Abstract
The CD1 family consists of lipid antigen-presenting molecules, which include group I CD1a, CD1b, and CD1c and group II CD1d proteins. Topologically, they resemble the classical peptide antigen-presenting MHC molecules except that the large, exclusively nonpolar and hydrophobic, antigen-binding groove of CD1 has evolved to present cellular and pathogen-derived lipid antigens to specific T lymphocytes. As an approach to understanding the biochemical basis of lipid antigen presentation by CD1 molecules, we have characterized the natural ligands associated with mouse CD1d1 as well as human CD1b and CD1d molecules. We found that both group I and II CD1 molecules assemble with cellular phosphatidylinositol (PI), which contains heterogeneous fatty acyl chains. Further, this assembly occurs within the endoplasmic reticulum. Because the structures of the antigen-binding grooves of CD1a and CD1c closely resemble those of CD1b and CD1d, we conclude that the assembly of CD1 molecules with PI in the endoplasmic reticulum is evolutionarily conserved. These findings suggest that PI plays a chaperone-like role in CD1 assembly, possibly to preserve the integrity of the antigen-binding groove until CD1 binds antigenic lipids in the endocytic pathway.
Lipids play crucial roles in normal functioning of a cell. To impart these functions, lipids depend on interactions with proteins because of their propensity to aggregate and form micelles. The CD1 family consists of lipid antigen-presenting molecules. It includes group I CD1a, CD1b, and CD1c and group II CD1d molecules. Most mammals express both group I and II CD1 molecules (human), but some express only group II (mice and rats) molecules. Current evidence indicates that group I CD1 proteins present self and bacterial lipids to specific T cells, whereas group II molecules display self lipids for an appraisal by a specialized subset of T lymphocytes called natural T cells (reviewed in ref. 1).
The solution of the 3D structures of CD1a, CD1b, and CD1d reveals that they resemble the classical peptide antigen-presenting MHC molecules (2–4). In contrast to MHC molecules, the antigen-binding groove of CD1 is large, exclusively nonpolar, and hydrophobic (2–4) and hence has evolved to chaperone lipid antigens to the cell surface. The majority of antigenic lipid binding to CD1 occurs within endocytic compartments (5–8). Because CD1 biosynthesis occurs in the endoplasmic reticulum (ER), the question arises as to how the large hydrophobic groove is maintained during folding and intracellular traffic to the endosomes before antigen binding. At least a subset of human CD1b (hCD1b) and CD1d molecules assemble with the MHC class II-associated invariant chain (Ii) in the ER (8–10). It has been suggested that binding of Ii to CD1d maintains the large hydrophobic groove through assembly and intracellular traffic in a form receptive to antigen in the late endosomes (9). Whereas the CD1–Ii complex formation could partly solve the biosynthetic assembly problem in cells that express Ii (dendritic cells, macrophages, and B cells), the problem remains in cells that express CD1 but not Ii (e.g., CD4+8+ thymocytes and hepatocytes).
Our previous studies revealed that mouse CD1d1 (mCD1d1) associates with cellular phosphatidylinositol (PI) and PI-glycans in cells that do not express Ii and provided indirect evidence that this CD1–lipid interaction occurs in the ER (10, 11). Here we demonstrate that group I hCD1b, group II hCD1d, and mCD1d1 assemble with PI. Further, we provide direct evidence that, at least, hCD1d assembles with PI within the ER. Thus, assembly of CD1 with cellular lipids in the ER is an evolutionarily conserved feature suggesting a chaperone-like role for lipids.
Materials and Methods
Cell Lines. The generation and maintenance of Oβ, Oβ/sCD1d1, and Db-sol (secreting H2Db) cell lines have been described (11–13). The cDNA encoding the leader and the three extracellular domains of hCD1b was amplified by PCR by using hCD1b cDNA (pDOI-5-hCD1b PCR-cloned from 7-day CSF-2- and IL-4-stimulated human peripheral blood cells) template and forward 5′-CGGGATCCCGAAATGGTGCTGCTGCCA-3′ and reverse 5′-TCCCCCGGGGGAtca-[ATGGTGATGGTGATGGTG]CCAGTAGAGGATGAT-3′ primers. The reverse primer contains a sequence that encodes the hexa-histidine tag [bracketed], which is followed by a stop codon (lower case). The PCR product was digested with BamHI and SmaI (underlined within primer sequences), cloned into BamHI and EcoRV cleaved pCR3, and the authenticity of the PCR product confirmed by dideoxy-chain termination sequencing using Sequenase kit (Stratagene) according to the manufacturer's protocol. An authentic pCR3-hCD1b was linearized with ScaI, transfected into Oβ cells by electroporation (Bio-Rad Gene Pulsar), and selected in 0.8 mg/ml G418 as described (13). The cell line secreting the highest level of hCD1b was identified (data not shown), expanded, and used as the source of soluble CD1.
