Abstract
The presenilin (PS) proteins are polytopic integral membrane proteins that are critically involved in the development of Alzheimer's disease. The topography of the PS molecule in the endoplasmic reticulum membrane is widely accepted as exhibiting eight-hydrophobic-transmembrane (8-TM) helices. We have previously provided evidence, however, that the intact PS molecule is also present in the cell surface where it exhibits exclusively a 7-TM topography, which differs in significant structural features from the 8-TM model. This evidence, however, has been disparaged and generally rejected by researchers in Alzheimer's disease. The 7-TM model is definitively demonstrated in the present study for PS-1 at the surfaces of PS-1-transfected cells and for endogenous PS-1 at the surfaces of untransfected cells, by immunofluorescence studies using mAbs. These studies force substantial revision of current views of the structural and functional properties of the PS proteins.
Keywords: Alzheimer's disease, protein topology, seven-transmembrane
In 1995, two previously unknown and ubiquitous integral membrane proteins, presenilin-1 and -2 (PS-1 and PS-2), were discovered through familial and molecular genetic studies that implicated them in the etiology of Alzheimer's disease (AD) (1–3). They have subsequently been shown (4–6) to be critical to the proteolytic processing in the brain of the membrane-anchored β-amyloid precursor protein to form β-amyloid (Aβ), a family of oligopeptides which, by an as yet not fully established neurotoxic process, contributes to the development of AD. In addition, the PS proteins are essential for a similar proteolytic processing of the developmentally important Notch protein (7) in various organisms, as well as of other membrane proteins (8).
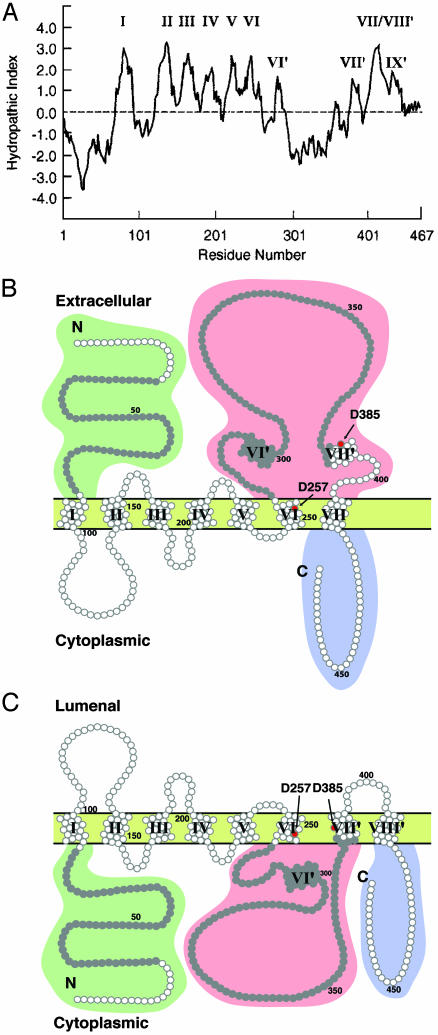
Knowledge of the detailed three-dimensional structure of the PS proteins in their native conformation is vital to an understanding of the molecular mechanisms of their functions and may provide important leads to therapeutic intervention in AD. In the absence of a high-resolution x-ray crystallographic analysis of conformationally intact PS in appropriate crystals of detergent micelles, less detailed structural information may nevertheless be useful. From the amino acid sequences of PS-1 and PS-2, as deduced from their gene sequences, the two proteins are closely related integral membrane proteins that span the membrane multiple times; from their hydropathy plots (ref. 9 and Fig. 1A), the discoverers of PS suggested that they spanned the membrane seven times [seven-transmembrane (7-TM)] (1–3).
Fig. 1.
PS hydropathy plots and derived membrane topographies. (A) The Kyte-Doolittle plot for PS-1 (9) by using a window of 15 residues. The roman numerals correspond to the hydrophobic sequences serving as TM-spanning stretches in B and C.(B) The topography of the proposed 7-TM model of PS-1. The N-terminal domain and the hydrophilic loop between TM VI and VII are located in the exoplasm. This determines that the orientation of each of the first six TM-spanning helices is the opposite of the corresponding helix in the 8-TM spanning model (C). Moderately hydrophobic stretches VI′, VII′, and IX′ (A) do not span the membrane in the 7-TM model. Of the two critical Asp residues (red dots) suggested to be implicated in the γ-cleavage of β-amyloid precursor protein by PS, only Asp-257 resides in a membrane-spanning domain (VI). Asp-385 resides in the extracellular loop between TM domains VI and VII. The residues in gray represent the oligopeptides used to generate the two mAbs, 1563 for the N-terminal domain and 5232 for the loop region. (C) The topography of the proposed 8-TM model of PS-1. The N-terminal domain, hydrophilic loop, and C-terminal tail are all located in the cytoplasm. The moderately hydrophobic stretch labeled VII′ in A as well as the stretch labeled VII/VIII′ are threaded successively through the membrane. Both Asp-275 and Asp-385 are located in the membrane in this model (shown by red dots). In the 6-TM model of PS-1 (not shown), the topography is the same as in the 8-TM model up to and including helix VI, with all following amino acid residues through to the C terminus located in the cytoplasmic domain.
