chosen expression vector rat NOV-1. The suppression of CCN3/NOV was associated with an increase of α-SMA, collagen type I and to a smaller extent CCN2/CTGF mRNA expression (cf Fig. 3a). Western blot analysis confirmed the induction of α-SMA, collagen type I, CCN2/CTGF and TIMP-1 expression in cells that were transfected for 96 h with siRNAs targeting CCN3/NOV. Contrarily, the transfection with the CCN3/NOV expression vector (rat NOV-1) was able to inhibit the expression of respective fibrogenic genes in CFSC (cf Fig. 4d). Although CCN3/NOV expression is correlated with CCN2/CTGF during HSC transdifferentiation in vitro and in experimental fibrotic models in vivo, we hypothesized in the initial stage that CCN3/NOV gene knock down would, or could, attenuate liver fibrosis, but surprisingly CCN3/NOV siRNA showed a clear upregulation of several fibrotic genes instead. CCN3/NOV functions in fibrogenesis seem to be antipodal in part to what we found in the ccn3/nov gene expression. This corresponds with a recent finding in kidney mesangial cells showing that cultured rat mesangial cells express CCN3/NOV mRNA and protein, and TGF-β treatment reduced CCN3/NOV expression levels while increasing CCN2/CTGF and collagen type I activities (Riser et al. 2009). Conversely, the addition of CCN3/NOV or CCN3/NOV overexpression produced a marked down-regulation of CCN2/CTGF followed by virtual blockade of collagen type I transcription and its accumulation, pointing at yin/yang regulation of extracellular matrix metabolism by CCN2/CTGF and CCN3/NOV. It further indicates that CCN3/NOV is a factor that naturally limit fibrosis in vivo by showing increased CCN3/NOV mRNA presence in kidney and urine protein in the late stages of diabetic mice, whereas healthy mice had little or no measurable urinary CCN3/NOV, yet all of the diabetic mice with kidney fibrosis showed strongly CCN3/NOV positive (Riser et al. 2010). This might further indicate that the higher levels of CCN3/NOV are a natural compensation to protect against, or counter balance pathological organ fibrosis.
Fig. 4.
Suppression of CCN3/NOV by siRNA technology and fibrogenic gene expression in CFSC. a CFSC were transfected with an unspecific siRNA control, siRNAs (siNOV3, siNOV4, siNOV5, siNOV6) targeting CCN3/NOV expression, or an expression vector for rat NOV (rat NOV-1) and maintained in 0.5% FCS for 48 h. RNA was isolated from respective cells and subjected to qRT-PCR revealing that CCN3/NOV siRNA increased α-SMA, collagen type I and CCN2/CTGF mRNA expression, while CCN3/NOV overexpression showed no significant effects. b Quantitative RT-PCR showed that the transfection with siRNA targeting ccn3/nov gene expression resulted in decreased amounts of ccn3/nov mRNAs while the expression was significantly increased after transfection with expression vector rat NOV-1.c CCN3/NOV knock down efficacy was further demonstrated in ELISA testing using cell supernatants from the same experiment. d CFSC were transfected for 96 h with CCN3/NOV siRNAs and rat NOV-1 expression vector. Cell extracts were prepared and analysed by Western blot for expression of Fibronectin, collagen type I, α-SMA, CCN2/CTGF, TIMP-1 and β-Actin
Ccn3/Nov adenovirus gene transfer in primary HSC To confirm the fibrogenic effects CCN3/NOV from siRNA gene knock down, we over-expressed CCN3/NOV using Ad5-CMV-NOV. Primary rat HSC were isolated and seeded 1 × 106 cells/6 cm diameter uncoated culture disc and maintained in DMEM containing 10% FCS. Medium was changed to 4 ml of 4% FCS after 24 h, the plating efficiency evaluated and adenovirus gene transduction achieved by 24 h incubation with 2 × 108 virions/ml of Ad5-CMV-NOV, Ad5-CMV-CTGF and Ad5-CMV-Luc as a control virus. Following adenoviral transduction, HSC were either maintained in 10% FCS under normal growth condition for the next 48 h or starved for 16 h and 24 h stimulation with TGF-β. Thereafter, the cells were harvested and analysed for expression of respective fibrogenic genes. The result of HSC maintained in medium with 10% FCS showed that