Constructs encoding soluble hCD1d molecules were generated by PCR based on the vector CD1d/pSRα-neo (gift from S. A. Porcelli). For generating soluble hCD1d, the sequence encoding the signal peptide and luminal domain of hCD1d was cloned between XhoI and BamHI sites of the vector pcDNA3.1-PAC. Similarly, ER-retained soluble hCD1d (hCD1d-er) was cloned between the NotI and BamHI sites of pIRESneo (Clontech) with a sequence encoding the ER-retrieval signal KDEL (5′-AAAGATGAGTTG-3′) incorporated at the 3′ end. For detection, a sequence encoding a FLAG tag (DYKDDDDK) was included at the 3′ end of the soluble secreted CD1d and inserted between the 3′ end of the hCD1d sequence and the 5′ end of the KDEL-encoding sequence for hCD1d-er. The constructs were transfected into the HLA class I-negative .221 B cell line by electroporation at 210 V and 960 mF and selected with either 750 ng/ml puromycin for soluble hCD1d-expressing .221.SECCD1d.f cells or 0.6 mg/ml G418 for the hCD1d-er-expressing .221.ERCD1d.f cells. They were screened for hCD1d expression by Western blotting. Cells were maintained in Iscove's medium (Invitrogen) containing 5% bovine calf serum or 10% fetal bovine serum.
Affinity Purification. Soluble mCD1d1 and hCD1b secreted into 10 liters of cell culture medium were concentrated to ≈0.5 liter by tangential flow filtration against a 30,000 Mr membrane (Pall). Soluble CD1 was purified by Ni chromatography (HiTrap chelating column) by using an FPLC unit (Amersham Pharmacia Biotech). The column was eluted with 0.3 M imidazole (Sigma) in PBS. After removing imidazole by dialysis against 1,000-fold excess PBS, purity was ascertained by Coomassie (Brilliant blue R250) staining of the eluted material separated by 15% reducing SDS/PAGE; purity was >90%. The identity of hCD1b was ascertained by Western blot analysis with hexa-histidine-specific antibody (Roche Biochemicals) by using an enhanced chemiluminescence kit according to the manufacturer's protocol (Amersham Pharmacia Biotech). Protein concentration was determined by using the BCA method according to the manufacturer's protocol (Pierce). BSA from tissue culture medium used to grow Db-sol cells was similarly purified.
Soluble hCD1d molecules were purified from the supernatant of .221.SECCD1d.f and hCD1d-er from the lysate of .221.ERCD1d.f. .221.ERCD1d.f cells were frozen and thawed on ice in 10 mM Tris·HCl, pH 7.4, containing PMSF and iodoacetamide. Cells were pelleted twice to remove nuclei and debris at 600–1,000 × g for 10 min. The supernatants were sonicated to disrupt the organelles and centrifuged at 100,000 × g for 1 hr at 4°C. The supernatant and the soluble fractions were passed through mouse IgG precolumn followed by an hCD1d/β2-microglobulin-specific monoclonal antibody (CD1d.51)-coupled affinity column. The columns were washed separately with 10 mM Tris·HCl/150 mM NaCl, pH 7.4, and CD1d molecules were eluted with buffer containing 3.5 M MgCl2 and 10 mM phosphate, pH 7.4, or 0.1 N acetic acid. The fractions containing proteins were pooled and dialyzed against 50 mM ammonium acetate and lyophilized.
Lipid Extraction. Lipids were extracted by the Bligh–Dyer method (14) with two times the sample volume of chloroform/methanol (CM, 2:1), as described (12). After thorough mixing, the top aqueous and the bottom organic phases were allowed to separate. The organic phase was carefully transferred into another tube. The aqueous phase was extracted one more time, as above. The resulting organic phase was pooled with the first and dried under a gentle stream of nitrogen gas. The dried extract was dissolved in 100 μl of mobile phase A (constitution described below) for MS.