The membrane topography of PS has since been investigated directly, but only in a few articles, each using only one technique. Briefly, one method used was the construction, expression, and analysis of mostly truncated fusion hybrids of the PS proteins and of the homologous protein SEL-12 from Caenorhabditis elegans (10, 11). From one group of such studies (10), the authors concluded that the PS proteins were absent from the cell surface and confined to intracellular membranes, and that in the endoplasmic reticulum (ER) membranes, the PS proteins spanned the membrane eight times (8-TM). In another similar study (12), however, the conclusion derived was that they spanned the ER membranes six times (6-TM). In a more recent work, Nakai et al. (13), using fusion hybrids between truncated PS-1 and as the reporter group, the Escherichia coli signal peptidase, came to the conclusion that PS-1 has a 7-TM topography, different, however, from the 7-TM model advanced by Dewji et al. (refs. 14 and 15 and this study).
In our previous articles (14, 15), by immunofluorescence labeling of human PS-transfected live as well as of permeabilized cells, intact PS proteins were found to be present not only intracellularly but also expressed at the cell surface. This was also the case for endogenous PS on the surfaces of untransfected human neuronal cells (15). Furthermore, with a battery of six independently prepared polyclonal antipeptide Ab, directed against specific extramembranous domains of the human PS proteins that were predicted to protrude from one or the other side of the membrane in different models (Fig. 1 B and C), strong evidence was obtained that the PS proteins in the cell surface spanned the membrane seven times, with either the 6-TM or 8-TM topographies ruled out. Another subsequent study (16) was consistent with our proposed exoplasmic location of the N-terminal domain of cell-surface PS.
Until now, this discordance of conclusions about the membrane topography of PS has not been satisfactorily resolved. Instead, the results supporting a 7-TM topography have been arbitrarily dismissed as artifactual on the basis of alleged Ab heterogeneity (17, 18), and the 8-TM model has been nearly universally adopted among AD investigators as correct.
The question of the TM topography of PS is not trivial; on the contrary, it is of first-order importance to the understanding of PS functions and the interaction of PS with other proteins. This is because even at this relatively crude level of structure determination, the 7-TM model is significantly different from either the 6-TM or 8-TM models (Fig. 1 B and C). Given the complete disregard by the AD community for the 7-TM model, we felt it necessary to revisit our immunofluorescent labeling investigations, this time employing mAb of unambiguous specificities to two extramembranous domains of PS-1, as well as using PS-1–/– and PS-2–/– double-null mouse embryonic stem (ES) cells (19), with or without transfection with human PS-1, reagents and cells that have become available since our original studies. In addition, we used deconvolution immunofluorescence microscopy to study the small amounts of endogenous PS-1 on human DAMI cells. With these tools, we herein provide compelling evidence supporting the 7-TM, as opposed to either the 6-TM or 8-TM, topography of both endogenous and transfected PS-1 molecules in cell-surface membranes. Among the consequences, the possibility is briefly examined that the PS proteins are members of the superfamily of 7-TM heterotrimeric G protein-coupled receptors (GPCRs).
Materials and Methods
Cell Culture and Transfections. ES PS-1–/– and PS-2–/– double-null cells were the kind gift of Drs. Dorit Donoviel and Alan Bernstein (Mount Sinai Hospital and The Samuel Lunenfeld Research Institute, Toronto) and were cultured as described (19). Cells were plated overnight and transfected with a pcDNA3 construct of full-length human PS-1 cDNA using Lipofectamine (Invitrogen) according to the manufacturer's protocols. DAMI cells were cultured as already described (14, 15) and used untransfected.
Abs. Primary Abs. Rat anti-human PS-1 mAb 1563 directed to the N-terminal domain was raised to a fusion protein antigen containing part of the N-terminal domain of human PS-1 (residues 21–80) fused to GST, and mouse mAb (5232) reactive to the human PS-1 loop domain was raised to an oligopeptide containing amino acids 263–378 of PS-1 fused to GST. Both mAb were purchased from Chemicon. Affinity-purified polyclonal Ab was raised in rabbits to the synthetic peptide sequence STDNLVRPF (within the C-terminal domain of human PS-2), which was conjugated to keyhole limpet hemocyanin (14). AbC1 cross-reacted strongly with the closely homologous C-terminal domain of human PS-1. Rabbit polyclonal Ab raised to chicken actin conjugated to keyhole limpet hemocyanin (AB978) was purchased from Chemicon. Mouse mAb (Ab-1, CP07) to chicken β-tubulin was purchased from Oncogene Research (San Diego). Affinity-purified polyclonal Ab (C-20) raised in a goat against a peptide located in the C-terminal domain of human lamin A and another goat affinity-purified polyclonal Ab (M20) raised to a peptide mapping to the C-terminal domain of mouse lamin B1 were purchased from Santa Cruz Biotechnology.
Secondary Abs. FITC-conjugated affinity-pure donkey anti-mouse IgG and FITC-conjugated goat anti-rat IgG, tetramethylrhodamine B isothiocyanate-conjugated affinity-pure donkey antirabbit IgG, and tetramethylrhodamine B isothiocyanate-conjugated donkey anti-mouse IgG secondary Abs were purchased from Jackson ImmunoResearch.
