Abstract
We investigated the functional effects of microRNA-34a (miR-34a) on c-Myc transcriptional complexes in renal cell carcinoma. miR-34a down-regulated expression of multiple oncogenes including c-Myc by targeting its 3′ untranslated region, which was revealed by luciferase reporter assays. miR-34a was also found to repress RhoA expression by suppressing the c-Myc–Skp2–Miz1 transcriptional complex that activates RhoA. Overexpression of c-Myc reversed miR-34a suppression of RhoA expression and inhibition of cell invasion, suggesting that miR-34a inhibits invasion by suppressing RhoA through c-Myc. miR-34a was also found to repress the c-Myc–P-TEFb transcription elongation complex, indicating one of the mechanisms by which miR-34a has profound effects on cellular functions. Our results demonstrate that miR-34a suppresses assembly and function of the c-Myc complex that activates or elongates transcription, indicating a novel role of miR-34a in the regulation of transcription by c-Myc.
Introduction
MicroRNAs (miRNAs) are highly conserved, single stranded, non-coding RNAs of ∼22 nucleotides that regulate gene expression by posttranscriptional silencing of specific target messenger RNAs (mRNAs) by repressing translation or cleaving RNA transcripts (1). miRNAs regulate diverse cellular processes, such as cell-cycle progression, proliferation apoptosis and development. miRNAs have been shown to function as oncogenes or tumor suppressor genes (2). p53 has been found to target the microRNA-34 (miR-34) family (3–5) and the ectopic expression of miR-34 genes has drastic effects on cell proliferation and survival. Ectopic miR-34a causes cell-cycle arrest in the G1 phase (5,6), increased apoptosis (6,7) and inhibits metastasis (8). It has also been suggested that a positive feedback loop exists between p53 and miR-34a as miR-34a activates p53 by inhibiting silent information regulator 1 (9). Implications of miR-34a in renal cancer suppression and progression have been reported (10,11), however, no functional study has been conducted.
The proto-oncogene c-Myc regulates cell proliferation and transformation both transcriptionally and non-transcriptionally and is frequently de-regulated in human cancers (12,13). c-Myc is a basic helix–loop–helix and leucine zipper transcription factor, which binds to Enhancer Box elements (E-boxes) and activates the transcription of genes which stimulate cell-cycle progression and cell growth. c-Myc was reported to activate MiR-17-92, a polycistronic miRNA cluster consisting of miR-17, -18a, -19a, -20a, -19b and -92a (14,15). miR-19 was found to be the principal oncogenic component of this cluster, targeting the tumor suppressor (16). miR-34c has been shown to negatively regulate c-Myc in response to DNA damage and to inhibit c-Myc-induced DNA synthesis (17). During oncogene-induced senescence, miR-34a was also found to target c-Myc (18).
Rho GTPases are small G proteins that regulate various cellular processes, including cytoskeletal dynamics, migration, vesicle trafficking, cell proliferation, apoptosis and transcription (19–21). Rho GTPases, their regulators and their effectors have been suggested to control tumor formation and progression. RhoA has been found to control cancer metastasis and progression (22–24). Recently, the c-Myc–Skp2–Miz1 complex was shown to activate the RhoA gene (25).
The positive transcription elongation factor b (P-TEFb) regulates the promoter-proximal pause release of the elongation phase of transcription by RNA polymerase II (Pol II) (26) and integrates mRNA synthesis with histone modification, pre-mRNA processing and mRNA export (27). P-TEFb is composed of cyclin (CycT1, CycT2a, CycT2b or CycK) and cyclin-dependent kinase 9 (Cdk9) (26). c-Myc interacts with cyclin T1 (CycT1), the regulatory component of P-TEFb and controls the elongation phase of transcription of Pol II (28–30).
We report here the effects of miR-34a on the two c-Myc transcriptional complexes. Via targeting c-Myc, miR-34a reduced the c-Myc–Miz–Skp2 complex, which induces RhoA transcription and inhibited cell invasion, showing miR-34a indirectly regulates RhoA. miR-34a also suppressed c-Myc–P-TEFb complex that plays a key role in controlling the elongation phase of transcription by Pol II, indicating one of mechanisms by which miR-34a has dramatic effects on cellular function. Our results demonstrate that miR-34a directly suppresses formation and function of the c-Myc complex that activates or elongates transcription, revealing a novel role of miR-34a in the regulation of transcription by c-Myc.
Materials and methods
Cell culture and transfection
Human renal carcinoma cells, A498 [primary renal cell carcinoma (RCC)] and 769P cells were purchased from The American Type Culture Collection (Manassas, VA). A498 and 769P cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum.
Cells in six-well plates were transfected with 30 nM pre-miR negative control (NC) or pre-miR-34a (Applied Biosystems, Foster City, CA) using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions.
Transwell invasion assay
A498 or 769P cells were grown in Dulbecco's modified Eagle’s medium containing 10% fetal bovine serum. Culture inserts of 8 μm pore size (Transwell; Costar) were coated with Matrigel (BD Biosciences, San Jose, CA) (100 μg per well) and placed into the wells of 24-well culture plates. In the lower chamber, 500 μl of Dulbecco's modified Eagle’s medium containing 10% fetal bovine serum was added and 1 × 105 cells were seeded to the upper chamber. After 48 h of incubation at 37°C with 5% CO2, the number of cells that had migrated through the pores was fixed with 10% formalin and stained with 0.05% Crystal Violet. Crystal Violet was solubilized with methanol and absorbance at 540 nm was measured by a kinetic microplate reader (Spectra MAX 190; Molecular Devices Co., Sunnyvale, CA). Data are the mean ± standard deviation of three independent experiments.