MS. PI (1-stearoyl-2-arachidonyl-sn-glycerol-3-phospho-myo-inositol) standard (Sigma) and ligands extracted from soluble CD1 and BSA were separated by RP-HPLC by using self-packed 1.0 × 150-mm POROS 10R2 (mCD1d1, hCD1d, and BSA; Applied Biosystems), Zorbax Eclipse XDB-C8 (hCD1b; Agilent Technologies, Palo Alto, CA), or Jupiter (hCD1d-er; Phenomenex, Belmont, CA) columns. Lipids were applied to the column by using mobile phase A (vol/vol): 10:90 acetonitrile/water (PI, mCD1d1, and BSA) or 69:30:1 water/methanol/acetic acid (hCD1b and hCD1d-er), 5:95:0.5 acetonitrile/water/acetic acid (hCD1d) and eluted with a gradient of mobile phase B (vol/vol): 45:50:5 acetonitrile/isopropanol/water (PI, mCD1d1, BSA), 99:1 methanol/acetic acid (hCD1b), 45:45:10:0.5 acetonitrile/isopropanol/water/acetic acid (hCD1d), or 99:1 methanol/acetic acid (hCD1d-er) established over 20 min at a flow rate of 200 μl per min (PI, mCD1d1, hCD1d-er, and BSA) or 35 min at a flow rate of 100 μl per min (hCD1b and hCD1d). Mobile phases A and B also contained 10 mM ammonium acetate. MS analysis was performed by using an LCQ ion trap instrument (ThermoFinnigan, San Jose, CA) operating in negative and positive ion modes. The instrument was operated in full scan mode from m/z 200–2,000. An electrospray ionization (ESI) source was fitted with a 100-μm internal diameter deactivated fused silica capillary; nitrogen was used for both sheath and auxiliary gas. The ESI needle was maintained at 3.5 kV (–ESI) and 4–4.5 kV (+ESI); the heated capillary was operated at 32 V and 230°C, and the tube lens was set at 0 V. Data acquisition and analysis were performed by using xcalibur Ver. 1.2 software (ThermoFinnigan).
Results
Studies of lipid–protein interactions are hampered by the poor sensitivity of routine lipid analysis techniques. The extraction of integral membrane proteins such as CD1 with detergents, which have the potential of binding the protein (3) or even exchanging with the bound ligand (S.J., unpublished data), also impedes analysis. To overcome these limitations, we expressed soluble forms of mCD1d1, hCD1d (including an ER-retained version, hCD1d-er), and hCD1b, and sufficient quantities of these molecules were purified for mass spectrometric analyses of the associated lipid ligands. Biochemical characterization of the various purified CD1 proteins and verification of the ER retention of hCD1d-er by pulse–chase analysis are presented in Fig. 6, which is published as supporting information on the PNAS web site.
mCD1d1-Associated PI Contains Heterogeneous Fatty Acyl Chains. We previously reported that both wild-type and soluble mCD1d1 assemble with PI and PI-glycans in vivo (11, 12). However, in vitro, CD1 appears to bind a wide variety of lipids that vary in the chemical character of the moiety that directly interacts with the antigen-binding groove (11, 15–26). Therefore, our previous findings (11, 12) may have been biased by the techniques used for the extraction, purification, and analyses of mCD1d1-associated ligands. In the first report (11), the ligand(s) was isolated by acid denaturation, centricon filtration, and RP-HPLC by using a solid phase consisting of a mixture of C18 and cation exchange matrix, which was eluted with a gradient of acetonitrite/trifluoroacetic acid solution. The isolated ligand(s) was then analyzed by matrix-assisted laser desorption ionization (MALDI)-MS. In the second approach (12), a method was developed for lipid extraction from mCD1d1 affinity-purified from cells radiolabeled with precursors that are incorporated into lipids. Analysis by thin-layer chromatography permitted tentative identification of associated ligands. In this study, we have isolated ligands associated with affinity-purified soluble mouse and human CD1 molecules by lipid extraction by using CM phase separation and analyzed them by tandem RP-HPLC/ESI-MS.
RP-HPLC/ESI-MS analysis of material eluted from ≈200 μg of affinity-purified soluble mCD1d1 revealed the presence of negatively charged compounds, which elute between ≈15 and 18 min (Fig. 1A). To determine whether the negatively charged compounds were bound specifically to soluble CD1d1, the nonspecific lipid-binding protein, BSA, a common contaminant copurifying during Ni-affinity chromatography, was purified from spent tissue culture medium in which cells expressing soluble mouse H2Db class I molecules were grown. Note that soluble H2Db does not contain an hexa-histidine tag and hence does not bind Ni-affinity column. Lipids were extracted from purified BSA and analyzed by RP-HPLC/ESI-MS in negative ion mode. The total ion chromatogram (TIC) revealed that lipids, if bound to BSA, were undetectable under the conditions of extraction and analysis described here (Fig. 1B). Thus, the compounds extracted from soluble mCD1d1 appear specifically bound to this lipid antigen-presenting molecule.
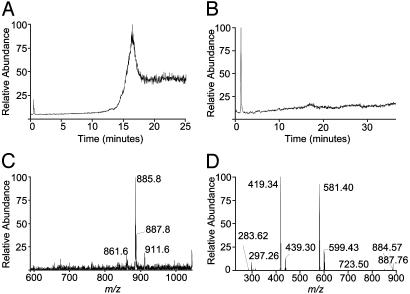
Fig. 1.