Immunofluorescence Labeling. Cells plated on polylysine-coated coverslips remained impermeable to protein after fixation with 4% paraformaldehyde in PBS for 10 min. The cells were rendered permeable, if desired, by further treatment of the fixed cells with 0.2% Triton X-100 for 5 min. After the subsequent double-immunolabeling reaction, cells were washed with PBS, and coverslips were mounted onto slides in the presence of mounting medium (Vector Laboratories). The immunolabeled cells were examined with a Zeiss Photoscope III fluorescence microscope by using FITC and tetramethylrhodamine B isothiocyanate filters, or images were captured with a DeltaVision deconvolution microscope system (Applied Precision, Issaquah, WA). The system includes a CoolSnap digital camera mounted on a Nikon TE-200 inverted epifluorescence microscope. In general, 5–30 optical sections spaced 0.1–0.5 μm apart were captured. Exposure times were set such that the camera response was in the linear range for each fluorophore and kept constant when imaging the same fluorophore within comparative experiments. Lenses included ×100 (n.a. 1.4), ×60 (n.a. 1.4), and ×40 (n.a. 1.3). The data sets were deconvolved and analyzed by using softworx software (Applied Precision) on a Silicon Graphics Octane workstation. Only single optical sections are shown (see Figs. 4 and 6).
Fig. 4.
Endogenous PS-1 N-terminal domain immunolabeling. Double immunolabeling of fixed but not permeabilized untransfected DAMI cells using deconvolution microscopic imaging. Primary rat mAb (1563) to PS-1 N-terminal domain (image 1) and affinity-purified goat polyclonal Abs to lamin (image 2) were the primary Abs followed by the appropriate secondary Abs. Image 1 shows that the immunolabeling of the endogenous PS-1 N terminus appears confined to the exterior cell surface; image 2 shows that lamin is not accessible to its Abs in these fixed, nonpermeabilized cells. If untransfected DAMI cells are permeabilized after fixation, the immunolabeling of the N-terminal domain of PS-1 is much more intense and diffused throughout the cell (image 3) than is the surface labeling of the impermeable cells (image 1). Intranuclear lamin in the same cells (image 4) can now be immunolabeled, demonstrating that these cells are permeable.
Fig. 6.
Endogenous PS-1 loop domain immunolabeling. Deconvolution microscopic imaging of doubly immunofluorescently labeled endogenous PS-1 in DAMI cells. Fixed but not permeabilized untransfected DAMI cells were immunolabeled for the PS-1 loop (image 1) and for lamin (image 2), followed by the appropriate secondary Abs. Image 1 shows that the immunolabeling of the PS-1 loop is confined to the exterior of the cell surface because image 2 shows that lamin is not accessible to its Abs in these impermeable cells. Image 3 indicates that if untransfected DAMI cells are permeabilized after fixation, the immunolabeling of the large loop domain of PS-1 is much more intense and diffused throughout the cell compared to the surface labeling of the impermeable cells (image 1). Nuclear lamin in the same cells (image 4) can be immunolabeled, demonstrating that these cells are indeed permeable.
FLAG Fusion and Expression of the Fusion Proteins. The cDNA encoding the entire N-terminal domains of PS-1 and PS-2 were obtained by PCR and cloned into the Tth111I and Xho-1 sites of the FLAG expression vector to produce fusion proteins with FLAG at the N-terminal and either PS-1- or -2 N-terminal domain at the C terminus. The FLAG fusion kit was obtained from Sigma. The FLAG fusion proteins were grown in DH5α bacteria and affinity-purified according to the manufacturer's protocols. The purified recombinant proteins were checked by Western blots by using Abs to both FLAG and N-terminal domains of either PS-1 or PS-2.
Results
Immunofluorescence experiments are reported herein by using as the primary anti-PS-1 Ab, mAbs directed either to the N-terminal domain of human PS-1 (rat mAb 1563), or the large extramembranous loop region of human PS-1 following TM helix VI (mouse mAb 5232; see Fig. 1 B and C), or polyclonal rabbit antipeptide antiserum (AbC1; ref. 14), directed to the C-terminal domain of human PS-1. With PS-1-transfected cells, double immunofluorescence labeling was performed with one primary Ab directed to a PS-1 determinant and a second primary Ab directed either to tubulin or actin, the latter two to confirm that there was no access of these Abs to the cytoplasm of fixed (but not permeabilized) cells, but only to cells fixed and permeabilized with Triton X-100, as already described (14). Following the primary Abs, the appropriate fluorescent-tagged secondary Ab reagents were then applied. One cell type used in these new studies was ES cells from PS-1–/– and PS-2–/– double-null mice (19), which were examined either untransfected or after transfection with PS-1.
