Abstract
With the arrival of low-cost, next-generation sequencing, a multitude of new plant genomes are being publicly released, providing unseen opportunities and challenges for comparative genomics studies. Here, we present PLAZA 2.5, a user-friendly online research environment to explore genomic information from different plants. This new release features updates to previous genome annotations and a substantial number of newly available plant genomes as well as various new interactive tools and visualizations. Currently, PLAZA hosts 25 organisms covering a broad taxonomic range, including 13 eudicots, five monocots, one lycopod, one moss, and five algae. The available data consist of structural and functional gene annotations, homologous gene families, multiple sequence alignments, phylogenetic trees, and colinear regions within and between species. A new Integrative Orthology Viewer, combining information from different orthology prediction methodologies, was developed to efficiently investigate complex orthology relationships. Cross-species expression analysis revealed that the integration of complementary data types extended the scope of complex orthology relationships, especially between more distantly related species. Finally, based on phylogenetic profiling, we propose a set of core gene families within the green plant lineage that will be instrumental to assess the gene space of draft or newly sequenced plant genomes during the assembly or annotation phase.
Thanks to recent advances in sequencing technologies (Martinez and Nelson, 2010), the price per base pair has dropped sharply (Schuster, 2008). Therefore, genome sequencing is no longer restricted to model organisms, and a variety of species of ecological, agricultural, or economical importance are sequenced by several laboratories around the world (Jaillon et al., 2007; Sato et al., 2008; Velasco et al., 2010). Recently, resequencing additional genomes of a reference species has become feasible as well (Garris et al., 2005), improving the understanding of genomic variation. Whereas a single genome provides a basic catalog of all genes it encodes, comparison of genomes gives insights into the evolution and adaptation of species to specific environments (Dassanayake et al., 2011). However, comparative genomics studies come at an extra cost: as the number of available genomes increases, large-scale analyses become increasingly difficult for nonexperts, whereas the computational requirements to extract biological information grow rapidly. Furthermore, biological variation between species and differences in sequence quality enhance the complexity of evolutionary analyses. Therefore, platforms for comparative genomics (Lyons et al., 2008; Proost et al., 2009; Kersey et al., 2010; Rouard et al., 2011) that take care of some of these challenges are valuable resources for experimental biologists.
A key challenge in comparative genomics is reliably grouping homologous genes (derived from a common ancestor) and orthologous genes (homologs separated by a speciation event) into gene families (Fitch, 1970; Chen et al., 2007; Gabaldón, 2008; Kuzniar et al., 2008). Orthology is generally considered a good proxy to identify genes performing a similar function in different species (Koonin, 2005). Consequently, orthologs are frequently used as a means to transfer functional information from well-studied model systems, such as Arabidopsis (Arabidopsis thaliana) or rice (Oryza sativa), to nonmodel organisms. In plants, the utilization of orthology is not trivial, due to a wealth of paralogs (homologous genes created through a duplication event) in almost all plant lineages. Ancient duplication events preceding speciation led to outparalogs, which are frequently considered as subtypes within large gene families. In contrast to these are inparalogs, genes that originated through duplication events occurring after a speciation event (Fitch, 1970). Besides continuous duplication events (for instance, via tandem duplication), many plant paralogs are remnants of whole genome duplications (WGDs). In flowering plants, the frequent WGDs in several lineages (Van de Peer et al., 2009) result in the establishment of one-to-many and many-to-many orthologs (or co-orthologs). Other modes of duplication, such as retrotransposition, also introduce co-orthologous relationships, but the duplicated copy ends up in a different genomic context and is probably regulated differently due to the absence of its original promoter. As such, the transfer of functional information between organisms is a nontrivial operation (Jensen et al., 2008). Various algorithms for orthology detection have been developed and benchmarked (Trachana et al., 2011) and, overall, can be cataloged as graph-based and tree-based methods, with the latter closer to the original orthology definition (Fitch, 1970), because they are based on the reconciliation of a family tree with a species tree.
PLAZA, an online resource for plant genomics, had been developed to integrate and distribute comparative genomics data for both computational and experimental plant biologists (Proost et al., 2009). The first release, based on nine sequenced plant genomes, included various tools to easily retrieve specific data types, such as gene families, multiple sequence alignments, phylogenetic trees, and genomic homology. To accommodate the evolutionary analysis of an increasing number of available plant genomes, more powerful and streamlined computational pipelines were required, as well as new tools to visualize genome information from multiple species. Here, we present version 2.5 of PLAZA, a major update of the comparative genomics platform, which currently hosts 25 species together with a variety of new tools to browse gene families, study functional clustering, and explore multispecies colinearity data. In addition to the development of a new tool to identify complex gene orthology relationships, different prediction methods were also evaluated by means of expression context conservation.
RESULTS AND DISCUSSION
Gene Annotation and Gene Families
Parsing the 25 genomes present in PLAZA 2.5 resulted in 909,850 genes, covering 85.8% protein-coding genes, 13.7% transposable elements, 0.3% RNA genes, and 0.1% pseudogenes (Table I). Besides nuclear gene annotations, chloroplast and/or mitochondrial gene information was included as well, when available. In total, 13 eudicots, five monocots (Liliopsida), one lycopod, one moss, and five algae were integrated, of which 16 are new species compared with the previous release. The functional annotation pipeline resulted in 462,958 (419,028 without Gene Ontology [GO] projection) genes with at least one associated GO term and 519,047 protein-coding genes with at least one InterPro domain (Table I). Overall, projected functional information inferred through sequence orthology (Proost et al., 2009) covered 10% of the available gene-GO annotations (43,930 genes from 18 different species have only GO annotations based on projection).