Soluble mCD1d1-associated ligands. (A) TIC of compounds associated with soluble mCD1d1 revealed on tandem RP-HPLC/ESI-MS analysis of the organic phase CM extracted from affinity-purified CD1 molecule. (B) TIC of compounds associated with BSA. (C) Molecular [M-H]– ion chromatogram of compounds extracted from soluble mCD1d1. (D) Daughter ion chromatogram resulting from CID of the 885.8 [M-H]– molecular ion.
To characterize the compounds bound to soluble mCD1d1, the mass spectrometer was scanned from m/z 200–2,000. A closer examination revealed an [M-H]– ion of m/z 885.8 (Fig. 1C), which was observed in our previously reported MALDI-MS analysis (11). Additionally, [M-H]– ions of m/z 861.6, 887.8, and 911.6 were also observed (Fig. 1C). The [M-H]– ion of 885.8 was previously identified as PI (C38:4), which consists of stearoyl and arachidonyl acyl chains (11). To confirm the identity of this compound, 1-stearoyl-2-arachidonyl-sn-glycero-3-phosphoinositol (C38:4 PI) isolated from bovine liver was analyzed by RP-HPLC/ESI-MS. This PI, used as a standard, eluted at ≈16.49 min (Fig. 2A) and consisted of [M-H]– ions of 885.73, 887.67, and 911.67 that are accompanied by an ion of m/z 861.73 (Fig. 2B). The [M-H]– ion 887.67 most likely results from the reduction of 885.73 and hence the loss of an unsaturated bond (C38:3).
Fig. 2.
Tandem RP-HPLC/ESI-MS and MS/MS analyses of PI standard. (A) TIC of PI purified from bovine liver (Sigma) that is used as a standard. (B) Molecular [M-H]– ion chromatogram of PI standard. (C) Structure of PI standard whose negative ion m/z = 885.5. The structure shows labile bonds and their m/z.(D) Daughter ion chromatogram of the 885.73 [M-H]– ion after CID and MS/MS analysis.
Tandem MS (MS/MS) analysis of [M-H]– 885.73 after collision-induced dissociation (CID) revealed daughter ions characteristic of labile components of PI (Fig. 2C). The [M-H]– ions 601.23 and 581.2 (Fig. 2D) are lyso-PI that lacks either stearate or arachidonate ions (Fig. 2C), respectively. A further loss of m/z 162, that of a hexose, results in [M-H]– 439.23 and 419.21 (Fig. 2 C and D). Analysis of the 885.8 [M-H]– ion extracted from soluble mCD1d1 yielded a daughter ion chromatogram (Fig. 1D) similar to the one described for PI standard (Fig. 2D). Additionally, a 723.5 [M-H]– ion was observed, which reflects the loss of ≈162 mass unit hexose from the 885.8 molecular ion (Fig. 1D). Thus, soluble mCD1d1 associates with PI.
CID of 861.73 [M-H]– ion of the PI standard (Fig. 7, which is published as supporting information on the PNAS web site) revealed that it consists of C36:2 PI, which contains fatty acyl groups of heterogeneous chain lengths (C16:0+C20:2, C18:0+C18:2, and C18:1+C18:1; see ref. 27 for [M-H]– ions of PI varying in fatty acyl chains). The 911.6 [M-H]– ion observed in both mCD1d1-associated compounds and PI standard may be C40:5 PI (27), which needs to be confirmed.
The [M+H]+ TIC obtained from RP-HPLC/MS analysis of soluble mCD1d1-associated ligands revealed a dominant peak, which elutes between 16 and 18 min (Fig. 3A). This dominant peak contained a dominant [M+H]+ ion of m/z 903.93 and a few minor ions of m/z 853.93, 879.80, and 929.93 (Fig. 3B). These [M+H]+ ions correspond to C38:5 ([M+H]+ = 885), C34:2 ([M+H]+ = 835), C36:3 ([M+H]+ = 861), and C40:6 ([M+H]+ = 911) PI (see ref. 27), respectively, coordinated with a molecule of 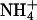 (m/z 18). CID of the dominant 903.3 [M+H]+ ion resulted in an ion of m/z 885.3 and two major daughter ions of m/z 604.4 and 643.4 (Fig. 3C). The daughter ions of m/z 604.4 and 643.4 reflect the loss of a C18:1 acyl chain and phosphoinositol, respectively, from the 885.3 [M+H]+ ion. Thus, soluble mCD1d1-associated PI contains heterogeneous fatty acyl chains, which includes C36:2 (C16:0+C20:2, C18:0+C18:2, and C18:1+C18:1), C38:5 (C18:1+C20:4), C38:4 (C18:0+C20:4) identified based on CID analysis of the molecular ions and possibly C34:2, C36:3, C38:3, C40:5, and C40:6 deduced from the masses of the respective molecular ions.