both Ad5-CMV-NOV and Ad5-CMV-CTGF induced MMP-9 expression and decreased expression of TIMP-1 and collagen type I, while there was no significant difference in α-SMA and Vimentin expression (Fig. 5a). Interestingly, HSC that overexpressed CCN2/CTGF showed significant lower expression of CCN3/NOV (Fig. 5b), contrasted by HSC that were infected with a virus overexpressing CCN3/NOV showing a slight increase in CCN2/CTGF expression (Fig. 5c), a phenomenon that become more obvious when the experiment was performed in medium containing only 0.5% FCS (Fig. 5d). Moreover, HSC overexpressing CCN3/NOV showed slightly decreased TGF-β- induced expression of collagen type I and α-SMA, and both CCN2/CTGF and CCN3/NOV increased expression of MMP9 and down regulation of TIMP-1 (Fig. 5e). These findings point at the complex interaction of CCN proteins. CCN2/CTGF and CCN3/NOV can act both antagonistic as well as synergistic to each other. Although CCN2/CTGF is a well established fibrotic marker but by itself does not act as a strong fibrotic inducer like TGF-β. In our experiments, the overexpession of CCN2/CTGF in HSC in vitro only slightly enhanced collagen type I and α-SMA expression, a finding that is in line with a previous report showing that elevated CCN2/CTGF levels in hepatocytes of transgenic mice in vivo does not cause hepatic injury or fibrosis per se but renders the liver more susceptible to the injurious actions of other fibrotic stimuli (Tong et al. 2009).
Fig. 5.
CCN3/NOV adenovirus gene transfer in primary HSC. a Primary HSC were transduced with 2 × 108 virions/ml of Ad5-CMV-rNOV, Ad5-CMV-CTGF, and Ad5-CMV-Luc as a control and maintained in either 10% FCS for 48 h, or 16 h starvation followed by 24 h TGF-β stimulation. Cell lysates were prepared and analysed by Western blot. The analysis revealed that HSC maintained in 10% FCS and overexpression CCN2/CTGF and CCN3/NOV showed increased MMP9 expression, while the expression of TIMP-1 and collagen type I was decreased. b, c Overexpression of CCN2/CTGF in HSC resulted in CCN3/NOV down regulation, but overexpression of CCN3/NOV induced enhanced CCN2/CTGF production. d HSC maintained in 0.5% FCS showed upregulation of CCN2/CTGF when cells were infected with an adenovirus driving expression of CCN3/NOV. Please note that the stimulation with TGF-β induced CCN2/CTGF expression in HSC e CCN3/NOV overexpression slightly decreased TGF-β-induced collagen type I and α-SMA expression, while remarkably both CCN2 and CCN3 significantly reduced TIMP-1
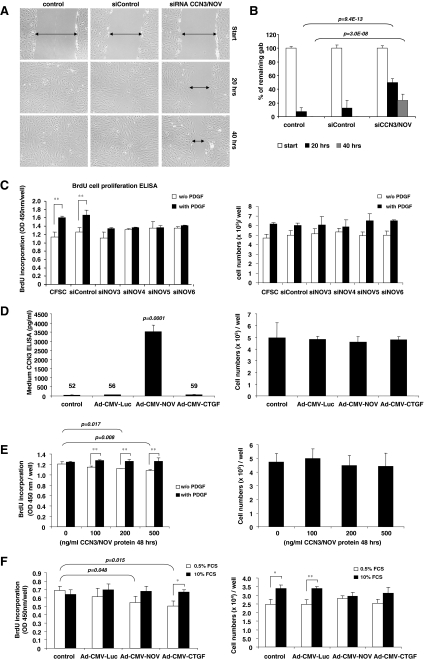
CCN3/NOV effects on cell migration and proliferation CFSC were transfected with a control siRNA and siRNAs specifically targeting CCN3/NOV. Since serum contains CCN3/NOV (Thibout et al. 2003), we examined the effects of CCN3/NOV siRNA in 0.5% FCS and applied a scratch-induced migration assay in CCN3/NOV siRNA-transfected CFSC. Upon monolayer scratching with a small pipette tip across the cultured plates, cells polarized perpendicularly to the scratch to migrate for wound closing. All CCN3/NOV siRNA-transfected CFSC reflected significantly delayed gab closing (Fig. 6a and b). It has been well established that overexpression of CCN3 promotes cell migration in different cancer cells (Benini et al. 2005) and that CCN3/NOV increases the activity of the small GTPase Rac1, revealing a pathway directly linking Cx43 to actin reorganization (Sin et al. 2009). CCN3/NOV siRNA is able to attenuate gap closing in monolayer cell scratch-induced migration indicating that NOV/CCN3 promotes cell migration. These effects on migration are in line with our in vivo findings showing that various CCN3/NOV expressing cells migrate into hepatic areas that are prone to injury and regeneration. Foregoing may indicate that overexpression of both CCN2/CTGF and CCN3/NOV in HSC enhances MMP9 activity and inhibits TIMP-1 resulting in enhanced cell migration both in vitro and in vivo. For CFSC proliferation assay, the cells were starved for 24 h and stimulated with PDGF-B in the presence of BrdU. This analysis