Table I. Data content for PLAZA 2.5.
PLAZA 2.5 is available from http://bioinformatics.psb.ugent.be/plaza and is free for academic use.
| Name | Genesa | Scaffoldsb | GOc | InterProd | Version | Reference |
| % | ||||||
| Arabidopsis lyrata | 32,670 (100%) | 8+1 (429) | 53.8 (65.6) | 72.1 | JGI 1.0e | Hu et al. (2011) |
| Arabidopsis | 33,602 (81.6%) | 5C,M | 77.3 (80.2) | 78.3 | TAIR10 | Arabidopsis Genome Initiative (2000) |
| B. distachyon | 26,678 (99.8%) | 5+1 (15)C | 56.6 (66.9) | 78.2 | MIPS 1.2e | International Brachypodium Initiative (2010) |
| Carica papaya | 28,072 (99.8%) | 4,635C | 43.4 (49.6) | 58 | HI ARC | Ming et al. (2008) |
| Chlamydomonas reinhardtii | 16,841 (99.7%) | 88C,M | 50.7 (50.7) | 53.4 | JGI 4.0 | Merchant et al. (2007) |
| Fragaria vesca | 34,809 (100%) | 7+1 (1,080) | 43.5 (49) | 61.4 | Strawberry Genome 1.0e | Shulaev et al. (2011) |
| Glycine max | 46,509 (99.9%) | 20+1 (97)C | 61.3 (70.2) | 82.9 | JGI 1.0e | Schmutz et al. (2010) |
| L. japonicus | 69,647 (61.9%) | 6+1 (22,048)C | 42.2 (45.8) | 57.3 | Kazusa 1.0e | Sato et al. (2008) |
| Malus domestica | 95,230 (66.7%) | 17+1 (23,653) | 61.8 (66.4) | 69.3 | IASMAe | Velasco et al. (2010) |
| Manihot esculenta | 30,800 (99.8%) | 3,142C | 57.6 (66.5) | 78.8 | Cassava4e | http://www.phytozome.net/cassava |
| M. truncatula | 57,587 (78.5%) | 8+1 (145)C | 35.4 (39.5) | 48.7 | Mt3.5e | Young et al. (2011) |
| Micromonas sp. RCC299 | 10,276 (99.3%) | 17C,M | 58.3 (58.3) | 69.8 | JGI 3.0e | Worden et al. (2009) |
| Rice indica | 59,430 (82.8%) | 12+1 (2,217)C,M | 44.1 (53.9) | 59.6 | 9311_BGF_2005e | Yu et al. (2002) |
| Rice japonica | 57,874 (72.9%) | 12C,M | 55.2 (58.6) | 58.6 | MSU RGAP 6.1 | Ouyang et al. (2007) |
| Ostreococcus lucimarinus | 7,805 (100%) | 21 | 60.7 (60.7) | 74.4 | JGI 2.0 | Palenik et al. (2007) |
| Ostreococcus tauri | 8,116 (98.5%) | 20C,M | 49.6 (49.6) | 63.7 | Ghent Universitye | Derelle et al. (2006) |
| Physcomitrella patens | 36,137 (77.8%) | 1,121C,M | 47.8 (47.8) | 57.9 | JGI 1.1; cosmoss.org 1.2 | Rensing et al. (2008) |
| P. trichocarpa | 41,521 (99.9%) | 19+1 (957)C | 54.6 (61.8) | 73.7 | JGI 2.0 | Tuskan et al. (2006) |
| Ricinus communis | 31,221 (100%) | 4,962 | 48.3 (54.1) | 65 | JCVI 1.0e | Chan et al. (2010) |
| S. moellendorffii | 22,285 (100%) | 361 | 55.7 (55.7) | 71.8 | JGI 1.0e | Banks et al. (2011) |
| S. bicolor | 34,686 (99.8%) | 10+1 (207)C,M | 54.8 (62.1) | 71.1 | JGI 1.4 | Paterson et al. (2009) |
| Theobroma cacao | 46,269 (62.4%) | 11C | 50.7 (57.7) | 69.4 | CocoaGen v1.0e | Argout et al. (2011) |
| Vitis vinifera | 26,644 (99.5%) | 19+1 (14)C,M | 72.6 (76.4) | 71.8 | Genoscope_v1 | Jaillon et al. (2007) |
| Volvox carteri | 15,544 (100%) | 762 | 39.1 (39.1) | 54.1 | JGI 1.0e | Prochnik et al. (2010) |
| Z. mays | 39,597 (99%) | 11C,M | 48.1 (55.9) | 65.6 | Version 5.60e | Schnable et al. (2009) |
Numbers in parentheses refer to the fraction of protein-coding genes.
Numbers in parentheses refer to the number of genomic sequences in the original annotation (assembly) containing genes. The “+1” tag indicates the creation of a virtual chromosome zero to group scaffolds together, whereas “C” and “M” indicate the inclusion of chloroplast and mitochondrial genomes, respectively.
Percentage of coding genes with an associated GO term. The fraction after the GO projection is displayed in parentheses.
Percentage of coding genes with an associated InterPro domain.
New species compared with PLAZA 1.0.