(m/z 18). CID of the dominant 903.3 [M+H]+ ion resulted in an ion of m/z 885.3 and two major daughter ions of m/z 604.4 and 643.4 (Fig. 3C). The daughter ions of m/z 604.4 and 643.4 reflect the loss of a C18:1 acyl chain and phosphoinositol, respectively, from the 885.3 [M+H]+ ion. Thus, soluble mCD1d1-associated PI contains heterogeneous fatty acyl chains, which includes C36:2 (C16:0+C20:2, C18:0+C18:2, and C18:1+C18:1), C38:5 (C18:1+C20:4), C38:4 (C18:0+C20:4) identified based on CID analysis of the molecular ions and possibly C34:2, C36:3, C38:3, C40:5, and C40:6 deduced from the masses of the respective molecular ions.
Fig. 3.
RP-HPLC/ESI-MS analysis of soluble mCD1d1-associated ligands in positive ion mode. (A) TIC of compounds associated with soluble mCD1d1 revealed on tandem RP-HPLC/ESI-MS analysis in positive ion mode. (B) Molecular [M+H]+ ion chromatogram of compounds CM extracted from soluble mCD1d1. (C) CID and MS/MS analysis of the 903.3 [M+H]+ ion.
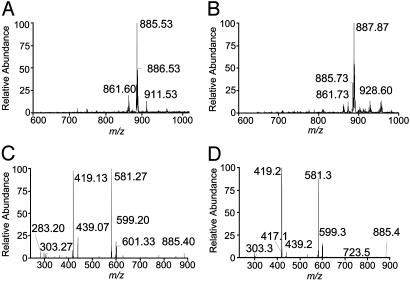
hCD1b and CD1d Are Associated with PI. To determine whether binding cellular lipids is a common feature of CD1 molecules, we extracted ≈200 μg of affinity-purified secreted soluble hCD1b and hCD1d with CM and subjected the compounds in the organic phase to tandem RP-HPLC/ESI-MS analysis, as described for ligands eluted from soluble mCD1d1. A scan of the resulting TIC (data not shown) revealed [M-H]– ions of m/z 861.6, 885.53, 886.53, and 911.53 (Fig. 4A) eluted from hCD1b. hCD1d-associated ligands have m/z 861.73, 885.73, and 888.87 negative ions (Fig. 4B). Thus mouse and human CD1, including group I and II molecules, bind similar cellular lipids.
Fig. 4.
Soluble hCD1b- and hCD1d-associated ligands. (A and B) Molecular [M-H]– ion chromatograms of compounds CM extracted from soluble hCD1b (A) and hCD1d (B). (C and D) Daughter ion chromatograms of 885.53 (hCD1b) and 885.73 (hCD1d) [M-H]– ion subjected to CID and MS/MS analysis.
CID analysis of the 885.53 and 885.73 [M-H]– ions extracted from hCD1b and hCD1d, respectively, resulted in daughter ion chromatograms (Fig. 4 C and D) very similar to the one obtained from the analysis of 885.8 [M-H]– ion associated with soluble mCD1d1 (see Fig. 1D). Phosphatidylserine, β-d-galactosylceramide, the ganglioside GM3, and sphingomyelin standards bound, eluted, and ionized under the conditions of RP-HPLC/ESI-MS described in Materials and Methods. However, none of these lipids were detected in CD1 eluates (data not shown). Thus, the binding of cellular lipids, predominantly if not exclusively PI, is a common feature of CD1 molecules.
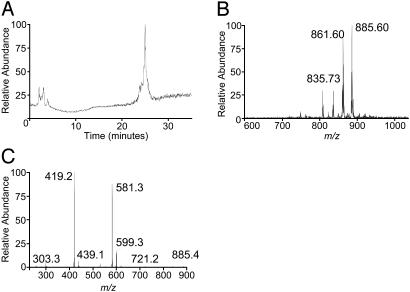
hCD1d Assembles with PI in the ER. The biosynthesis of PI and the biosynthetic assembly of CD1 molecules occur in the ER (reviewed in ref. 28). Further, our preliminary evidence using an ER-retained form of soluble mCD1d1 revealed that this CD1 molecule binds PI in an early secretory compartment (12). To confirm this finding, the soluble form of hCD1d was tagged at its C terminus with an ER-retention/retrieval signal (hCD1d-er; Fig. 6). Cells expressing hCD1d-er were lysed by freeze–thaw in hypotonic buffer. This method avoids the use of detergent and hence is less likely to alter the composition of compounds assembled with hCD1d-er. Affinity-purified hCD1d-er was CM extracted and analyzed as described above. The [M-H]– TIC (Fig. 5A) was similar to the one obtained with PI standard (see Fig. 2 A). Furthermore, as with the ligands associated with soluble CD1 molecules, hCD1d-er associates with compounds that have [M-H]– ions of m/z 861.6, 885.6, and also 835.73 but not the 911.6 anion (Fig. 5B). The 835.73 [M-H]– ion (Fig. 5B) may be C34:1 PI, an assignment that needs confirmation. CID of the 885.6 [M-H]– ion yielded a daughter ion chromatogram (Fig. 5C) very similar to PI (Fig. 2D) and the same ion extracted from soluble forms of mCD1d1 (Fig. 1D), hCD1b (Fig. 4C), and hCD1d (Fig. 4D). Thus, the apparently exclusive association of PI with CD1d occurs during biosynthetic assembly in the ER.