revealed that all CCN3/NOV siRNAs tested showed efficient inhibition of PDGF-induced BrdU incorporation (Fig. 6c, left panel). Since it was already reported that CCN3/NOV-induced inhibition of cell growth is caused by prolonged G2/M cell cycle transition (Bleau et al. 2007), we performed cell counts to confirm the results of BrdU incorporation. However, we noticed no significant differences in cell numbers of CCN3/NOV siRNA transfected cells compared to controls (Fig. 6c, right panel), pointing at a significant role of CCN3/NOV in CFSC cell migration rather than proliferation. We next investigated the direct effects of CCN3/NOV in PDGF signalling but found no alterations in PDGF signalling after CFSC transfection with CCN/NOV siRNAs (Suppl. Fig. 2A and B).Actually, the impact of CCN3/NOV activity on cell proliferation is controversially discussed since overexpression of CCN3/NOV inhibits proliferation in most types of tumours such as glioblastoma and Ewing sarcoma (Benini et al. 2005; Gupta et al. 2001). CCN3/NOV expression in fibroblasts was found to either negatively influence chick embryo fibroblasts or positively regulate 3T3 cell proliferation (Joliot et al. 1992; Liu et al. 1999). In our analysis, we demonstrated that CCN3/NOV siRNA inhibits PDGF-induced DNA synthesis in CFSC without influencing PDGF signalling and cell numbers (cf. Fig. 6c and Suppl. Fig. 2).Additionally, we applied adenoviral gene transfer (Ad5-CMV-NOV) and recombinant CCN3/NOV protein in CFSC cultures. Ad5-CMV-NOV transduction showed high levels of CCN3/NOV secreted into culture medium reaching concentrations of ~3,500 pg/ml (Fig. 6d, left panel). In line with our previous experiments, we again found no differences in cell numbers compared to the uninfected control cultures, or cultured that were infected with Ad5-CMV-Luciferase or Ad5-CMV-CTGF (Fig. 6d, right panel). Application of the exogenous CCN3/NOV protein showed significantly decreased BrdU incorporation at concentrations of 200 ng/ml (p = 0.017) and 500 ng/ml (p = 0.008), respectively (Fig. 6e, left panel). No significant differences in regard to BrdU incorporation and cell counts (Fig. 6e, right panel) were noticed when cells were stimulated for 24 h with 20 ng/ml PDGF-B.Since the biological responses of immortalized cells and primary cells might be different, we used adenovirus gene transfers for CCN proteins in primary HSC. We seeded 100,000 cells/well on 12-well plates and infected them at the next day with different adenoviruses as mention above. Plating efficiency after 24 h viral incubation was approximately 35% (35,000 HSC/well). HSC were maintained in medium containing 0.5% or 10% FCS for 48 h in the presence (BrdU assay) or absence (cell count) of BrdU and harvested 24 h later for respective analysis. Both Ad5-CMV-CTGF (p = 0.015) and Ad5-CMV-NOV (p = 0.048) significantly decreased BrdU incorporation in the group of HSC maintained in 0.5% FCS, but no significant differences were observed when cells were cultured in 10% FCS (Fig. 6f, left panel). Cell counts confirmed the tendency that in both Ad5-CMV-CTGF- and Ad5-CMV-NOV-transduced HSC cell growth was attenuated in medium containing 10% FCS (Fig. 6f, right panel). All these findings indicate that CCN3/NOV by itself did not induce CFSC or HSC proliferation but rather inhibits proliferation instead.In summary, although expression of both CCN2/CTGF and CCN3/NOV overlap in culture-activated HSC and upregulate in vivo during ongoing fibrogenesis, their activities appear to counterbalance each other. Despite the fact that CCN2/CTGF is known as a prominent profibrogenic mediator in liver, we found CCN3/NOV to counteract fibrogenic marker gene expression in our experiments. Overexpression of CCN2/CTGF by Ad5-CMV-CTGF down regulates CCN3/NOV, but contrastingly overexpression of CCN3/NOV by Ad5-CMV-NOV and CCN3/NOV gene knock down both enhance CCN2 expression, showing that the CCN protein interplay is highly complex and extends beyond classic Yin/Yang interaction and raising additional questions to be answered.