Protein clustering based on all-against-all sequence similarity searches resulted in 32,294 gene families, covering 87.8% of all the protein-coding genes, and 22,350 multispecies gene families, covering 82.6% of all protein-coding genes (Supplemental Table S1), with a gene family defined as a cluster of two or more homologous genes. This coverage represents a considerable increase compared with PLAZA 1.0, in which only 77.6% and 68.1% of the coding genes were assigned to gene families and multispecies gene families, respectively. Multispecies gene families are commonly applied for improving, through homology, the structural annotation of gene models (Meyer and Durbin, 2004). The increase in gene number assigned to both classes of gene families demonstrates the importance of sequencing additional species to obtain a better gene coverage within specific phylogenetic clades.
Reliable transfer of known functional descriptions from the gene level to the gene family level was achieved by calculating GO enrichment statistics for each family (see “Materials and Methods”). Through the Web site, this functional information, together with protein domain information, is displayed per family. Although this family GO annotation procedure yielded information for only 8,606 gene families and 28,281 subfamilies, they cover more than 70% of the protein-coding genes present in gene families (Supplemental Fig. S1).
Core Plant Gene Families and Detection of Clade-Specific or Expanded Gene Families
Most new genome sequences generated by next-generation sequencing methods do not provide the full genomic sequence (Al-Dous et al., 2011) but rather aim at providing sequences containing the majority of the proteome, potentially missing noncoding genes or intergenic regions. The extremely large genome sizes associated with some organisms prevent full-genome sequencing and enforce the application of transcriptome sequencing to build gene catalogs (Bennett and Leitch, 2005). A key challenge in comparative gene family analysis is discerning whether the absence of a species within a gene family is functionally and evolutionarily relevant or rather an artifact from the assembly and/or annotation procedures. As a consequence, the reliable assessment of the gene space provides an important measure to determine the quality of genome sequencing and annotation projects.
Based on families conserved in a specific set of species, core gene families were created by means of PLAZA 2.5. Families were selected on the basis of their gene content in phylogenetic subclades from the PLAZA species tree, tolerating missing homologs in a small subset of species (see “Materials and Methods”; Supplemental Method S1). Three sets of core gene families were built based on the subclades rosids, monocots, and green plants. This phylogenetic approach resulted in 6,316, 7,076, and 2,928 core gene families for the rosids, monocots, and green plants, respectively (Supplemental Tables S2–S4). As expected, the core gene families cover, among others, housekeeping genes and genes involved in primary metabolism. For each gene family, a representative gene was selected from the rosids and monocots (with a preference for genes from Arabidopsis and rice, respectively) that could be used as a probe to quantify genome completeness. Assessment of the gene space of each species included in the platform using the weighted core gene family scores revealed relatively low gene coverage for some species (Supplemental Fig. S2). Especially Lotus japonicus and Medicago truncatula within the eudicot species, and Selaginella moellendorffii within the primitive land plants, showed high numbers of potentially missing genes. We recommend these lists of core gene families as a reference set to quantify the gene space in future genome projects.
Whereas core gene families are a useful tool for asserting proteome completeness, the study of species-specific or lineage-specific (expanded) gene families is equally important to understand how species can adapt to particular niches (Supplemental Tables S5 and S6). Tandem gene duplications are a known mechanism used by plants to rapidly increase the expression rate of a gene (Hanada et al., 2008), instead of the transcription rate. Two new tools were implemented to facilitate the detection of gene families based on phylogenetic profiles (presence or absence of a gene family in a species) or expansion statistics. Whereas the Gene Family Finder tool enables the identification of (expanded) gene families specific to one or more species, the Gene Family Expansion Plot displays gene family expansion patterns between two (sets of) organisms (Supplemental Fig. S3).
Integrative Orthology Viewer: An Ensemble Approach to Detect Orthology Relationships
Several methods for finding orthologs between two or more species have been described, each with its own strengths and weaknesses (Gabaldón, 2008). Whereas reciprocal best BLAST hit detection (Huynen and Bork, 1998) between closely related species provides a practical solution to identify orthologs, it cannot deal with complex one-to-many or many-to-many orthologous relationships between more distantly related species. Although the construction of phylogenetic trees (Page and Charleston, 1997; Zmasek and Eddy, 2001) should offer the highest confidence to identify speciation events in gene family trees, it has a relatively low gene coverage compared with sequence-based clustering methods, as trees could not be generated for all gene families. In PLAZA 2.5, 46,651 phylogenetic trees were constructed covering 81% of all protein-coding genes assigned to gene families. Besides heavy computational requirements, the method is also hampered by its sensitivity to differences in the topology of the gene tree compared with the species tree, which are used for reconciliation (Hahn, 2007).
To detect orthologous gene relationships in plants with an enhanced robustness, an integrative approach was developed to identify orthologs on a gene-by-gene basis. The developed ensemble approach consists of four distinct orthology prediction methods: orthologous gene families inferred through sequence-based clustering with OrthoMCL (Li et al., 2003; including modeling of reciprocal best BLAST hit orthology and inparalogy), reconciled phylogenetic trees, colinearity information, and multispecies best hits and inparalogs (BHI) families. The latter are based on the best BLAST hit for each species, extended with the inparalogous genes in each species (Linard et al., 2011). The integration of gene colinearity facilitates the detection of positional orthologs, namely genes with conserved genome organization between species. The combination of different methods for orthology detection, as implemented in the PLAZA platform, allows for the more accurate selection of orthologs, for example using majority voting (Pryszcz et al., 2011) or through the application of a weighted voting scheme based on the sensitivity of individual tools. Other plant comparative genomics databases like GreenPhylDB (Rouard et al., 2011) and Phytozome (Goodstein et al., 2012) only group homologous genes into families using clustering, the latter also including synteny information to identify putative positional orthology. PlantEnsembl (Kersey et al., 2010) performs orthology and paralogy predictions solely based on reconciled gene family phylogenetic trees.