Fig. 5.
Ligands associated with ER-retained hCD1d. (A) TIC of hCD1d-er-associated ligands, which were extracted and analyzed as in Fig. 1 A. (B) Molecular [M-H]– ion chromatogram of ligands associated with hCD1d-er. (C) Daughter ion chromatogram of the 885.6 [M-H]– ion subjected to CID and MS/MS analysis.
Discussion
In summary, group I hCD1b, group II hCD1d, and mCD1d1 molecules all assemble with a cellular phospholipid. Further, our data revealed that this assembly occurs within the ER. These data are consistent with our original reports, which demonstrate that group II mCD1d1 molecules assemble with PI in an early secretory compartment (11, 12). Further, these data are consistent with the finding that T cells recognize hCD1b- and mCD1d1-restricted nonself and self PI (1, 15, 29). Because the antigen-binding grooves of hCD1a and hCD1c closely resemble those of CD1b and CD1d (2–4), we conclude that the assembly of CD1 molecules with PI in the ER is evolutionarily conserved.
Our previous MALDI-MS of mCD1d1-associated ligand revealed an 885.3 [M-H]– molecular ion, which on fragmentation resulted in the 705.6 [M-H]– ion. The loss of m/z ≈180 was assigned to the characteristic loss of the inositol head group (11). Fragmentation of PI between the C and O atoms within the phosphoinositol bond yields a characteristic loss of m/z 162 (see Fig. 2C). A plausible explanation for why the previous MALDI-MS (11) and the current CID ESI-MS/MS yield different fragmentation patterns is that the acidic ammonium sulfate, used to supplement the matrix (saturated α-cyno-4-hydroxycinnamic acid in 45% ethanol containing 8.8% formic acid) to enhance ion signal during MALDI-MS (30), may have induced hydrolysis between the P and O atoms of the phosphoinositol bond in addition to the conventional fragmentation of PI between the C and O atoms of this bond.
We also reported that PI-glycans associate with mCD1d1 (11, 12). However, PI-glycans were not observed in the current analysis. The methods used for the isolation and characterization of CD1-associated ligands in these reports are distinct. Therefore, we predict that the conditions of RP-HPLC as well as the sensitivity of the ESI-MS analysis may partly explain why PI-glycans were not detected in the current analysis.
In vitro biochemical and functional studies indicate that group I and II CD1 molecules bind a wide variety of cellular and bacterial lipids. These lipids include cellular phospholipids and sphingolipids as well as mycobacterial phospholipids and mycolates (15–26). Remarkably, however, CD1 molecules appear to associate with a restricted variety of lipids in vivo. Our studies have consistently revealed that only PI and PI-based glycolipids bind CD1 in vivo (this report and refs. 11 and 12). Perhaps these cellular lipids may be the major CD1-associated natural ligands. Thus other lipids, if bound, probably form minor components of CD1-associated natural ligands. The low yields of the natural T cell lipid antigen(s) that can be extracted from mCD1d1 (J.-J.P. and S.J., unpublished data) support the idea that this and other cellular lipids form minor CD1 ligands.
hCD1d (31), and perhaps hCD1b (32, 33), assemble in the ER with the assistance of calnexin, calreticulin, and ERp57. Interestingly, hCD1d heavy chain folds first with the ER chaperones and then binds β2m (31). These initial steps differ strikingly from MHC class I assembly, in which the heavy chain and β2m associate before they bind the assembly complex consisting of calreticulin, ERp57, tapasin, and TAP (34). Perhaps, after CD1 heavy chain-β2m assembly in Ii-positive cells, CD1b and CD1d bind Ii (8–10). Whether the formation of a CD1–Ii complex inhibits CD1 assembly with PI in the ER remains unknown. The .221 cell line used to express the hCD1d species studied here is Ii-positive, but it is possible that a subset of CD1 may assemble with Ii and the remainder with PI. Alternatively, Ii might bind CD1 without occluding the antigen-binding groove, hence permitting PI to bind CD1 during biosynthetic assembly.