RNA extraction and quantitative real-time PCR
RNA was isolated using the RNeasy Mini kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Reverse transcription reactions were performed using a Reverse Transcription System Kit (Promega, Madison, WI). Quantitative real-time polymerase chain reaction (PCR) analysis was performed in triplicate with an Applied Biosystems Prism7500 Fast Sequence Detection System using TaqMan universal PCR master mix according to the manufacturer’s protocol (Applied Biosystems). Levels of RNA expression were determined using the 7500 Fast System SDS software version 1.3.1 (Applied Biosystems).
miRNA extraction and quantitative real-time PCR
Total RNA was isolated using the miRNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. Reverse transcription reactions were performed using a Reverse Transcription System Kit (Applied Biosystems). Quantitative real-time PCR analysis was performed as described above.
Plasmid construction
Putative target sites of the 3′ untranslated region (UTR) were cloned into the PmeI–XbaI site of the dual luciferase pmirGLO vector (Promega). For mismatch constructs, six mismatches were introduced in the putative target site.
A human c-Myc expression vector was constructed by subcloning the full-length complementary DNA of c-Myc (Invitrogen, Carlsbad, CA) into the HindIII–XhoI site of the pCMV6-ENTRY vector (Origene).
Generation of stable miR-34a cell lines
HIV-based lentiviral packaging system including a plasmid expressing miR-34a or vector control was purchased from GeneCopoeia (Rockville, MD). A498 cells were infected with the HIV-based lentivirus expressing miR-34a or vector control, and the infected A498 cells were selected with puroc-Mycine.
Western blot
Protein extracts were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes (Hybond-P; GE Healthcare, Piscataway, NJ), followed by incubation with the indicated primary and secondary antibodies conjugated to horseradish peroxidise (GE Healthcare). Signals were detected using the enhanced chemiluminescence detection system (Amersham ECL plus Western Blotting detection system, Fairfield, CT). Antibodies against c-Myc, RhoA, Skp2 and GAPDH were purchased from Cell Signaling Technology (Danvers, MA). Antibodies against Miz1, Max, Cdk9, ARHGEF3 and ARHGEF18 were purchased from GeneTex Irvine, CA). Antibodies against BCL2and CycT1 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). An antibody against Rock1 was purchased from BD Biosciences.
Luciferase reporter assay
Cells in 24-well plates were transfected with 30 nM pre-miR NC or pre-miR-34a (Applied Biosystems) using Lipofectamin 2000 (Invitrogen), according to the manufacturer’s instructions. Transfections of plasmids were performed using FuGENE HD (Roche Diagnosis, Basel, Switzerland) according to the manufacturer’s instructions. Luciferase activity was assayed 48 h after transfection, using a dual-luciferase reporter assay system (Promega). All transfection experiments were performed in triplicate.
Chromatin immunoprecipitation
Chromatin immunoprecipitations (ChIPs) were performed using the Chromatin Immunoprecipitation Assay Kit (Epigentek, Brooklyn, NY) according to manufacturer’s instructions. DNA was sheared by sonication. A 1% portion of the sheared DNA–protein complex was used for an input DNA sample. Antibodies against c-Myc or Pol II (Millipore, Billerica, MA) or normal mouse/rabbit IgG (Millipore) was used for immunoprecipitation (IP). Real-time PCR quantitation of ChIP was performed in triplicate, normalized by input and expressed as a fold increase over the control. Primers used for real-time PCR were as follows: RhoA E-box5/6, 5′-CTTCGCGTGCGTGAAGAGTTG-3′and 5′-CATCCACTATTGCTCAGGAGC-3′; β-actin promoter, 5′-TCGATATCCACGTGACATCCA-3′ and 5'-GCAGCATTTTTTTACCCCCTC-3'.
Immunoprecipitation
Cells were lysed in buffer containing 250 mM NaCl, 50 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.5, 0.1% Nonidet P40, 5 mM ethylenediaminetetraacetic acid and a protease inhibitor cocktail (Sigma–Aldrich, St Louis, MO). Protein A/G–Sepharose beads (Santa Cruz Biotechnology) were added to the lysate for 60 min at 4°C for preclearance. The precleared lysate was incubated with an anti-c-Myc antibody (Cell Signaling Technology) and protein A/G–Sepharose beads overnight at 4°C. The protein A/G–Sepharose beads were washed with the lysis buffer. The proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and analyzed by western blot.
Statistical analysis
Data are shown as mean values ± standard deviation. The Student’s t-test was used to compare the two different groups. P values of <0.05 were regarded as statistically significant (n = 3).