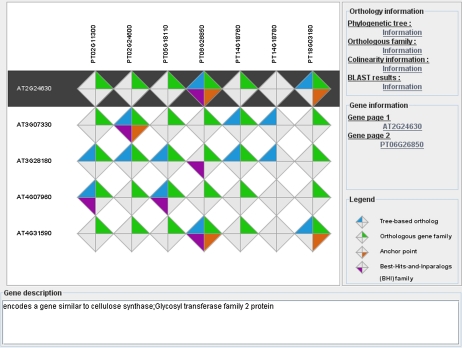
The Integrative Orthology Viewer displays for a query gene and its predicted inparalogs the associated orthologs, including support from the different orthology methods (Fig. 1). In addition, all links are provided to explore the supporting evidence and specific details of the individual predictions. For instance, the phylogenetic trees that served as the primary data source for the tree-based orthologs can be viewed and the user can evaluate the support of a specific speciation node.
Figure 1.
Integrative Orthology Viewer. Orthology overview for the Arabidopsis gene AT2G24630 and its paralogs and orthologs in Populus trichocarpa. The selected query gene is marked with a black background.
To compare the performance of individual methods, as well as of an integrative approach, we first generated basic statistics about the number of inferred orthology relationships. With focus on the model species Arabidopsis as query species and any other species as target, the gene coverage was highest for the BHI families and OrthoMCL (25,862 and 23,932 genes with at least one ortholog, respectively). As expected, reconciled phylogenetic trees only provided orthology information for 18,415 Arabidopsis genes. To evaluate the quality of these predictions, the percentage of orthologous gene pairs with conserved expression was determined by using the expression context conservation (ECC; Movahedi et al., 2011). The expression context was based on the expression similarity between a query gene and all other genes in that species (gene-centric coexpression cluster). The ECC was obtained by starting from a predicted orthologous gene pair, retrieving all coexpressed genes per species, and calculating how many homologs were coexpressed in both species. Significant ECC values indicate that the orthologous genes share coexpression with several other genes in both species. Consequently, conserved ECC gene pairs can be used as a proxy to measure conserved gene functions between putative orthologs, based on spatiotemporal expression information.
Based on a random sample of 9,319 orthologous Arabidopsis-rice gene pairs, ECC scores for the different orthology prediction methods indicated that gene pairs uniquely predicted by individual methods overall contain fewer gene pairs with conserved coexpression compared with predictions supported by multiple tools: 44%, 41%, and 41% for OrthoMCL, BHI families, and trees, respectively, versus 60% (supported by OrthoMCL and BHI families), 41% (supported by BHI families and trees), 57% (supported by OrthoMCL and trees), and 68% (supported by OrthoMCL, BHI families, and trees). Although these results indicated that multiple forms of evidence increase the reliability of orthology prediction, application of a majority voting system (i.e. only selecting orthologs with the highest number of forms of evidence) could miss true orthologs with fewer support types (i.e. false negatives). To compare the performance of a majority-voting protocol with a selection procedure only requiring two support types, we evaluated ECC scores for orthologous gene pairs supported by only two forms of evidence with those confirmed by three prediction methods. Despite majority voting orthologs having a higher fraction of ECC conserved genes (66%), 51% of the gene pairs with only two forms of evidence also showed conserved expression between Arabidopsis and rice (based on the same reference query Arabidopsis gene set). Therefore, we retained all orthologous predictions supported by two or more forms of evidence in the integrative orthology method (Supplemental Method S2).
Although OrthoMCL has been shown to have a good tradeoff between false positives and false negatives (Chen et al., 2007), we observed that 3,506 Arabidopsis genes (13% of the proteome) had a predicted orthologous rice gene based on the integrative method, whereas no ortholog was found using OrthoMCL. Of the 3,506 Arabidopsis genes having one or more rice orthologs (covering 3,874 rice genes in total), 40% exhibited conserved expression conservation. This result indicates that a considerable fraction of gene pairs not reported by OrthoMCL represents conservatively coexpressed orthologs, revealing the complementary nature of both approaches.
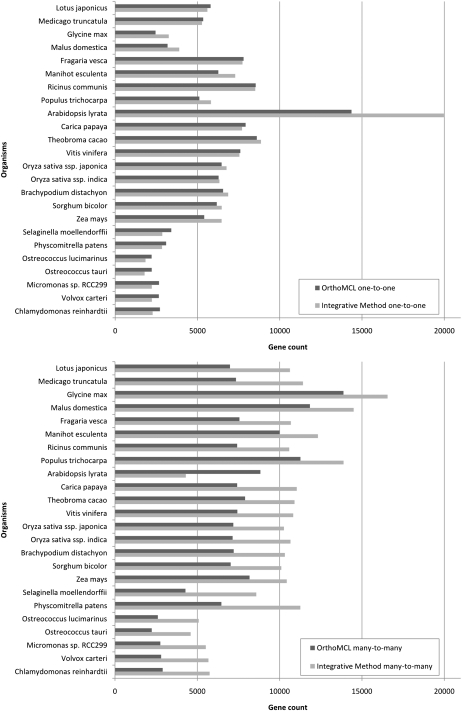
Application of the integrative method (requiring at least two support types) to predict orthologs from Arabidopsis in other species, revealed overall 30% more predictions compared with OrthoMCL (Fig. 2). Although the difference in the number of one-to-one orthologs is minor for most species, the number of complex orthology relationships (one-to-many and many-to-many orthologs) is higher for the integrative method. The frequent occurrence of WGD is an important driver responsible for the high frequency of complex orthology gene relationships in plant genomes.