The meaning of the evolutionarily conserved biosynthetic assembly of CD1 with PI in the ER remains unknown. However, a likely hypothesis is that the cellular phospholipid plays a chaperone-like role in CD1 assembly, preserving the integrity of the large nonpolar and hydrophobic antigen-binding groove and allowing CD1 molecules to avoid chaperone interactions and consequent ER retention and degradation. Exchange of PI for antigenic lipids in the endocytic pathway might then allow their display on the cell surface for appraisal by specific T lymphocytes.
Supplementary Material
Acknowledgments
We thank Dr. S. A. Porcelli (Albert Einstein College of Medicine, Bronx, NY) for CD1d/pSRα-neo and the hybridoma CD1d.51, Dr. L. Van Kaer for critical reading of the manuscript, A. J. Joyce for technical assistance, and M. H. Henderson and N. Dometios for secretarial assistance. We acknowledge support by the National Institutes of Health (Grant AI42284 to S.J.), the Human Frontier Science Program (to S.J.), the Associazione Italiana Ricerca sul Cancro (to G.C.), and the Howard Hughes Medical Institute (to P.C.).
Abbreviations: Ii, invariant chain; ER, endoplasmic reticulum; PI, phosphatidylinositol; ESI, electrospray ionization; MALDI, matrix-assisted laser desorption ionization; CM, chloroform/methanol; TIC, total ion chromatography; CID, collision-induced dissociation; mCD1d1, mouse CD1d1; hCD1d-er, ER-retained soluble hCD1d; MS/MS, tandem MS; hCD1b/d, human CD1b/d.
References
- 1.Moody, D. B. & Porcelli, S. A. (2003) Nat. Rev. Immunol. 3, 11–22. [DOI] [PubMed] [Google Scholar]
- 2.Zeng, Z., Castano, A. R., Segelke, B. W., Stura, E. A., Peterson, P. A. & Wilson, I. A. (1997) Science 277, 339–345. [DOI] [PubMed] [Google Scholar]
- 3.Gadola, S. D., Zaccai, N. R., Harlos, K., Shepherd, D., Castro-Palomino, J. C., Ritter, G., Schmidt, R. R., Jones, E. Y. & Cerundolo, V. (2002) Nat. Immunol. 3, 721–726. [DOI] [PubMed] [Google Scholar]
- 4.Zajonc, D. M., Elsliger, M. A., Teyton, L. & Wilson, I. A. (2003) Nat. Immunol. 4, 808–815. [DOI] [PubMed] [Google Scholar]
- 5.Sugita, M., Grant, E. P., van Donselaar, E., Hsu, V. W., Rogers, R. A., Peters, P. J. & Brenner, M. B. (1999) Immunity 11, 743–752. [DOI] [PubMed] [Google Scholar]
- 6.Chiu, Y. H., Jayawardena, J., Weiss, A., Lee, D., Park, S. H., Dautry-Varsat, A. & Bendelac, A. (1999) J. Exp. Med. 189, 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts, T. J., Sriram, V., Spence, P. M., Gui, M., Hayakawa, K., Bacik, I., Bennink, J. R., Yewdell, J. W. & Brutkiewicz, R. R. (2002) J. Immunol. 168, 5409–5414. [DOI] [PubMed] [Google Scholar]
- 8.Sugita, M., Cao, X., Watts, G. F., Rogers, R. A., Bonifacino, J. S. & Brenner, M. B. (2002) Immunity 16, 697–706. [DOI] [PubMed] [Google Scholar]
- 9.Jayawardena-Wolf, J., Benlagha, K., Chiu, Y. H., Mehr, R. & Bendelac, A. (2001) Immunity 15, 897–908. [DOI] [PubMed] [Google Scholar]
- 10.Kang, S. J. & Cresswell, P. (2002) EMBO J. 21, 1650–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joyce, S., Woods, A. S., Yewdell, J. W., Bennink, J. R., De Silva, A. D., Boesteanu, A., Balk, S. P., Cotter, R. J. & Brutkiewicz, R. R. (1998) Science 279, 1541–1544. [DOI] [PubMed] [Google Scholar]
- 12.De Silva, A. D., Park, J.-J., Matsuki, N., Stanic, A. K., Brutkiewicz, R. R., Medof, M. E. & Joyce, S. (2002) J. Immunol. 168, 723–733. [DOI] [PubMed] [Google Scholar]