Results
miR-34a targets oncogenes
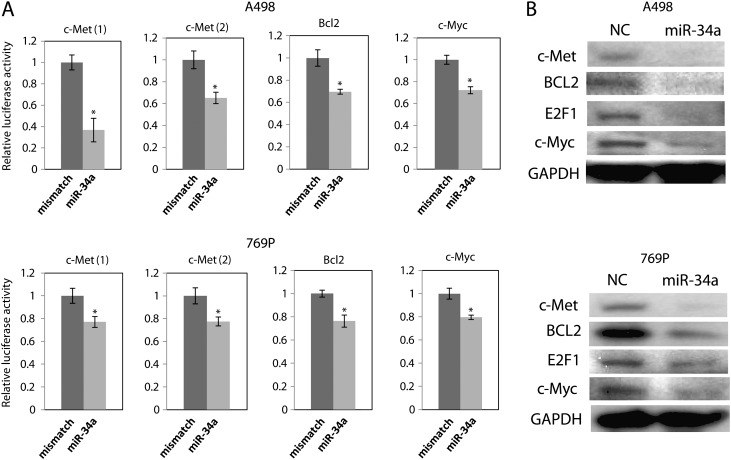
To study the target genes of miR-34a in RCCs, we transiently transfected A498 and 769P cells with pre-miR NC or pre-miR-34a. The transient transfection of pre-miR-34a increased miR-34a levels in A498 and 769P cells (Supplementary Figure S1, available at Carcinogenesis Online). Oncogenes, c-Met, c-Myc and BCL2, have predicted binding sites for miR-34a in their 3′ UTRs (Supplementary Figure S2, available at Carcinogenesis Online). c-Met has two predicted binding sites for miR-34a in its 3′ UTR (Supplementary Figure S2, available at Carcinogenesis Online). We cloned the putative miR-34a targets in the 3′ UTRs into a luciferase construct. Luciferase reporter assays with miR-34a-expressing A498 and 769P cells showed that miR-34a repressed luciferase activity. Mutation of the putative miR-34a target sites in these UTRs decreased the response to miR-34a. These results indicate that miR-34a binds to the 3′ UTRs of these genes (Figure 1A).
Fig. 1.
miR-34a targets oncogenes in A498 and 769P cells. (A) A498 or 769P cells were transfected with pre-miR NC or pre-miR-34a or pre-miR-con for 8 h, followed by transfection with control reporter plasmids or 3′ UTR plasmids for 48 h. 3′ UTR reporter activity was measured with a luciferase assay and normalized to activity of Renilla luciferase. *P < 0.05 compared with control. (B) A498 or 769P cells were transfected with pre-miR NC or pre-miR-34a or pre-miR-con for 72 h. Protein expression level was analyzed by western blot.
Subsequently, we found that transfection of pre-miR-34a reduced the protein levels of c-Met, c-Myc and BCL2 protein in A498 and 769P cells (Figure 1B). The protein expression levels were quantified and shown in Supplementary Figure S3, available at Carcinogenesis Online. These results indicate that miR-34a directly targets c-Met and c-Myc via binding to the 3′ UTRs in A498cells. miR-34a also reduced transcription factor E2F1, which regulates the cell cycle, DNA replication and cell proliferation (Figure 1B). Compared with the results of 3 ′UTR reporter assays (Figure 1A), the reduction of the protein level was significant, suggesting translational regulation. Other signaling pathways on which miR-34a has effects also may reduce c-Myc protein expression because miR-34a targets many genes. Further studies are required to account for the differences in these assays.
miR-34a inhibits A498 and 769P cell invasion
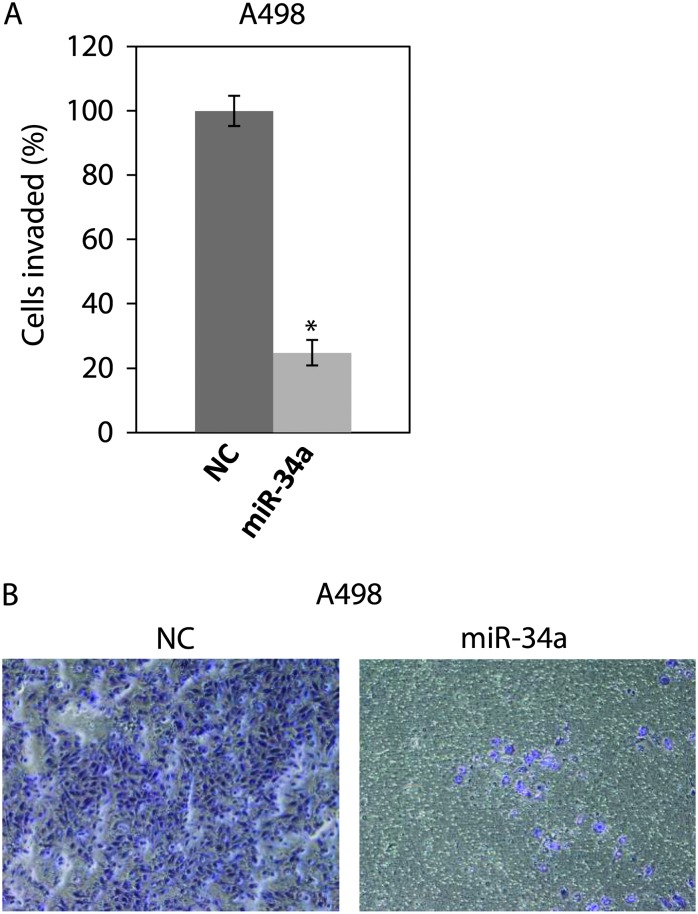
The c-Myc–Skp2–Miz1 transcriptional complex was found to activate RhoA gene and to be critical for cell invasion and cancer metastasis (25). Since we found that c-Myc is a target of miR-34a, we studied effect of miR-34a on RhoA through the c-Myc–Skp2–Miz1 transcriptional complex. We first performed a transwell invasion assay using Matrigel to investigate the effect of miR-34a on the invasion of A498 cells. We transiently transfected the cells with pre-miR NC or pre-miR-34a and performed transwell invasion assay. miR-34 reduced invasion to about 80% of that of NC in A498 cells (Figure 2). We also investigated the effect of miR-34a on the invasion of 769P cells. miR-34a reduced invasion to only about 20% of that of NC in 769P cells (Supplementary Figure S4, available at Carcinogenesis Online).