Figure 2.
Quantification of Arabidopsis orthologs. Summary of the different orthologous relationships predicted by the PLAZA integrative method and OrthoMCL between Arabidopsis and all other PLAZA 2.5 species, respectively. The integrative method requires at least two support types to retain orthologous genes. Species are ordered per phylogenetic clade. The top panel displays results for one-to-one orthologs, and the bottom panel shows many-to-many orthologs.
Clusters of Functionally Related Genes in Eukaryotic Genomes
Whereas in many prokaryotic genomes genes are organized in operons, this is relatively rare in eukaryotes (Osbourn and Field, 2009). However, the overall absence of polycistronic mRNAs in eukaryotic genomes does not imply a random gene organization within chromosomes (Hurst et al., 2004; Michalak, 2008; Koonin, 2009; Osbourn, 2010). In several eukaryotic species clusters, with genes sharing similar expression patterns, members of the same pathway or genes with related functions have been described, indicating that the null hypothesis of random gene order is incorrect (Hurst et al., 2004). Recent studies have suggested that the chromatin state, either euchromatin or heterochromatin, is one of the contributing factors to the coexpression of neighboring genes (Hurst et al., 2004; Michalak, 2008) and bidirectional promoters as well (Fabry et al., 1995).
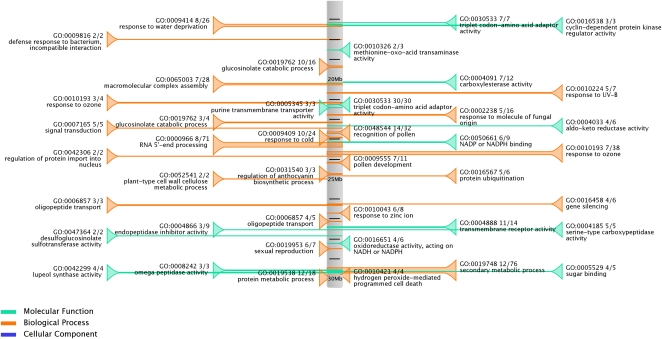
To study the clustering of functionally related genes, the C-Hunter program (Yi et al., 2007) was used for a genome-wide analysis. This tool detects statistically significant clusters of neighboring genes based on the similarity of GO annotations. The standard C-Hunter run (no tandem gene removal, minimum genes two, maximum genes 30) resulted in 5,408 significant clusters covering 34,407 genes from 25 different species. As the majority of these clusters (68%) are composed uniquely of tandemly duplicated genes, an extra data set was created to detect clustering of nonhomologous genes (Michalak, 2008). In this data set, every set of tandem genes was represented by a single gene representative (see “Materials and Methods”). The number of clusters varied widely among the different species (Supplemental Fig. S4A), suggesting that both the quality and quantity of the structural and GO annotations of genes played a major role, as well as the assembly of scaffolds in the chromosomes. More compact genomes, such as those of the algal species, had a smaller number of functional clusters, whereas the number of detected functional clusters in larger genomes correlated with the number of genes per scaffold and the number of genes with a GO term (Supplemental Fig. S4B). The resulting clusters are included in the database and can be browsed from both gene and GO pages on the PLAZA Web site. Furthermore, a visualization presenting the significant functional clusters per chromosome (Fig. 3) was created with a GO domain-based coloring.
Figure 3.
Functional clusters based on GO annotation. Functional clusters in Arabidopsis chromosome 1, detected with the C-Hunter software package, are shown. Data in the text fields include GO term and description, cluster size, and the number of genes within a cluster with a specific GO term.
Colinearity-Based Genome Analysis
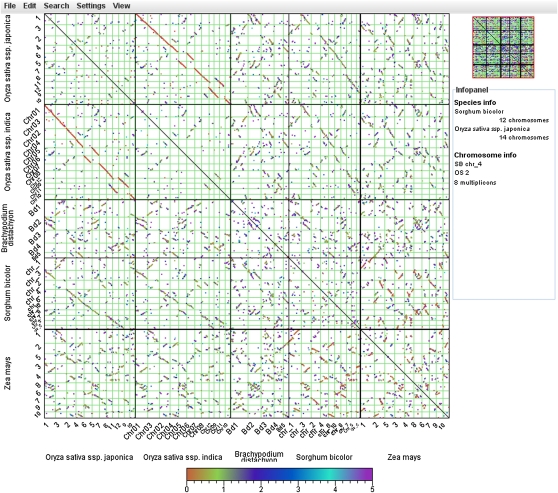
As a means to study genome organization and evolution, i-ADHoRe (Proost et al., 2011) is used to discover genomic homology based on gene colinearity within and among species. Colinearity information can be applied to analyze segmental and WGD events, whereas cross-species genome conservation facilitates the analysis of chromosomal rearrangements, such as inversions, chromosomal fissions/fusions, and translocations. As the increase in the number of species resulted in more complex genomic homology relationships, two new tools, the WGDotplot applet (Fig. 4) and the Circle Plot (Supplemental Fig. S5), were developed to provide more advanced and configurable visualizations. For both tools, the dating of colinear regions, based on the fraction of synonymous substitutions over all synonymous sites, is visualized by color coding.