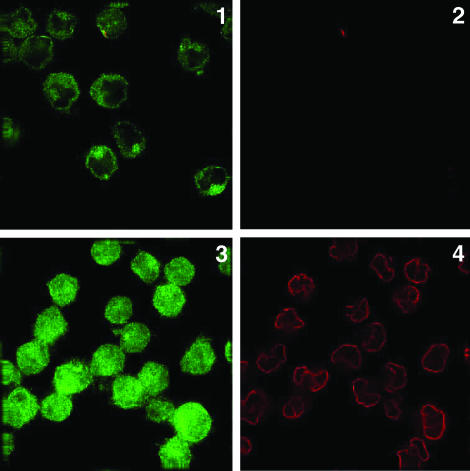
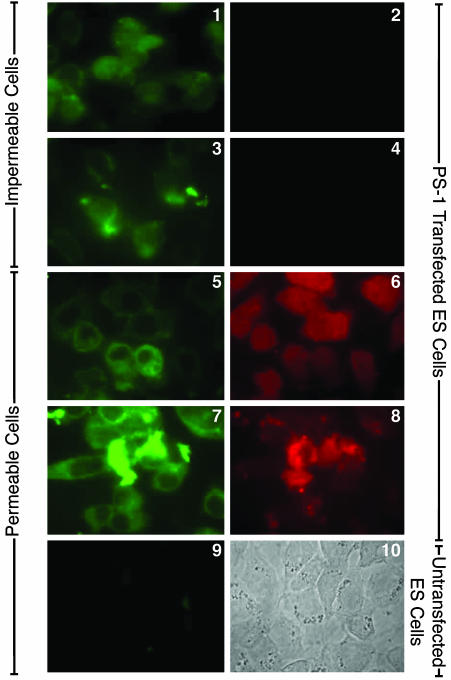
The N-Terminal Domain of Transfected Surface PS-1 Faces the Cell Exterior. Representative results with human PS-1-transfected ES double-null cells are presented in Fig. 2. The rat mAb 1563 directed to the N-terminal domain of PS-1 immunolabeled fixed, but not permeabilized, PS-1-transfected ES cells (Fig. 2, image 1), which were not labeled for tubulin (Fig. 2, image 2), showing that these cells were indeed impermeable to Ab and, therefore, that the labeling in Fig. 2, image 1, was on the cell surface. Untransfected ES double-null cells that were similarly treated were not labeled by rat mAb 1563 (Fig. 2, image 7). Furthermore, the surface labeling of the PS-1 N-terminal domain by rat mAb (1563) was inhibited in the presence of an excess (25 μg) of the specific fusion protein N-terminal domain of PS-1-FLAG (Fig. 2, image 3) but was unaffected by a similar excess of the nonspecific fusion protein N-terminal domain of PS-2-FLAG (Fig. 2, image 5), demonstrating the specificity of the labeling in Fig. 2, image 1. Fig. 2, image 6, shows that the cells examined in Fig. 2, image 5, did not stain for tubulin and were therefore indeed impermeable. The results shown in Fig. 2 collectively demonstrate that the N-terminal domain of human PS-1 is exposed on the extracellular surface of PS-1-transfected ES cells.
Fig. 2.
PS-1 N-terminal domain immunolabeling in transfected cells. Immunofluorescence microscopic labeling with a Zeiss Photoscope III instrument of PS-1-transfected PS-1/PS-2 double-null ES cells. Images 1 and 2 show double immunolabeling of the same fixed, impermeable cells with primary rat mAb (1563) (image 1) and mouse mAb to chicken β-tubulin (image 2), followed by the appropriate secondary Abs. Image 1 shows cell-surface immunolabeling of PS-1 N-terminal because image 2 shows that intracellular tubulin is not accessible to its Ab in these fixed, nonpermeabilized cells. Image 3 shows double immunolabeling of fixed, impermeable PS-1-transfected ES cells with rat mAb 1563, which had first been treated with an excess of 25 μg of PS-1 N-terminal-FLAG fusion protein, and also with mouse mAb to β-tubulin (not shown) followed by appropriate secondary Abs. Image 4 is the Nomarski image of image 3. Image 3 shows the inhibition of immunolabeling of PS-1 N-terminal in the presence of the specific fusion protein; the same fixed, nonpermeabilized cells were not accessible to the antitubulin Abs (data not shown). Images 5 and 6 show fixed, impermeable PS-1 transfected ES null cells double-immunolabeled with rat mAb 1563, which was first treated with an excess of 25 μg of the nonspecific N-terminal of PS-2–FLAG fusion protein (image 5), and with antitubulin (image 6) followed by appropriate secondary Abs. No significant inhibition of Ab labeling to the PS-1 N-terminal domain was induced by the nonspecific fusion protein (image 5). Image 7 shows untransfected fixed ES cells show no immunolabeling with rat mAb 1563. Image 8 shows a Nomarski image of same field as in image 7.