Fig. 2.
miR-34a inhibits invasion of A498 cells. (A) A498 cells were transfected with pre-miR NC or pre-miR-34a for 48 h. The cells were harvested and subjected to transwell invasion assay. The values are normalized to those of NC. *P < 0.05 compared with control. (B) Representative images of the invaded A498 cells.
miR-34a inhibits RhoA and suppresses assembly of the c-Myc transcriptional complex
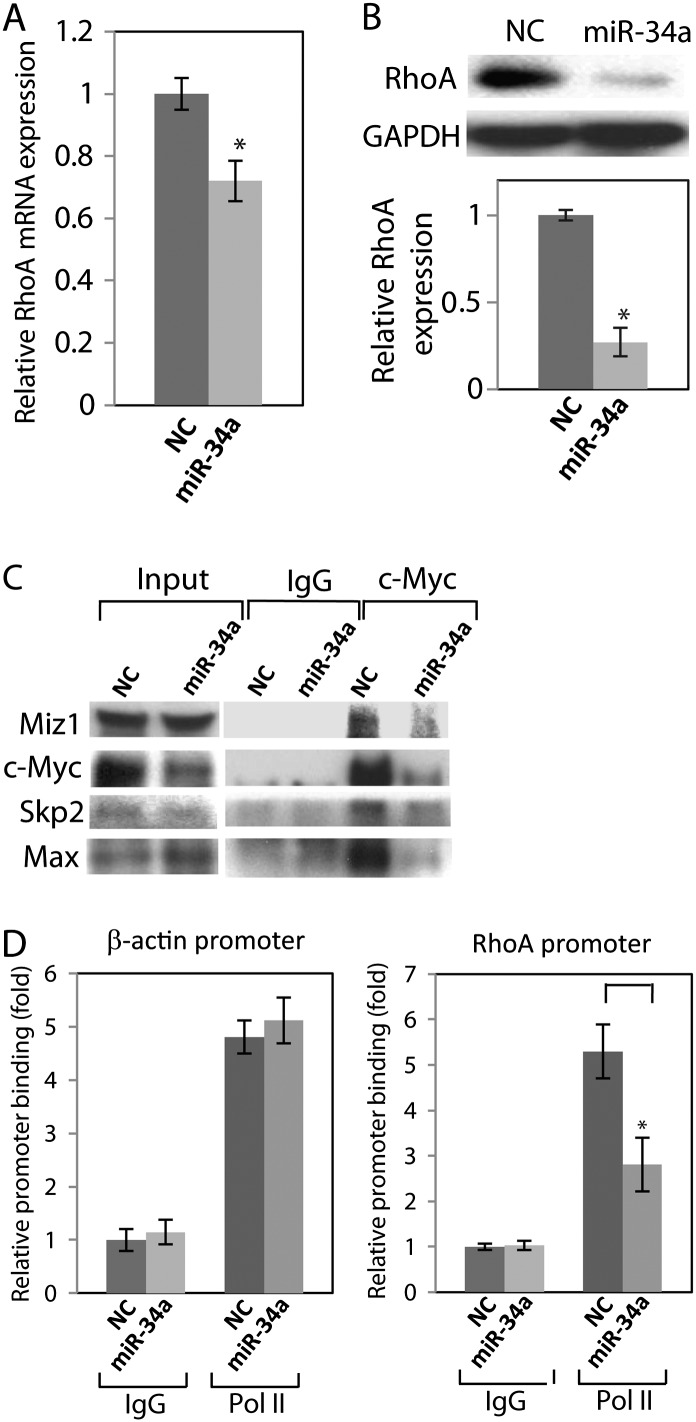
Since we found that miR-34a down-regulated the expression of c-Myc by targeting its 3′ UTR (Figure1), we examined whether the down-regulation of c-Myc by miR-34a results in reduction of RhoA expression through decreasing the assembly of the c-Myc–Skp2–Miz1 complex. We observed a large-inhibition effect of miR-34a on invasion in A498 cells, thus we used A498 cells for further invasion study. Real-time PCR shows that miR-34a decreased RhoA mRNA levels (Figure 3A) and western blot revealed that miR-34a reduced RhoA protein expression (Figure 3B).
Fig. 3.
miR-34a suppresses the c-Myc transcriptional complex of RhoA. miR-34a reduces RhoA expression in A498cells. A498cells were transfected with pre-miR NC or pre-miR-34a for 72 h. (A) RhoA mRNA level was analyzed by real-time PCR. (B) RhoA protein expression was analyzed by western blot. (C) c-Myc pulls down endogenous Miz1, Skp2 and Max in A498 cells. A498 cells were transfected with pre-miR NC or pre-miR-34a for 72 h and total cell lysates were immunoprecipitated with c-Myc antibody, followed by western blot analysis. (D) miR-34a decreases the recruitment of c-Myc to the RhoA promoter. A498 cells were transfected with pre-miR NC or pre-miR-34a for 72 h and ChIP assays were performed. P < 0.05 compared with control.