- 13.Joyce, S. (1997) J. Mol. Biol. 267, 993–1001. [DOI] [PubMed] [Google Scholar]
- 14.Bligh, E. G. & Dyer, W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917. [DOI] [PubMed] [Google Scholar]
- 15.Sieling, P. A., Chatterjee, D., Porcelli, S. A., Prigozy, T. I., Mazzaccaro, R. A., Soriano, T., Bloom, B. R., Brenner, M. B., Kronenberg, M., Brennan, P. J., et al. (1995) Science 269, 227–230. [DOI] [PubMed] [Google Scholar]
- 16.Moody, D. B., Reinhold, B. B., Guy, M. R., Beckman, E. M., Frederique, D. E., Furlong, S. T., Ye, S., Reinhold, V. N., Sieling, P. A., Modlin, R. L., et al. (1997) Science 278, 283–286. [DOI] [PubMed] [Google Scholar]
- 17.Kawano, T., Cui, J., Koezuka, Y., Toura, I., Kaneko, Y., Motoki, K., Ueno, H., Nakagawa, R., Sato, H., Kondo, E., et al.. (1997) Science 278, 1626–1629. [DOI] [PubMed] [Google Scholar]
- 18.Brossay, L., Chioda, M., Burdin, N., Koezuka, Y., Casorati, G., Dellabona, P. & Kronenberg, M. (1998) J. Exp. Med. 188, 1521–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brossay, L., Naidenko, O., Burdin, N., Matsuda, J., Sakai, T. & Kronenberg, M. (1998) J. Immunol. 161, 5124–5128. [PubMed] [Google Scholar]
- 20.Earnst, W. A., Maher, J., Cho, S., Niazi, K. R., Chatterjee, D., Moody, D. B., Besra, G. S., Watanabe, Y., Jensen, P. E., Porcelli, S. A., et al. (1998) Immunity 8, 331–340. [DOI] [PubMed] [Google Scholar]
- 21.Shamshiev, A., Donda, A., Carena, I., Mori, L., Kappos, L. & De Libero, G. (1999) Eur. J. Immunol. 29, 1667–1675. [DOI] [PubMed] [Google Scholar]
- 22.Miyamoto, K., Miyake, S. & Yamamura, T. (2001) Nature 413, 531–534. [DOI] [PubMed] [Google Scholar]
- 23.Moody, D. B., Ulrichs, T., Muhlecker, W., Young, D. C., Gurcha, S. S., Grant, E., Rosat, J.-R., Brenner, M. B., Costello, C. E., Besra, G. S., et al. (2000) Nature 404, 884–888. [DOI] [PubMed] [Google Scholar]
- 24.Shamshiev, A., Gober, H. J., Donda, A., Mazorra, Z., Mori, L. & De Libero, G. (2002) J. Exp. Med. 195, 1013–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moody, D. B., Briken, V., Cheng, T. Y., Roura-Mir, C., Guy, M. R., Geho, D. H., Tykocinski, M. L., Besra, G. S. & Porcelli, S. A. (2002) Nat. Immunol. 3, 435–442. [DOI] [PubMed] [Google Scholar]
- 26.Cantu, C., 3rd, Benlagha, K., Savage, P. B., Bendelac, A. & Teyton, L. (2003) J. Immunol. 170, 4673–4682. [DOI] [PubMed] [Google Scholar]
- 27.Murphy, R. C. (1993) Mass Spectrometry of Lipids (Plenum, New York).
- 28.Joyce, S. (2001) Cell. Mol. Life Sci. 58, 442–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gumperz, J. E., Roy, C., Makowska, A., Lum, D., Sugita, M., Podrebarac, T., Koezuka, Y., Porcelli, S. A., Cardell, S., Brenner, M. B., et al. (2000) Immunity 12, 211–221. [DOI] [PubMed] [Google Scholar]
- 30.Woods, A. S., Huang, A. Y. C., Cotter, R. J., Pasternack, G. R., Pardoll, D. M. & Jaffee, E. M. (1995) Anal. Biochem. 226, 15–25. [DOI] [PubMed] [Google Scholar]
- 31.Kang, S. J. & Cresswell, P. (2002) J. Biol. Chem. 277, 44838–44844. [DOI] [PubMed] [Google Scholar]
- 32.Sugita, M., Porcelli, S. A. & Brenner, M. B. (1997) J. Immunol. 159, 2358–2365. [PubMed] [Google Scholar]
- 33.Huttinger, R., Staffler, G., Majdic, O. & Stockinger, H. (1999) Int. Immunol. 11, 1615–1623. [DOI] [PubMed] [Google Scholar]
- 34.Dick, T. P., Bangia, N., Peaper, D. R. & Cresswell, P. (2002) Immunity 16, 87–98. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.