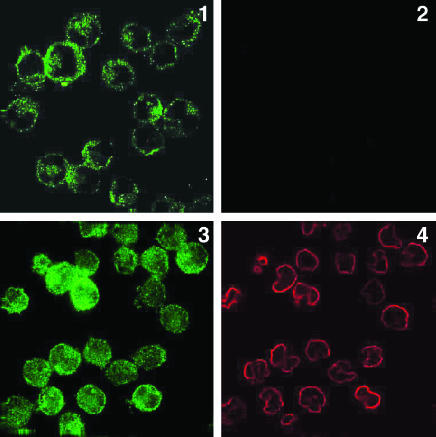
The Large Loop Region Following TM Helix VI of Transfected PS-1 Faces the Cell Exterior, and the C-Terminal Domain Faces the Cell Interior. The mouse mAb (5232) directed to the large loop region of human PS-1 likewise labeled the surfaces of fixed, but not permeabilized, PS-1-transfected ES double-null cells (Fig. 3, images 1 and 3), which were not labeled either for actin (Fig. 3, image 2) or for polyclonal rabbit Ab C-1 (Fig. 3, image 4), respectively. Therefore, the C-terminal domain of PS-1 is located, as previously found (14), on the side of the surface membrane opposite to the loop region of PS-1, with the loop region and the N-terminal domain on the same exterior side of the membrane. Cytoplasmic labeling of fixed and permeabilized PS-1-transfected ES cells was observed for actin (Fig. 3, image 6) and for AbC1 (Fig. 3, image 8). In a double-labeling experiment, untransfected fixed, but impermeable ES double-null cells were not labeled with the mAb (5232) to the loop region of PS-1 (Fig. 3, image 9) or with AbC1 (not shown).
Fig. 3.
PS-1 loop domain immunolabeling in transfected cells. Immunofluorescence microscopic labeling using the Zeiss Photoscope III instrument of PS-1-transfected PS-1/PS-2 double-null ES cells. Images 1 and 2 show double immunolabeling of fixed, impermeable cells with mouse mAb 5232 (image 1) and with rabbit polyclonal Ab to chicken actin (image 2), followed by appropriate secondary Abs. Image 1 shows immunolabeling of PS-1 loop region at the exterior cell surface because image 2 shows the same cells are not accessible to the anti-actin Abs. Images 3 and 4 show double immunolabeling of fixed, impermeable PS-1-transfected ES null cells with mouse mAb 5232 (image 3) and with rabbit polyclonal Ab C1 to the C-terminal domain of PS-1 (image 4), followed by appropriate secondary Abs. Image 3 shows cell-surface immunolabeling of PS-1 loop region, whereas image 4 shows that in the same cells, the PS-1 C-terminal domain is not accessible to its Abs in fixed, nonpermeabilized cells. Images 5 and 6 show double immunolabeling of fixed and permeabilized PS-1-transfected ES null cells with mouse mAb 5232 (image 5) and with rabbit polyclonal Ab to chicken actin (image 6), followed by the appropriate secondary Abs. Both Abs are labeling the cell interior. Images 7 and 8 show double immunolabeling of fixed and permeabilized PS-1-transfected ES null cells with mouse mAb 5232 (image 7) and rabbit polyclonal Ab C1 to PS-1 C-terminal domain (image 8) and appropriate secondary Abs. Both Abs labeled the interior of the permeabilized cells. Image 9 shows double immunolabeling of fixed and permeabilized untransfected ES double-null cells with mouse mAb 5232 (image 9) and rabbit polyclonal Ab C1 to PS-1 C-terminal domain (data not shown), followed by appropriate secondary Abs. Image 9 shows an absence of labeling of the permeabilized untransfected cells by the PS-1 N-terminal mAb. Image 10 shows a Nomarski image of the same field as in image 9.
The N-Terminal Domain of Endogenous Surface PS-1 Faces the Cell Exterior. Because our monoclonal primary Abs to PS-1 were directed to domains of the human protein, the endogenous PS-1 was examined with human DAMI instead of mouse ES cells. In addition, the deconvolution microscope was used for most of the experiments instead of the Zeiss Photoscope III for immunofluorescence detection. Also, the primary Ab directed to an intracellular antigen to detect whether fixed cells were impermeable to Ab reagents was an equal-part mixture of two affinity-purified goat polyclonal Abs to nuclear lamins A and B1 (20, 21).
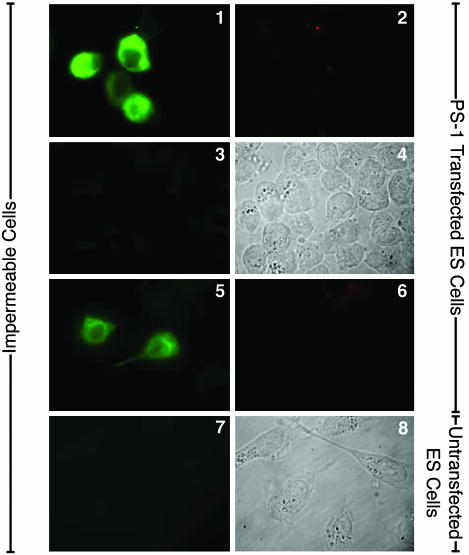
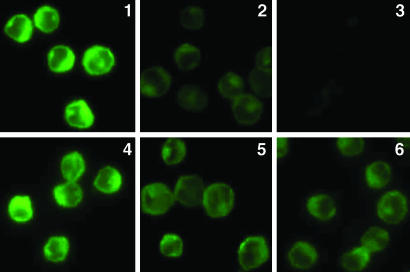
In double-immunolabeling studies, fixed but not permeabilized untransfected DAMI cells became labeled for their endogenous PS-1 N-terminal domain (Fig. 4, image 1) but not for lamin (Fig. 4, image 2). Therefore, the N-terminal domain of endogenous surface PS-1 faces the cell exterior. Consistent with this, the labeling of the N-terminal domain (Fig. 4, image 1) appeared largely confined to the cell surface in such a single optical section. If untransfected DAMI cells were permeabilized after fixation, the immunolabeling for the N-terminal domain (Fig. 4, image 3) was much more intense than for the impermeable cell and was diffused throughout the cell instead of confined to the cell surface. This is due to the PS-1 that is Ab accessible within intracellular membranes in these cells, demonstrated to be permeable by the immunolabeling of their nuclear lamin (Fig. 4, image 4). Furthermore, the immunolabeling of the N-terminal domain of endogenous PS-1 in fixed, impermeable cells (Fig. 5, image 1) was specifically inhibited by the fusion protein N-terminal domain of PS-1-FLAG (Fig. 5, images 2 and 3), the degree of inhibition increasing, as expected, with increase in the concentration of the specific fusion protein. However, in the presence of the nonspecific fusion protein N-terminal domain of PS-2-FLAG (Fig. 5, images 5 and 6), even at 10 times higher concentration than with the specific fusion protein, the immunolabeling of endogenous PS-1 was not inhibited. Therefore, these combined results show that the N-terminal domain of endogenous PS-1 is specifically immunolabeled by mAb 1563 and is located on the exterior side of the surface membrane of the DAMI cell.