We performed IP to study the assembly of c-Myc–Skp2–Miz1 transcriptional complex in miR-34a-transfected cells. IP revealed that endogenous c-Myc immunocomplex contained Miz1, Skp2 and Max, indicating that c-Myc–Skp2–Miz1 complex was assembled in A498 cells and miR-34a decreased these components in the c-Myc immunocomplex (Figure 3C).
We performed ChIP assays to examine whether miR-34a reduces binding of c-Myc to the RhoA promoter region of E-boxes 5 and 6, which is a primary binding site of c-Myc (25). The ChIP assay showed that endogenous c-Myc binds to E-boxes 5 and 6 and miR-34a reduced the binding. Pol II bound to the β-actin promoter was not altered in control experiments (Figure 3D). These results suggested that miR-34a reduced the recruitment of c-Myc–Skp2–Miz1 complex to the RhoA promoter reducing RhoA expression.
Overexpression of c-Myc reverses inhibition of invasion
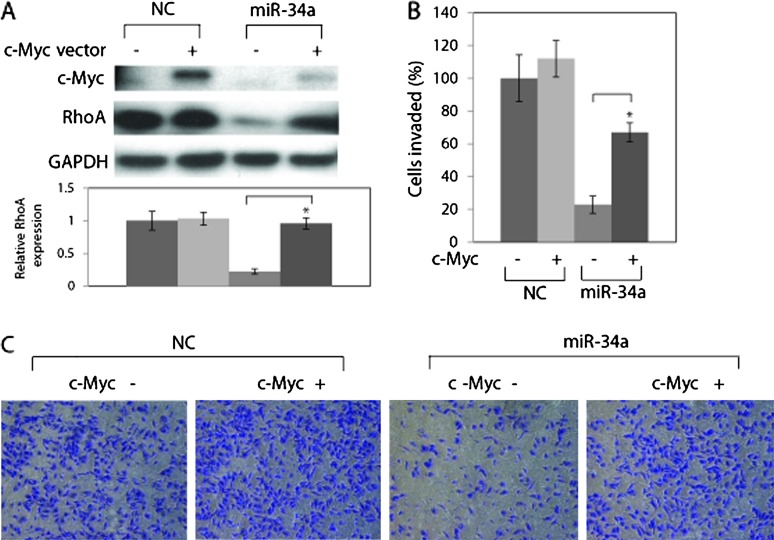
To examine whether inhibition of invasion by miR-34 could be reversed via restoration of c-Myc levels, we transfected A498 cells with c-Myc expressing plasmid together with pre-miR-34a. c-Myc overexpression partially rescued RhoA expression (Figure 4A) and miR-34-induced suppression of invasion (Figure 4B).
Fig. 4.
c-Myc overexpression partially rescues RhoA suppression by miR-34a. (A) A498 cells were transfected with pCMV6-ENTRY only (control) or pCMV6-ENTRY–c-Myc for 8 h followed by transfection with pre-miR NC or pre-miR-34a for 72 h. Protein level was analyzed by western blot. (B) c-Myc expression partially rescued inhibition of invasion induced by miR-34a. A498 cells were transfected with pCMV6-ENTRY only (control) or pCMV6-ENTRY–c-Myc for 8 h followed by transfection with pre-miR NC or pre–miR-34a for 48 h. Cells were harvested and subjected to transwell invasion assay. P < 0.05 compared with control. (B) Representative images of the invaded cells.
We also transfected a stable miR-34a A498 cell line with c-Myc expressing plasmid. To express miR-34a stably, we employed the HIV lentiviral system to infect A498 cells with the virus expressing miR-34a or vector control and the infected cells were selected with puromycin. The infection increased miR-34a levels by 65-fold in A498 cells (Supplementary Figure S5, available at Carcinogenesis Online). c-Myc overexpression also partially rescued RhoA expression and miR-34-induced suppression of invasion in the stable miR-34a A498 cell line (Supplementary Figure S6, available at Carcinogenesis Online). These results suggest that miR-34a inhibits invasion at least partially via RhoA reduction by targeting c-Myc.
miR-34a suppresses assembly of the c-Myc–P-TEFb complex
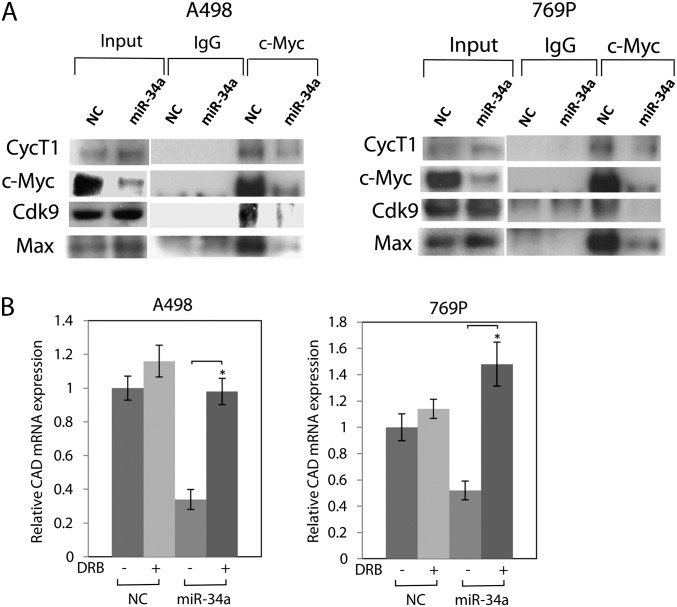
The P-TEFb plays a key role in controlling the elongation phase of transcription by Pol II (26). It is a Cdk comprised of Cdk9 and Cyc. c-Myc has been shown to interact with P-TEFb through CycT1 and regulate transcription elongation (28–30). Since miR-34a targets c-Myc, we performed IP to study the assembly of c-Myc–P-TEFb transcription elongation complex in mir-34a-transfected A498 and 769P cells. IP revealed that endogenous c-Myc pulled down CycT1, Cdk9 and Max and interacted with P-TEFb. miR-34a decreased the presence of P-TEFb in the c-Myc immunocomplex in A498 and 769P cells (Figure 5A).