Figure 4.
WGDotplot applet. Visualization of the colinearity between five monocot species: Oryza sativa ssp. japonica, Oryza sativa ssp. indica, Brachypodium distachyon, Sorghum bicolor, and Zea mays. Green lines indicate different chromosomes, and the colors of the colinear regions reflect the Ks values (fraction of synonymous substitutions over all synonymous sites).
The WGDotplot applet was implemented in Java and designed to be an interactive extension of the static visualizations present in PLAZA 1.0, also allowing the visualization of colinear regions between more than two species. At the same time, the functionalities were extended to encompass a rich palette of visualization options, such as hiding chromosomes and rearranging chromosomal positions, adapting color usage, and using stepless zoom features to browse the genomic colinearity between multiple species (Fig. 4).
The Circle Plot tool was developed as a lightweight and interactive circular visualization tool, similar to the popular Circos software (Krzywinski et al., 2009). Fully written in Javascript, this program runs natively on most modern browsers. Whereas the primary use of the Circle Plot is the study of intraspecies colinear regions, the ability to map interspecies colinear regions on the circumference of the plot closes the gap between the capabilities of the Circle Plot and the WGDotplot applet (Supplemental Fig. S5). Extra features, such as coding gene density and InterPro and GO terms, can also be displayed on the circumference of the Circle Plot. Another main difference is the mode of chromosome size determination. Whereas the Circle Plot uses nucleotide-based coordinates, the WGDotplot applet uses genes as the smallest units (retaining protein-coding genes only). Consequently, the former can display information in low-coding regions, such as centromeres or telomeres, and the latter facilitates the comparison of colinear regions from species with differences in gene density.
User Interactivity via Workbench and Bulk Downloads
PLAZA offers a versatile resource for easy data mining of homologous genes, sequence alignments and phylogenetic trees, genome organization, and functional annotation in different plants. However, large-scale analyses with a Web-based user interface quickly become tedious and time consuming. To overcome this problem, a user-oriented workbench was implemented in which specific gene sets can be analyzed. Different collections of user-provided gene lists are stored as separate experiments and genes can be added to an experiment based on internal/external gene identifiers or sequence similarity searches (see the Tutorial on the PLAZA Web site), providing a versatile approach to map genes across species or to summarize sequencing data on a reference genome annotation. Whereas the initial workbench contained tools to explore the functional annotation of sets of genes, in PLAZA 2.5, multiple improvements were made for an easier and more comprehensive user experience. The GO enrichment tool is extended (Supplemental Fig. S6), bulk detection of orthologs on a gene-by-gene basis is possible, and multiple workbench experiments can be compared. In addition, based on the outcome of a first analysis (such as gene filters in an experiment with GO), a new workbench experiment can easily be created or, conversely, genes can be removed from the initial experiment. The export function allows the user to retrieve general gene information (functional annotation, gene family data, orthologs, duplication data) as well as various sequence features (e.g. coding sequence, intron, and upstream and downstream sequences) for large gene sets covering all 25 genomes. Overall, the workbench offers a user-friendly solution for the efficient analysis of multiple data sets containing hundreds of genes. In addition, bulk downloads of most data sets in PLAZA are available through the FTP site.
In conclusion, the PLAZA platform is a user-friendly platform for small- and large-scale comparative sequence analyses of plant genomes. This new version includes 16 new genomes and implements new methods for colinearity and orthology detection.
MATERIALS AND METHODS
Gene Models and Gene Families
An overview of all primary sources supplying gene annotation data is presented in Table I. All genomes, and their associated gene models, were first parsed into a uniform format and stored in a relational database. The association of a gene model with one of the four different gene types (coding, RNA, transposable element, and pseudogene) was extracted from the primary data sources. For species lacking chloroplast and mitochondrial DNA sequences, organellar genomes, when available, were obtained from the European Bioinformatics Institute (http://www.ebi.ac.uk/genomes/organelle.html).
The gene models, DNA sequences, and protein sequences were tested for consistency, and irregular results (such as mismatches between translated DNA sequences and the provided protein sequences) were flagged in the database. The longest splicing variant was selected as representative for genes with alternative transcripts and, in turn, used in subsequent analyses focusing on gene family delineation and colinearity detection. Splice variants, if annotated, could be explored with the genome browsers AnnoJ (Lister et al., 2008) or GenomeView (Abeel et al., 2011). Gene families were delineated by first computing the protein sequence similarity through an all-against-all BLAST (e-value cutoff of 1e-05, retaining the top 500 hits) and then by applying TribeMCL (Enright et al., 2002) and OrthoMCL (Li et al., 2003) to cluster genes in families and subfamilies, respectively.
The PLAZA species tree was manually constructed using information from the National Center for Biotechnology Information taxonomy (Federhen, 2011) and the literature (Moore et al., 2010) to resolve trifurcations.