Fig. 5.
Specific inhibition of immunolabeling. Immunofluorescence microscopic imaging without deconvolution of the N-terminal domain of endogenous PS-1 in untransfected fixed and impermeable DAMI cells (image 1) in the presence of increasing concentrations of the specific inhibitor, the FLAG fusion protein of the PS-1 N-terminal domain (image 2, 1.0 mM; image 3, 2 mM). The Ab staining is specifically and increasingly inhibited with increasing concentration of the specific inhibitor. Similarly, immunolabeled fixed and impermeable untransfected DAMI cells (image 4) first treated with the nonspecific FLAG fusion protein of PS-2 N-terminal domain at the 10-fold higher concentrations, 10 mM (image 5) and 20 mM (image 6), show no significant inhibition of PS-1 N-terminal Ab labeling.
The Large Loop Region Following TM Helix VI of Endogenous PS-1 Faces the Cell Exterior. Fixed untransfected DAMI cells double-immunolabeled with the two primary Abs, one to the loop region of PS-1 and the other to nuclear lamin (Fig. 6, images 1 and 2, respectively), show that the cells were impermeable to anti-lamin Ab and therefore that the loop Ab labeling was on the exterior surface of the cells. Consistent with this, loop labeling was confined to the cell surface (Fig. 6, image 1). If the fixed untransfected DAMI cells were permeabilized and then similarly double-immunolabeled, intracellular nuclear lamin was labeled (Fig. 6, compare image 4 with image 2), and the labeling for the loop region was spread throughout the cell (Fig. 6, image 3). The loop region and the N-terminal domain of endogenous PS-1 are therefore both located on the exterior of the cell surface.
Discussion
The results reported in Figs. 2, 3, 4, 5, 6 are entirely consonant with the 7-TM topography (Fig. 1B) of both transfected and endogenous human PS-1 in the cell-surface membrane and completely contradict the predictions of the 8-TM or 6-TM topographies (Fig. 1C). There is no ambiguity about the specificities of the mAbs used, or of the specificity of the immunolabeling results for the particular PS-1 domains studied. The new data strongly confirm the 7-TM topography of cell-surface PS derived in our earlier study (14).
A 7-TM topography for PS with its N terminus on the extracellular side of the surface membrane conforms to a generally reliable topological rule for polytopic proteins of eukaryotic as well as bacterial membranes (22). That rule predicts that the orientation of the first TM helix in the ER membrane depends on the difference in the net charges of the first 15 residues flanking the two sides of TM helix I, with the more negative side facing the exoplasm in the case of the plasma membrane. For PS-1, the net charge on the N-terminal side to that on the C-terminal side of TM helix I is –3/+1, whereas for PS-2 it is 0/+2. For both PS-1 and PS-2, the rule therefore predicts that the N-terminal domain is located on the extracellular side of the surface membrane, as we have demonstrated experimentally.
What can be the explanation for the entirely different conclusions concerning an 8-TM rather than a 7-TM topography for PS in ER membranes (10, 11, 18)? A possibility is that the evidence for the 8-TM topography is flawed. The method of topographical analysis by the results of transfection of cells with fusion proteins containing either truncated fragments of the integral protein or even the entire molecule under consideration, from which the main evidence for the 8-TM topography of PS-1 in the ER membrane was derived (10, 11, 18), can sometimes give misleading results (14). This is because the structural features that determine the membrane intercalation of domains of polytopic integral proteins are not yet fully understood (23–25) and may be inadvertently violated by experimenters in producing the particular fusion proteins for their transfection studies. All of our experiments, by contrast, have involved the intact PS protein in the plasma membrane.