Fig. 5.
miR-34a suppresses the c-Myc–P-TEFb complex. (A) A498 or 769P cells were transfected with pre-miR NC or pre–miR-34a for 72 h and total cell lysates were immunoprecipitated with c-Myc antibody, followed by western blot analysis. (B) miR-34a suppresses cad expression. A498 or 769P cells were transfected with pre-miR NC or pre–miR-34a or pre-miR-con for 72 h. Protein expression level of cad was analyzed by western blot.
c-Myc has been shown to activate transcription of carbamoyl-phosphate synthase/aspartate transcarbamoylase/dihydroorotase (CAD), via recruitment of P-TEFb (28,31,32). We studied the effect of miR-34a on the expression level of CAD with 5,6-di-chloro-1-b-d-ribofuranosyl-bensimidazole that specifically suppresses expression of c-Myc-responsive CAD by inhibiting P-TEFb (32). We found that miR-34a decreased CAD mRNA expression in A498 and 769P cells, whereas 5,6-di-chloro-1-b-d-ribofuranosyl-bensimidazole restored the expression (Figure 5B). These results indicate that miR-34a repressed these genes by suppressing the c-Myc–P-TEFb complex.
Discussion
In this study, we demonstrated the functional effects of miR-34a on c-Myc transcriptional complexes in RCC. We showed that miR-34a inhibited c-Myc by binding to its 3′ UTR and subsequently suppresses the assembly and function of the c-Myc–Skp2–Miz1 complex that activates RhoA and the c-Myc–P-TEFb complex that elongates transcription of a variety of genes in RCC. Mutations of c-Myc are found in various cancers and its de-regulated expression causes the uncontrolled expression of many genes, some of which regulate cell proliferation and results in tumor development (33,34). Although Myc alone is not able to transform cells and requires a co-operating oncogene, such as ras oncogene (35), c-Myc is one of the major factors in tumor development. Thus, inhibition of c-Myc is thought to be a significant function of miR-34a. Our results also show that miR-34a strongly inhibits cell proliferation, in vivo xenograft tumor growth and cell invasion, indicating that miR-34a functions as a tumor suppressor in A498 and 769P cells.
Rho GTPases, a family of small G proteins, regulate intracellular actin dynamics, including cell polarity, migration, vesicle trafficking, cell proliferation, apoptosis and gene expression (20). RhoA, a member of the Rho family of GTPases, is implicated in transformation and metastasis. RhoA has been documented to be an important factor associated with various human cancers (36).
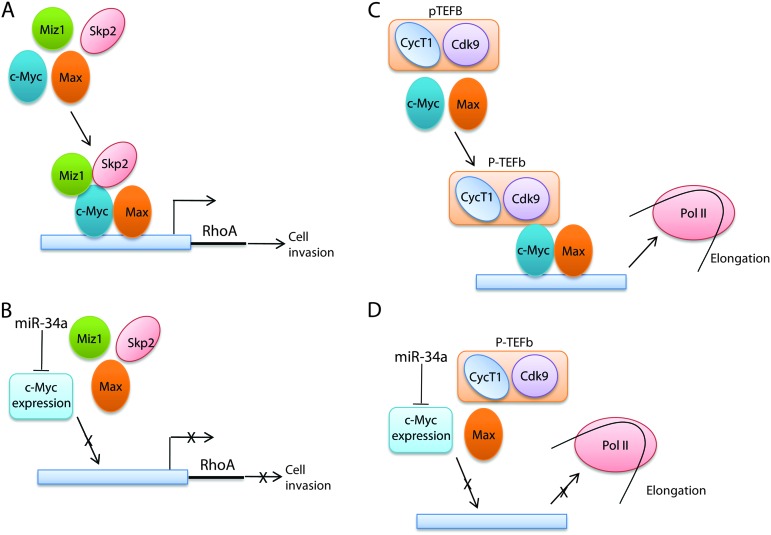
Cross talk between c-Myc and RhoA has been documented. Proteomic experiments have revealed that c-Myc controls the activity of RhoA-dependent routes (37). Transcriptomal profiling has revealed that c-Myc is important for Rho-mediated transformation (38). Ras activates Rho, which leads to activation of c-Myc through phosphorylation (39). c-Myc also inhibits the RhoA/Rock-dependent cytoskeleton (40). Existence of unknown pathways in association between c-Myc and RhoA has been implicated. As a transcription factor, c-Myc activates a variety of genes with its partner protein, Max and with other proteins (12). The c-Myc–Skp2–Miz1 transcriptional complex was shown to activate RhoA and to be essential for cell invasion and cancer metastasis (25). We found that c-Myc is a target of miR-34a in A498 cells, and we studied the effect of miR-34a on RhoA activation through the c-Myc–Skp2–Miz1 transcriptional complex. miR-34a reduced RhoA expression and overexpression of c-Myc reversed the reduction of RhoA and inhibition of cell invasion. ChIP assay revealed that miR-34a suppresses the recruitment of c-Myc to the RhoA promoter. We demonstrated that miR-34a targets c-Myc and suppressed RhoA transcription by reducing the c-Myc–Skp2–Miz1 transcriptional complex. These data document that miR-34a suppresses RhoA activation at the initiation of transcription via targeting c-Myc. Our results also demonstrate that miR-34a suppresses the assembly and function of the c-Myc complex that activates RhoA transcription. A schematic model of the mechanisms is shown in Figure 6A and B. Although miR-34a reduced c-Myc protein expression in 769P cells, RhoA expression was not significantly suppressed, which suggests that other mechanisms prevent reduction of RhoA expression. However, this result supports the small effect of miR-34a on 769P cell invasion and RhoA dependence of miR-34a regulation in A498 cell invasion.