Fig. 6.
CCN3/NOV effects on cell migration and proliferation. a, b Scratch-induced migration assay was performed in CCN3/NOV siRNA-transfected CFSC following a protocol outlined in the Material and Method section. All CCN3/NOV siRNA-transfected CFSC showed a significantly delayed wound gap closing after 20 and 40 h that become visible in light microscopic analysis (a) and was confirmed by gab closing quantification (b). c CFSC were transfected with control siRNA and siRNAs targeting CCN3/NOV. After 24 h starvation, cells were stimulated with PDGF-B in the presence of BrdU. The incorporation of BrdU was measured using an ELISA BrdU colorimetric assay kit. All CCN3/NOV siRNA showed inhibition of PDGF-induced cell proliferation (left panel), but no difference in cell count (right panel). d CFSC were infected with adenoviruses as indicated. Ad5-CMV-rNOV transduction induced high level expression of CCN3/NOV that was effectively secreted into culture medium (left panel), but that no impact on cell count compared to the control culture, control Ad5-CMV-Luciferase and Ad5-CMV-CTGF transduced CFSC (right panel). e CFSC were starved and incubated overnight with indicated doses of exogenous CCN3/NOV, followed by 24 h of 20 ng/ml PDGF-B and BrdU incubation. CCN3/NOV treated cells showed significantly decreased BrdU incorporation at concentrations of 200 and 500 ng/ml in 0.5% FCS (left panel), respectively. However, this analysis revealed no difference in cell numbers (right panel). f Primary HSC were infected with adenovirus as indicated and the medium changed after 24 h to 0.5% or 10% FCS for 48 h or 24 h in the presence of BrdU. Both Ad5-CMV-CTGF and Ad5-CMV-NOV significantly decreased BrdU incorporation in the group of HSC maintained in 0.5% FCS, but no significant difference in 10% FCS (left panel). Cell counts revealed that both Ad5-CMV-CTGF and Ad5-CMV-NOV but not the adenoviral control vector Ad-CMV-Luc attenuate cell growth when cells were cultured in medium containing 10% FCS (right panel)
Electronic supplementary material
Expression of CCN3/NOV in liver cell subpopulation and during transdifferentiation. (A) Quantitative RT-PCR analysis (TaqMan) from culture-activated HSC showing significant upregulation of CCN3/NOV, CCN2/CTGF and α-SMA that reach peak levels in fully transdifferentiated MFB. (B) No CCN3/NOV expression was found in naive hepatocytes that were cultured for indicated time intervals. (C) A standard RT-PCR for analysis of CCN3/NOV expression in different primary liver cells that were cultured for indicated time intervals was performed showing minimal expression in Kupffer cells (KC) and sinusoidal endothelial cells (SLEC) compared to culture-activated HSC and periportal miofibroblasts (pMF) in secondary culture (D) Double staining immunohistochemistry of CCN3/NOV (green) and ED2 (red). ED2, a specific Kupffer cell marker, showed no co-localization with CCN3/NOV (PPT 11.2 MB)
CCN3/NOV and PDGF signalling. (A) CFSC were transfected with control siRNA and CCN3/NOV siRNAs and starved for 24 h followed by stimulation with PDGF-B (20 ng/ml) for 15 min. Western blot analysis of respective cell lysates showed no effects of CCN3/NOV siRNA in PDGF-induced phosphorylation of PDGF receptors, AKT, ERK1/2, and p38. (B) The efficacy of CCN3/NOV knock down was confirmed in CCN3/NOV ELISA using cell supernatants taken from the same set of experiments (PPT 145 kb)
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (SFB/TRR57) to RW. We wish to extend our special thanks to U. Haas and G. Dietzel for their kind assistance.