The core gene families were selected by a phylogenetic approach: the clades with at least two nonleaf subclades were retained from the PLAZA species tree, and to be considered as a core family for these clades, at least one organism within each of the subclades had to possess a representative gene (Supplemental Method S1). This approach inferred, based on parsimony, that a gene family was present at ancestral nodes with a tolerance of potential annotation errors in a limited number of species. The total set of core gene families for a given clade consisted of the intersection gene family sets generated by subclades. For each core gene family, representative genes were selected, using BLASTP scores with other gene family members as an evaluation metric. To assess the gene space in available plant genomes, each core family was counted with a weight equal to one divided by the average family size. The average gene family size was defined by the total number of genes in a gene family divided by the number of species within that family. The weighting scheme corrected for the observation that the probability of finding a homolog is higher for large families compared with single-copy or small families.
Colinearity
Homologous genomic regions were detected with i-ADHoRe 3.0 (Fostier et al., 2011; Proost et al., 2011), which identified colinear regions based on conserved gene order and content. i-ADHoRe was run with the following settings: alignment_method gg2, gap_size 30, tandem_gap 30, cluster_gap 35, q_value 0.85, prob_cutoff 0.01, multiple_hypothesis_correction FDR, anchor_points 5, and level_2_only false. Tandem gene duplicates were also determined with i-ADHoRe, whereas the relative dating of duplicated genes in colinear regions was done with PAML (settings: verbose 0, noisy 0, runmode −2, seqtype 1, CodonFrEquation 2, model 0, NSites 0, icode 0, fix_alpha 0, fix_kappa 0, and RateAncestor 0).
Functional Annotation
GO annotation, when available, was downloaded along with the gene models. Furthermore, InterPro scan (Hunter et al., 2009) was run on all protein-coding gene models, and additional GO annotations were inferred with InterPro to GO mapping. Redundant GO annotations were merged according to the GO evidence code rank (Buza et al., 2008). To avoid the inclusion of obsolete GO terms, a filter was applied using the set of valid GO terms derived from http://geneontology.org (Ashburner et al., 2000). The GO annotation was also projected between orthologs from eudicots and monocots (Proost et al., 2009). GO enrichment was analyzed for each gene family, with only the organisms with genes in the gene family under investigation being used as the background model for the statistical analysis (hypergeometric distribution with Bonferroni correction for multiple testing). Only GO terms covering at least half of the annotated genes in a family and with corrected values of P < 0.05 were retained.
Functional Gene Clusters
Clusters of functionally related genes (functional clusters) were detected using C-Hunter (Yi et al., 2007) on two different data sets that differed by whether the tandemly duplicated genes had been collapsed to a single representative or not. Two different runs were performed on each data set, with different minimum (2/30) and maximum (10/150) cluster sizes. The e-value cutoff (0.001) and maximum cluster overlap (50%) were the same for the different runs. When multiple clusters spanning the same location were detected, because of GO term organization as a directed acyclic graph (Ashburner et al., 2000), only the most significant cluster was retained.
Orthology Prediction and Evaluation
The PLAZA integrative approach for orthology detection was based on four methods: orthologous gene families, phylogenetic trees, colinear regions, and multispecies best BLAST hits. For the gene families OrthoMCL clusters were used, the phylogenetic trees were constructed based on gene families inferred with TribeMCL, the colinear regions were detected with i-ADHoRe, and the best BLAST hits (with inparalogs), namely BHI families, were detected by an OrthoInspector-like approach (Linard et al., 2011). Briefly, interspecies best BLAST hits were first retrieved for each gene and in a second phase inparalogs were included, defined as the intraspecies BLAST hits that were more similar than the best interspecies BLAST hits.
For all gene families, phylogenetic trees were constructed with PhyML (Guindon and Gascuel, 2003) based on multiple sequence alignments generated by MUSCLE (Edgar, 2004). Duplication and speciation events in the phylogenetic trees were identified by applying the NOTUNG tree reconciliation method (Vernot et al., 2008). Based on a duplication consistency score, erroneous duplications due to incongruences between the gene family and species tree were determined (Proost et al., 2009).
The reliability of the different orthology predictions was scored with the ECC score (Movahedi et al., 2011). ECC compared the expression profile conservation between two species by a statistical framework evaluating shared homologous relationships between coexpressed genes. The retrieved expression compendia (Movahedi et al., 2011) consisted of 76 Arabidopsis (Arabidopsis thaliana) and 63 rice (Oryza sativa subspecies japonica) Affymetrix nonredundant microarray experiments. These expression data sets were constructed starting from 322 Arabidopsis and 203 rice experiments using data normalization, collapsing of redundant conditions, and removal of transgenic or mutant experiments. A total of 19,937 Arabidopsis and 32,004 rice genes were present on the microarrays for expression analysis (based on a custom Chip Description File [Movahedi et al., 2011]). Pearson correlation coefficient thresholds for Arabidopsis and rice were 0.48 and 0.41, respectively. While Movahedi et al. (2011) used one-to-one orthologous gene pairs as seeds, the evaluation of the different orthology predictions using ECC was performed using homologous gene relationships based on TribeMCL clusters.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Gene family coverage by GO enrichment, organized by gene family size.
Supplemental Figure S2. Core gene family coverage.
Supplemental Figure S3. Gene family expansion plot.
Supplemental Figure S4. Summary functional clusters.
Supplemental Figure S5. Circle Plot.
Supplemental Figure S6. GO enrichment graph.
Supplemental Table S1. Gene and gene family content for PLAZA 2.5.
Supplemental Table S2. List of rosid core gene families.
Supplemental Table S3. List of monocot core gene families.
Supplemental Table S4. List of green plant core gene families.
Supplemental Table S5. Species-specific gene families.
Supplemental Table S6. Clade-specific gene families.
Supplemental Method S1. Selection of core gene families.