Fig. 6.
Schematic model of miR-34a repression of c-Myc complexes. (A) The c-Myc–Skp2–Miz1 transcriptional complex activates RhoA. (B) miR-34a targets c-Myc and represses RhoA expression by suppressing the c-Myc–Skp2–Miz1 transcriptional complex. (C) The c-Myc–P-TEFb complex stimulates Pol II elongation. (D) miR-34a targets c-Myc and represses Pol II elongation by suppressing the c-Myc–P-TEFb elongation complex.
The P-TEFb regulates the promoter-proximal pause release of the elongation phase of transcription by Pol II (26). P-TEFb is composed of Cyc and Cdk9 (26). c-Myc interacts with CycT1, the regulatory component of P-TEFb, and regulates the elongation phase of transcription of Pol II (28–30). P-TEFb is globally involved in the generation of mRNAs and gene expression (30,41) and is recruited to genes by a variety of transcription factors (26,42). Our results demonstrate that miR-34a suppresses the assembly and function of the c-Myc complex that elongates transcription, which may affect global gene expression. The miR-34a-induced c-Myc immunocomplex may be heterogeneous since the ratios of the components were different from those in the control immunocomplexes. Since miR-34a affects wide variety of genes besides reducing c-Myc expression, the miR-34a-induced c-Myc immunocomplex may be heterogeneous.
P-TEFb has been shown to be involved in aberrant transcriptional elongation in mixed-lineage leukemia (43,44) and various tumor cell lines overexpress P-TEFb (45). We demonstrated here that miR-34a inhibits the assembly of the c-Myc–P-TEFb complex presumably by targeting c-Myc, which is of significance in cancer studies. Our results suggest that miR-34a suppresses by reducing c-Myc and affects Pol II pause release in tumor cells, which partly accounts for its dramatic tumor-suppressing effects. A schematic model of the mechanisms is shown in Figure 6C and D.
In this study, we show that miR-34a inhibits kidney cancer cell (A498 and 769P) cell invasion. We also show miR-34a inhibits c-Myc and its complexes, which we observed in prostate cancer cells (Yamamura,S, Saini,S, Majid,S, Hirata,H, Ueno,K, Deng,G and Dahiya,R, unpublished results). c-Myc activates a variety of genes through its protein complexes and also regulates transcriptional elongation. We demonstrate here that miR-34a suppresses assembly and function of the c-Myc–Skp2–Miz1 complex that activates RhoA and the c-Myc–P-TEFb complex that elongates transcription of various genes. Therefore, our study reveals a novel role of miR-34a in the regulation of transcription by c-Myc.
Supplementary material
Supplementary Figures S1–S6 can be found at http://carcin.oxfordjournals.org/.
Funding
National Institute of Health (R01CA138642, T32DK007790); Veterans Affairs Research Enhancement Award Program and Merit Review grants.