Glossary
- Ad5
adenovirus type 5
- α-SMA
α-smooth muscle actin
- BDL
bile duct ligation
- CCN2/CTGF
connective tissue growth factor
- CCN3/NOV
Nephroblastoma overexpressed gene
- CFSC
cirrhotic fat storing cells
- EMT
epithelial-to-mesenchymal transition
- HSC
hepatic stellate cells
- KC
Kupffer cell(s)
- LSEC
liver sinusoidal endothelial cell(s)
- MFB
myofibroblast(s)
- MMP
matrix metalloproteinase
- PDGF
platelet-derived growth factor
- PDGFRβ
platelet-derived growth factor receptor type β
- pMF
periportal myofibroblasts
- TGF-β
transforming growth factor-β
- TIMP-1
tissue inhibitor of metalloproteinases-1.
Contributor Information
Erawan Borkham-Kamphorst, Phone: +49-241-8088683, FAX: +49-241-8082512, Email: ekamphorst@ukaachen.de.
Ralf Weiskirchen, Phone: +49-241-8088683, FAX: +49-241-8082512, Email: rweiskirchen@ukaachen.de.
References
- Abreu JG, Ketpura NI, Reversade B, Robertis EM. Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-β. Nat Cell Biol. 2002;4:599–604. doi: 10.1038/ncb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias M, Sauer-Lehnen S, Treptau J, Janoschek N, Theuerkauf I, Buettner R, Gressner AM, Weiskirchen R. Adenoviral expression of a transforming growth factor-beta1 antisense mRNA is effective in preventing liver fibrosis in bile-duct ligated rats. BMC Gastroenterol. 2003;3:29. doi: 10.1186/1471-230X-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benini S, Perbal B, Zambelli D, Colombo MP, Manara MC, Serra M, Parenza M, Martinez V, Picci P, Scotlandi K. In Ewing’s sarcoma CCN3(NOV) inhibits proliferation while promoting migration and invasion of the same cell type. Oncogene. 2005;24:4349–4361. doi: 10.1038/sj.onc.1208620. [DOI] [PubMed] [Google Scholar]
- Bleau AM, Planque N, Lazar N, Zambelli D, Ori A, Quan T, Fisher G, Scotlandi K, Perbal B. Antiproliferative activity of CCN3: involvement of the C-terminal module and post-translational regulation. J Cell Biochem. 2007;101:1475–1491. doi: 10.1002/jcb.21262. [DOI] [PubMed] [Google Scholar]
- Bohr W, Kupper M, Hoffmann K, Weiskirchen R. Recombinant expression, purification, and functional characterisation of connective tissue growth factor and nephroblastoma-overexpressed protein. PLoS One. 2010;5(12):e16000. doi: 10.1371/journal.pone.0016000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P. The modular architecture of a new family of growth regulators related to connective tissue growth factor. FEBS Lett. 1993;327:125–130. doi: 10.1016/0014-5793(93)80155-N. [DOI] [PubMed] [Google Scholar]
- Borkham-Kamphorst E, Herrmann J, Stoll D, Treptau J, Gressner AM, Weiskirchen R. Dominant-negative soluble PDGF-β receptor inhibits hepatic stellate cell activation and attenuates liver fibrosis. Lab Invest. 2004;84:766–777. doi: 10.1038/labinvest.3700094. [DOI] [PubMed] [Google Scholar]
- Borkham-Kamphorst E, Stoll D, Gressner AM, Weiskirchen R. Inhibitory effect of soluble PDGF-β receptor in culture-activated hepatic stellate cells. Biochem Biophys Res Commun. 2004;318:451–462. doi: 10.1016/j.bbrc.2004.03.064. [DOI] [PubMed] [Google Scholar]
- Borkham-Kamphorst E, Roeyen CR, Ostendorf T, Floege J, Gressner AM, Weiskirchen R. Pro-fibrogenic potential of PDGF-D in liver fibrosis. J Hepatol. 2007;46:1064–1074. doi: 10.1016/j.jhep.2007.01.029. [DOI] [PubMed] [Google Scholar]