The 7-TM topography of the PS molecules is consistent with a number of functional predictions. A role for PS as a heterotrimeric GPCR is suggested. In vitro experiments (26) suggest that within the 39-aa residue C-terminal domain of PS-1 (located in the cytoplasm in all three topographic models of PS-1, Fig. 1), there exists a specific binding and regulating domain for the brain Go protein, but the possibility that PS-1 may be a GPCR has not been elaborated upon since that report. The GPCRs form one of the largest protein superfamilies found in nature (27), and all polytopic members of this superfamily are apparently exclusively 7-TM integral proteins. To the best of our knowledge, no case of an 8-TM or 6-TM GPCR has been firmly identified. Other than in their 7-TM topography, however, GPCRs are structurally very diverse. The PS proteins show no extensive amino acid homologies with any of the four subfamilies of GPCRs. However, the region of the C-terminal domain of PS-1 that binds Go shows significant local amino acid sequence homologies with the G-binding domains of the D2-dopaminergic and the 5HT-1B receptors, both of which are 7-TM GPCRs, as well as of the G protein-activating oligopeptide, mastoparan (26). These homologies strengthen the case that PS-1 and other PS proteins are 7-TM GPCRs, but in vivo experiments are required to establish this.
Although evidence exists that PS is essential for the protease activity that carries out the γ-cleavage of amyloid precursor protein (28) and Notch (29), whether PS is itself the γ-secretase enzyme or is instead only an essential structural component of the secretase complex, is unsettled (compare with ref. 30). If PS is indeed the γ-secretase enzyme, it has been suggested (31) that it is an aspartyl protease with two particular aspartyl residues, Asp-257 and Asp-385, that are critically involved in the cleavage mechanism. (This evidence, however, is not uncontroversial; see refs. 4 and 30). The two aspartyl proposal has used the schematic 8-TM topological representation of PS (Fig. 1C) to suggest that Asp-257 within helix VI and Asp-385 within the proposed helix VII of the 8-TM model are close to one another in the membrane interior (Fig. 1C), where they might cooperate to form a transition state complex with the susceptible peptide bond of amyloid precursor protein that is assumed to be conveniently located between them. However, in the 7-TM topography, Asp-385 is outside the membrane in the large loop region following helix VI, far from Asp-257 that lies within helix VI. The model of an intramembranous two-aspartyl transition complex is therefore not supported by the 7-TM topography. However, it is not altogether ruled out. In the actual 7-TM PS structure, the moderately hydrophobic domain labeled VII′ (Fig. 1 A and B) may be partially inserted into the extracellular half of the membrane, placing Asp-385 closer to Asp-257 than is schematically represented in Fig. 1B.
Finally, in view of our firm evidence supporting a 7-TM topography of the PS proteins, a variety of other proteins that have been suggested to be homologues of PS (32–34), with topographies analogous to the 8-TM model of PS, need to be reexamined. This will be done in detail elsewhere.
Acknowledgments
We thank Drs. Dorit Donoviel and Alan Bernstein for providing the PS-1–/–/PS-2–/– double-null ES cells, Ms. Julia Harwood and Mr. Kevin Ha for their excellent technical assistance, and Dr. J. Feramisco of the University of California San Diego Cancer Center Digital Imaging Shared Resource and Mr. Steve McMullen for help with deconvolution imaging. This work was supported by National Institutes of Health Grants 5 R01 NS 27580 and 1 R01AG 17888 (to N.N.D.).
Abbreviations: PS, presenilin; AD, Alzheimer's disease; TM, transmembrane; ER, endoplasmic reticulum; ES, embryonic stem; GPCR, G protein-coupled receptor.
See Commentary on page 905.
References
- 1.Sherrington, R., Rogaev, E. I., Liang, Y., Rogaeva, E. A., Levesque, G., Ikeda, M., Chi, H., Lin, C., Li, G., Holman, K., et al. (1995) Nature 375, 754–760. [DOI] [PubMed] [Google Scholar]
- 2.Rogaev, E. I., Sherrington, R., Rogaeva, E. A., Levesque, G., Ikeda, M., Liang, Y., Chi, H., Lin, C., Holman, K., Tsuda, T., et al. (1995) Nature 376, 775–778. [DOI] [PubMed] [Google Scholar]
- 3.Levy-Lahad, E., Wasco, W., Poorkaj, P., Romano, D. M., Oshima, J., Pettingell, W. H., Yu, C. E., Jondro, P. D., Schmidt, S. D., Wang, K., et al. (1995) Science 269, 973–977. [DOI] [PubMed] [Google Scholar]