Supplementary Material
Acknowledgments
We thank Dr Roger Erickson for his support and assistance with the preparation of the manuscript.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CAD
carbamoyl-phosphate synthase/aspartate transcarbamoylase/dihydroorotase
- ChIP
chromatin immunoprecipitation
- Cdk
cyclin-dependent kinase
- Cyc
cyclin
- CycT1
cyclin T1
- E-boxes
Enhancer Box elements
- IP
immunoprecipitation
- mRNA
messenger RNA; miR
- microRNA
miRNA
- microRNAs; miR-34a, microRNA-34a
NC
- negative control
PCR
- polymerase chain reaction
P-TEFb
- positive transcription elongation factor b
Pol II
- RNA polymerase II
RCC
- renal cell carcinoma; UTR
untranslated region
References
- 1.Brodersen P, et al. Revisiting the principles of microRNA target recognition and mode of action. Nat. Rev. Mol. Cell Biol. 2009;10:141–148. doi: 10.1038/nrm2619. [DOI] [PubMed] [Google Scholar]
- 2.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raver-Shapira N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 4.Chang TC, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69:7569–7576. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welch C, et al. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 8.Liu C, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat. Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamakuchi M, et al. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl Acad. Sci. USA. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H, et al. Identifying mRNA targets of microRNA dysregulated in cancer: with application to clear cell Renal Cell Carcinoma. BMC Syst. Biol. 2011;4:51. doi: 10.1186/1752-0509-4-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogt M, et al. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch. 2011;458:313–322. doi: 10.1007/s00428-010-1030-5. [DOI] [PubMed] [Google Scholar]
- 12.Adhikary S, et al. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 13.Cole MD, et al. Transcription-independent functions of MYC: regulation of translation and DNA replication. Nat. Rev. Mol. Cell Biol. 2008;9:810–815. doi: 10.1038/nrm2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Donnell KA, et al. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 15.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olive V, et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cannell IG, et al. p38 MAPK/MK2-mediated induction of miR-34c following DNA damage prevents Myc-dependent DNA replication. Proc. Natl Acad. Sci. USA. 2010;107:5375–5380. doi: 10.1073/pnas.0910015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christoffersen NR, et al. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ. 2010;17:236–245. doi: 10.1038/cdd.2009.109. [DOI] [PubMed] [Google Scholar]
- 19.Etienne-Manneville S, et al. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 20.Heasman SJ, et al. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 21.Jaffe AB, et al. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 22.Hodge JC, et al. Requirement of RhoA activity for increased nuclear factor kappaB activity and PC-3 human prostate cancer cell invasion. Cancer Res. 2003;63:1359–1364. [PubMed] [Google Scholar]
- 23.Cardone RA, et al. Protein kinase A gating of a pseudopodial-located RhoA/ROCK/p38/NHE1 signal module regulates invasion in breast cancer cell lines. Mol. Biol. Cell. 2005;16:3117–3127. doi: 10.1091/mbc.E04-10-0945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia M, et al. Tumor suppressor p53 restricts Ras stimulation of RhoA and cancer cell motility. Nat. Struct. Mol. Biol. 2007;14:215–223. doi: 10.1038/nsmb1208. [DOI] [PubMed] [Google Scholar]
- 25.Chan CH, et al. Deciphering the transcriptional complex critical for RhoA gene expression and cancer metastasis. Nat. Cell Biol. 2010;12:457–467. doi: 10.1038/ncb2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterlin BM, et al. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 27.Bres V, et al. The multi-tasking P-TEFb complex. Curr. Opin. Cell Biol. 2008;20:334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eberhardy SR, et al. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J. Biol. Chem. 2002;277:40156–40162. doi: 10.1074/jbc.M207441200. [DOI] [PubMed] [Google Scholar]
- 29.Kanazawa S, et al. c-Myc recruits P-TEFb for transcription, cellular proliferation and apoptosis. Oncogene. 2003;22:5707–5711. doi: 10.1038/sj.onc.1206800. [DOI] [PubMed] [Google Scholar]
- 30.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eberhardy SR, et al. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J. Biol. Chem. 2001;276:48562–48571. doi: 10.1074/jbc.M109014200. [DOI] [PubMed] [Google Scholar]
- 32.Gargano B, et al. P-TEFb is a crucial co-factor for Myc transactivation. Cell Cycle. 2007;6:2031–2037. doi: 10.4161/cc.6.16.4554. [DOI] [PubMed] [Google Scholar]
- 33.Meyer N, et al. Reflecting on 25 years with MYC. Nat. Rev. Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 34.Brooks TA, et al. The role of supercoiling in transcriptional control of MYC and its importance in molecular therapeutics. Nat. Rev. Cancer. 2009;9:849–861. doi: 10.1038/nrc2733. [DOI] [PubMed] [Google Scholar]
- 35.Land H, et al. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 36.Sahai E, et al. RHO-GTPases and cancer. Nat. Rev. Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 37.Shiio Y, et al. Quantitative proteomic analysis of Myc oncoprotein function. EMBO J. 2002;21:5088–5096. doi: 10.1093/emboj/cdf525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berenjeno IM, et al. Transcriptomal profiling of the cellular transformation induced by Rho subfamily GTPases. Oncogene. 2007;26:4295–4305. doi: 10.1038/sj.onc.1210194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watnick RS, et al. Ras modulates Myc activity to repress thrombospondin-1 expression and increase tumor angiogenesis. Cancer Cell. 2003;3:219–231. doi: 10.1016/s1535-6108(03)00030-8. [DOI] [PubMed] [Google Scholar]
- 40.Sauzeau V, et al. A transcriptional cross-talk between RhoA and c-Myc inhibits the RhoA/Rock-dependent cytoskeleton. Oncogene. 2010;29:3781–3792. doi: 10.1038/onc.2010.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chao SH, et al. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 2001;276:31793–31799. doi: 10.1074/jbc.M102306200. [DOI] [PubMed] [Google Scholar]
- 42.Zhou Q, et al. The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol. Mol. Biol. Rev. 2006;70:646–659. doi: 10.1128/MMBR.00011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mueller D, et al. Misguided transcriptional elongation causes mixed lineage leukemia. PLoS Biol. 2009;7:e1000249. doi: 10.1371/journal.pbio.1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin C, et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moiola C, et al. Cyclin T1 overexpression induces malignant transformation and tumor growth. Cell Cycle. 2010;9:3119–3126. doi: 10.4161/cc.9.15.12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.