- Borkham-Kamphorst E, Kovalenko E, Roeyen CR, Gassler N, Bomble M, Ostendorf T, Floege J, Gressner AM, Weiskirchen R. Platelet-derived growth factor isoform expression in carbon tetrachloride-induced chronic liver injury. Lab Invest. 2008;88:1090–1100. doi: 10.1038/labinvest.2008.71. [DOI] [PubMed] [Google Scholar]
- Bradham DM, Igarashi A, Potter RL, Grotendorst GR. Connective tissue growth factor: a cysteine-rich mitogen secreted by human vascular endothelial cells is related to the SRC-induced immediate early gene product CEF-10. J Cell Biol. 1991;114:1285–1294. doi: 10.1083/jcb.114.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner A, Chinn J, Neubauer M, Purchio AF. Identification of a gene family regulated by transforming growth factor-β. DNA Cell Biol. 1991;10:293–300. doi: 10.1089/dna.1991.10.293. [DOI] [PubMed] [Google Scholar]
- Fehrenbach H, Weiskirchen R, Kasper M, Gressner AM. Upregulated expression of the receptor for advanced glycation end products in cultured rat hepatic stellate cells during transdifferentiation to myofibroblasts. Hepatology. 2001;34:943–952. doi: 10.1053/jhep.2001.28788. [DOI] [PubMed] [Google Scholar]
- Gao R, Ball DK, Perbal B, Brigstock DR. Connective tissue growth factor induces c-fos gene activation and cell proliferation through p44/42 MAP kinase in primary rat hepatic stellate cells. J Hepatol. 2004;40:431–438. doi: 10.1016/j.jhep.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Greenwel P, Schwartz M, Rosas M, Peyrol S, Grimaud JA, Rojkind M. Characterization of fat-storing cell lines derived from normal and CCl4-cirrhotic livers. Differences in the production of interleukin-6. Lab Invest. 1991;65:644–653. [PubMed] [Google Scholar]
- Greenwel P, Rubin J, Schwartz M, Hertzberg EL, Rojkind M. Liver fat-storing cell clones obtained from a CCl4-cirrhotic rat are heterogeneous with regard to proliferation, expression of extracellular matrix components, interleukin-6, and connexin 43. Lab Invest. 1993;69:210–216. [PubMed] [Google Scholar]
- Gupta N, Wang H, McLeod TL, Naus CC, Kyurkchiev S, Advani S, Yu J, Perbal B, Weichselbaum R. Inhibition of glioma cell growth and tumorigenic potential by CCN3 (NOV) Mol Pathol. 2001;54:293–299. doi: 10.1136/mp.54.5.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasaki S, Koide N, Ujike K, Shinji T, Tsuji T. Expression of Nov, CYR61 and CTGF genes in human hepatocellular carcinoma. Hepatol Res. 2001;19:294–305. doi: 10.1016/S1386-6346(00)00101-7. [DOI] [PubMed] [Google Scholar]
- Joliot V, Martinerie C, Dambrine G, Plassiart G, Brisac M, Crochet J, Perbal B. Proviral rearrangements and overexpression of a new cellular gene (nov) in myeloblastosis-associated virus type 1-induced nephroblastomas. Mol Cell Biol. 1992;12:10–21. doi: 10.1128/mcb.12.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaki H, Kubota S, Suzuki A, Lazar N, Yamada T, Matsumura T, Ohgawara T, Maeda T, Perbal B, Lyons KM, Takigawa M. Cooperative regulation of chondrocyte differentiation by CCN2 and CCN3 shown by a comprehensive analysis of the CCN family proteins in cartilage. J Bone Miner Res. 2008;23:1751–1764. doi: 10.1359/jbmr.080615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafont J, Laurent M, Thibout H, Lallemand F, Bouc Y, Atfi A, Martinerie C. The expression of novH in adrenocortical cells is down-regulated by TGFβ1 through c-Jun in a Smad-independent manner. J Biol Chem. 2002;277:41220–41229. doi: 10.1074/jbc.M204405200. [DOI] [PubMed] [Google Scholar]