- 4.De Strooper, B. & Annaert, W. (2001) J. Cell Biol. 152, 785–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang, Z., Nadeau, P., Song, W., Donoviel, D., Yuan, M., Bernstein, A. & Yankner, B. A. (2000) Nat. Cell Biol. 2, 463–465. [DOI] [PubMed] [Google Scholar]
- 6.Kamal, A., Almenar-Qurealt, A., Leblanc, J. F., Roberts, E. A. & Goldstein, L. S. B. (2001) Nature 414, 643–648. [DOI] [PubMed] [Google Scholar]
- 7.Mumm, J. S., Schroeter, E. H., Saxena, M. T., Griesemer, A., Tian, X., Pan, D. J., Ray, W. J. & Kopan, R. (2000) Mol. Cell 5, 197–206. [DOI] [PubMed] [Google Scholar]
- 8.Ni, C. Y., Murphy, M., Golde, T. E. & Carpenter, G. (2001) Science 294, 2179–2181. [DOI] [PubMed] [Google Scholar]
- 9.Kyte, J. & Doolittle, R. F. (1982) J. Mol. Biol. 157, 105–132. [DOI] [PubMed] [Google Scholar]
- 10.Doan, A., Thinakaran, G., Borchelt, D. R., Slunt, H. H., Ratovitsky, T., Podlisny, M., Selkoe, D. J., Seeger, M., Gandy, S. E., Price, D. L. & Sisodia, S. S. (1996) Neuron 17, 1023–1030. [DOI] [PubMed] [Google Scholar]
- 11.Li, X. & Greenwald, I. (1996) Neuron 17, 1015–1021. [DOI] [PubMed] [Google Scholar]
- 12.Lehmann, S., Chiesa, R. & Harris, D. A. (1997) J. Biol. Chem. 272, 12047–12051. [DOI] [PubMed] [Google Scholar]
- 13.Nakai, T., Yamasaki, A., Sakaguchi, M., Kosaka, K., Mihara, K., Amaya, Y. & Miura, S. (1999) J. Biol. Chem. 274, 23647–23658. [DOI] [PubMed] [Google Scholar]
- 14.Dewji, N. N. & Singer, S. J. (1997) Proc. Natl. Acad. Sci. USA 94, 14025–14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dewji, N. N., Do, C. & Singer, S. J. (1997) Proc. Natl. Acad. Sci. USA 94, 9926–9931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwarzman, A. L., Singh, N., Tsiper, M., Gregori, L., Dranovsky, A., Vitek, M. P., Glabe, C. G., St George-Hyslop, P. H. & Goldgaber, D. (1999) Proc. Natl. Acad. Sci. USA 96, 7932–7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boeve, C. M. A., Cupers, P., Van Leuven, F., Annaert, W. & DeStrooper, B. (2000) in Membrane Structure in Disease and Drug Therapy, ed. Zimmer, E. (Dekker, New York), pp. 353–368.
- 18.Li, X. & Greenwald, I. (1998) Proc. Natl. Acad. Sci. USA 95, 7109–7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donoviel, D. B., Hadjantonakis, A. K., Ikeda, M., Zheng, M., StG eorgeHyslop, P. H. & Bernstein, A. (1999) Genes Dev. 13, 2801–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moir, R. D., Spann, T. P. & Goldman, R. D. (1995) Int. Rev. Cytol. 162, 141–182. [DOI] [PubMed] [Google Scholar]
- 21.Rao, L., Perez, D. & White, E. (1996) J. Cell Biol. 135, 1441–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartmann, E., Rapaport, T. A. & Lodish, H. F. (1989) Proc. Natl. Acad. Sci. USA 86, 5786–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nilsson, I., Witt, S., Kiefer, H., Mingarro, I. & Von Heijne, G. (2000) J. Biol. Chem. 275, 6207–6213. [DOI] [PubMed] [Google Scholar]
- 24.Zen, K. H., Consler, T. G. & Kaback, H. R. (1995) Biochemistry 34, 3430–3437. [DOI] [PubMed] [Google Scholar]
- 25.Singer, S. J. (1990) Annu. Rev. Cell Biol. 247–296. [DOI] [PubMed]
- 26.Smine, A., Xu, X., Nishiyama, K., Katada, T., Gambetti, P., Yadav, S. P., Wu, X., Shi, Y. C., Yasuhara, S., Homburger, V. & Okamoto, T. (1998) J. Biol. Chem. 273, 16281–16288. [DOI] [PubMed] [Google Scholar]
- 27.Wess, J. (1998) Pharmacol. Ther. 80, 231–264. [DOI] [PubMed] [Google Scholar]
- 28.Wolfe, M. S. & Haass, C. (2001) J. Biol. Chem. 276, 5413–5416. [DOI] [PubMed] [Google Scholar]
- 29.Schroeter, E. H., Kissingler, J. A. & Kopan, R. (1998) Nature 393, 382–386. [DOI] [PubMed] [Google Scholar]
- 30.Sisodia, S. S., Annaert, W., Kim, S. H. & De Strooper, B. (2001) Trends Neurosci. 24, S2–S6. [DOI] [PubMed] [Google Scholar]
- 31.Shearman, M. S., Beher, D., Clarke, E. E., Lewis, H. D., Harrison, T., Hunt, P., Nadin, A., Smith, A. L., Stevenson, G. & Castro, J. L. (2000) Biochemistry 39, 8698–8704. [DOI] [PubMed] [Google Scholar]
- 32.Ponting, C. P., Hutton, M., Baker, M., Jansen, K. & Golde, T. E. (2002) Hum. Mol. Genet. 11, 1037–1044. [DOI] [PubMed] [Google Scholar]
- 33.Weinofen, A., Binns, K., Lemberg, M. K., Ashman, K. & Martoglio, B. (2002) Science 296, 2215–2218. [DOI] [PubMed] [Google Scholar]
- 34.Annaert, W. & DeStrooper, B. (2002) Annu. Rev. Cell Dev. Biol. 18, 25–51. [DOI] [PubMed] [Google Scholar]