- Leask A, Chen S, Pala D, Brigstock DR. Regulation of CCN2 mRNA expression and promoter activity in activated hepatic stellate cells. J. Cell Commun. Signal. 2008;2:49–56. doi: 10.1007/s12079-008-0029-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Seo GS, Park YN, Sohn DH. Nephroblastoma overexpressed gene (NOV) expression in rat hepatic stellate cells. Biochem Pharmacol. 2004;68:1391–1400. doi: 10.1016/j.bcp.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Lin CG, Chen CC, Leu SJ, Grzeszkiewicz TM, Lau LF. Integrin-dependent functions of the angiogenic inducer NOV (CCN3): implication in wound healing. J Biol Chem. 2005;280:8229–8237. doi: 10.1074/jbc.M404903200. [DOI] [PubMed] [Google Scholar]
- Liu C, Liu XJ, Crowe PD, Kelner GS, Fan J, Barry G, Manu F, Ling N, Souza EB, Maki RA. Nephroblastoma overexpressed gene (NOV) codes for a growth factor that induces protein tyrosine phosphorylation. Gene. 1999;238:471–478. doi: 10.1016/S0378-1119(99)00364-9. [DOI] [PubMed] [Google Scholar]
- Minamizato T, Sakamoto K, Liu T, Kokubo H, Katsube K, Perbal B, Nakamura S, Yamaguchi A. CCN3/NOV inhibits BMP-2-induced osteoblast differentiation by interacting with BMP and Notch signaling pathways. Biochem Biophys Res Commun. 2007;354:567–573. doi: 10.1016/j.bbrc.2007.01.029. [DOI] [PubMed] [Google Scholar]
- Nevzorova YA, Tschaharganeh D, Gassler N, Geng Y, Weiskirchen R, Sicinski P, Trautwein C, Liedtke C. Aberrant cell cycle progression and endoreplication in regenerating livers of mice that lack a single E-type cyclin. Gastroenterology. 2009;137:691–703. doi: 10.1053/j.gastro.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzani M, Gesualdo L, Sabbah GM, Abboud HE. Effects of platelet-derived growth factor and other polypeptide mitogens on DNA synthesis and growth of cultured rat liver fat-storing cells. J Clin Invest. 1989;84:1786–1793. doi: 10.1172/JCI114363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riser BL, Najmabadi F, Perbal B, Peterson DR, Rambow JA, Riser ML, Sukowski E, Yeger H, Riser SC. CCN3 (NOV) is a negative regulator of CCN2 (CTGF) and a novel endogenous inhibitor of the fibrotic pathway in an in vitro model of renal disease. Am J Pathol. 2009;174:1725–1734. doi: 10.2353/ajpath.2009.080241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riser BL, Najmabadi F, Perbal B, Rambow JA, Riser ML, Sukowski E, Yeger H, Riser SC, Peterson DR. CCN3/CCN2 regulation and the fibrosis of diabetic renal disease. J. Cell. Commun. Signal. 2010;4:39–50. doi: 10.1007/s12079-010-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydziel S, Stadmeyer L, Zanotti S, Durant D, Smerdel-Ramoya A, Canalis E. Nephroblastoma overexpressed (Nov) inhibits osteoblastogenesis and causes osteopenia. J Biol Chem. 2007;282:19762–18772. doi: 10.1074/jbc.M700212200. [DOI] [PubMed] [Google Scholar]
- Schäfer S, Zerbe O, Gressner AM. The synthesis of proteoglycans in fat-storing cells of rat liver. Hepatology. 1987;7:680–687. doi: 10.1002/hep.1840070411. [DOI] [PubMed] [Google Scholar]
- Sin WC, Tse M, Planque N, Perbal B, Lampe PD, Naus CC. Matricellular protein CCN3 (NOV) regulates actin cytoskeleton reorganization. J Biol Chem. 2009;284:29935–29944. doi: 10.1074/jbc.M109.042630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soret J, Dambrine G, Perbal B. Induction of nephroblastoma by myeloblastosis-associated virus type 1: state of proviral DNAs in tumor cells. J Virol. 1989;63:1803–1807. doi: 10.1128/jvi.63.4.1803-1807.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibout H, Martinerie C, Créminon C, Godeau F, Boudou P, Bouc Y, Laurent M. Characterization of human NOV in biological fluids: an enzyme immunoassay for the quantification of human NOV in sera from patients with diseases of the adrenal gland and of the nervous system. J Clin Endocrinol Metab. 2003;88:327–336. doi: 10.1210/jc.2002-020304. [DOI] [PubMed] [Google Scholar]
- Tong Z, Chen R, Alt DS, Kemper S, Perbal B, Brigstock DR. Susceptibility to liver fibrosis in mice expressing a connective tissue growth factor transgene in hepatocytes. Hepatology. 2009;50:939–947. doi: 10.1002/hep.23102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeyen CR, Eitner F, Scholl T, Boor P, Kunter U, Planque N, Gröne HJ, Bleau AM, Perbal B, Ostendorf T, Floege J. CCN3 is a novel endogenous PDGF-regulated inhibitor of glomerular cell proliferation. Kidney Int. 2008;73:86–94. doi: 10.1038/sj.ki.5002584. [DOI] [PubMed] [Google Scholar]
- Weiskirchen R. CCN proteins in liver health and disease. Front Biosci. 2011;16:1939–1961. doi: 10.2741/3832. [DOI] [PubMed] [Google Scholar]
- Weiskirchen R, Bister K. Suppression in transformed avian fibroblasts of a gene (crp) encoding a cysteine-rich protein containing LIM domains. Oncogene. 1993;8:2317–2324. [PubMed] [Google Scholar]
- Weiskirchen R, Kneifel J, Weiskirchen S, Leur E, Kunz D, Gressner AM. Comparative evaluation of gene delivery devices in primary cultures of rat hepatic stellate cells and rat myofibroblasts. BMC Cell Biol. 2000;1:4. doi: 10.1186/1471-2121-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng ZJ, Yang LY, Ding X, Wang W. Expressions of cysteine-rich 61, connective tissue growth factor and Nov genes in hepatocellular carcinoma and their clinical significance. World J Gastroenterol. 2004;10:3414–3418. doi: 10.3748/wjg.v10.i23.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of CCN3/NOV in liver cell subpopulation and during transdifferentiation. (A) Quantitative RT-PCR analysis (TaqMan) from culture-activated HSC showing significant upregulation of CCN3/NOV, CCN2/CTGF and α-SMA that reach peak levels in fully transdifferentiated MFB. (B) No CCN3/NOV expression was found in naive hepatocytes that were cultured for indicated time intervals. (C) A standard RT-PCR for analysis of CCN3/NOV expression in different primary liver cells that were cultured for indicated time intervals was performed showing minimal expression in Kupffer cells (KC) and sinusoidal endothelial cells (SLEC) compared to culture-activated HSC and periportal miofibroblasts (pMF) in secondary culture (D) Double staining immunohistochemistry of CCN3/NOV (green) and ED2 (red). ED2, a specific Kupffer cell marker, showed no co-localization with CCN3/NOV (PPT 11.2 MB)
CCN3/NOV and PDGF signalling. (A) CFSC were transfected with control siRNA and CCN3/NOV siRNAs and starved for 24 h followed by stimulation with PDGF-B (20 ng/ml) for 15 min. Western blot analysis of respective cell lysates showed no effects of CCN3/NOV siRNA in PDGF-induced phosphorylation of PDGF receptors, AKT, ERK1/2, and p38. (B) The efficacy of CCN3/NOV knock down was confirmed in CCN3/NOV ELISA using cell supernatants taken from the same set of experiments (PPT 145 kb)