Supplemental Method S2. Different strategies for the integrative orthology detection.
Supplementary Material
Acknowledgments
We thank Thomas Van Parys and Thomas Abeel for technical assistance with the GenomeView applet browser, Lieven Sterck and Jeffrey Fawcett for helpful suggestions about the platform, and Annick Bleys and Martine De Cock for help in preparing the manuscript.
References
- Abeel T, Van Parys T, Saeys Y, Galagan J, Van de Peer Y. (2012) GenomeView: a next-generation genome browser. Nucleic Acids Res (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Dous EK, George B, Al-Mahmoud ME, Al-Jaber MY, Wang H, Salameh YM, Al-Azwani EK, Chaluvadi S, Pontaroli AC, DeBarry J, et al. (2011) De novo genome sequencing and comparative genomics of date palm (Phoenix dactylifera). Nat Biotechnol 29: 521–527 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Argout X, Salse J, Aury JM, Guiltinan MJ, Droc G, Gouzy J, Allegre M, Chaparro C, Legavre T, Maximova SN, et al. (2011) The genome of Theobroma cacao. Nat Genet 43: 101–108 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000) Gene Ontology: tool for the unification of biology. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks JA, Nishiyama T, Hasebe M, Bowman JL, Gribskov M, dePamphilis C, Albert VA, Aono N, Aoyama T, Ambrose BA, et al. (2011) The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332: 960–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ. (2005) Nuclear DNA amounts in angiosperms: progress, problems and prospects. Ann Bot (Lond) 95: 45–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buza TJ, McCarthy FM, Wang N, Bridges SM, Burgess SC. (2008) Gene Ontology annotation quality analysis in model eukaryotes. Nucleic Acids Res 36: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AP, Crabtree J, Zhao Q, Lorenzi H, Orvis J, Puiu D, Melake-Berhan A, Jones KM, Redman J, Chen G, et al. (2010) Draft genome sequence of the oilseed species Ricinus communis. Nat Biotechnol 28: 951–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Mackey AJ, Vermunt JK, Roos DS. (2007) Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS ONE 2: e383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassanayake M, Oh DH, Haas JS, Hernandez A, Hong H, Ali S, Yun DJ, Bressan RA, Zhu JK, Bohnert HJ, et al. (2011) The genome of the extremophile crucifer Thellungiella parvula. Nat Genet 43: 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle E, Ferraz C, Rombauts S, Rouzé P, Worden AZ, Robbens S, Partensky F, Degroeve S, Echeynié S, Cooke R, et al. (2006) Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci USA 103: 11647–11652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. (2002) An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30: 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabry S, Müller K, Lindauer A, Park PB, Cornelius T, Schmitt R. (1995) The organization structure and regulatory elements of Chlamydomonas histone genes reveal features linking plant and animal genes. Curr Genet 28: 333–345 [DOI] [PubMed] [Google Scholar]
- Federhen S. (2012) The NCBI taxonomy database. Nucleic Acids Res 40: D136–D143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch WM. (1970) Distinguishing homologous from analogous proteins. Syst Zool 19: 99–113 [PubMed] [Google Scholar]
- Fostier J, Proost S, Dhoedt B, Saeys Y, Demeester P, Van de Peer Y, Vandepoele K. (2011) A greedy, graph-based algorithm for the alignment of multiple homologous gene lists. Bioinformatics 27: 749–756 [DOI] [PubMed] [Google Scholar]
- Gabaldón T. (2008) Large-scale assignment of orthology: back to phylogenetics? Genome Biol 9: 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S. (2005) Genetic structure and diversity in Oryza sativa L. Genetics 169: 1631–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Hahn MW. (2007) Bias in phylogenetic tree reconciliation methods: implications for vertebrate genome evolution. Genome Biol 8: R141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu SH. (2008) Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol 148: 993–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, et al. (2011) The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 43: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. (2009) InterPro: the integrative protein signature database. Nucleic Acids Res 37: D211–D215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Pál C, Lercher MJ. (2004) The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet 5: 299–310 [DOI] [PubMed] [Google Scholar]
- Huynen MA, Bork P. (1998) Measuring genome evolution. Proc Natl Acad Sci USA 95: 5849–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Julien P, Kuhn M, von Mering C, Muller J, Doerks T, Bork P. (2008) eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res 36: D250–D254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey PJ, Lawson D, Birney E, Derwent PS, Haimel M, Herrero J, Keenan S, Kerhornou A, Koscielny G, Kähäri A, et al. (2010) Ensembl Genomes: extending Ensembl across the taxonomic space. Nucleic Acids Res 38: D563–D569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. (2005) Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet 39: 309–338 [DOI] [PubMed] [Google Scholar]
- Koonin EV. (2009) Evolution of genome architecture. Int J Biochem Cell Biol 41: 298–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzniar A, van Ham RC, Pongor S, Leunissen JA. (2008) The quest for orthologs: finding the corresponding gene across genomes. Trends Genet 24: 539–551 [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linard B, Thompson JD, Poch O, Lecompte O. (2011) OrthoInspector: comprehensive orthology analysis and visual exploration. BMC Bioinformatics 12: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons E, Pedersen B, Kane J, Alam M, Ming R, Tang H, Wang X, Bowers J, Paterson A, Lisch D, et al. (2008) Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol 148: 1772–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez DA, Nelson MA. (2010) The next generation becomes the now generation. PLoS Genet 6: e1000906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer IM, Durbin R. (2004) Gene structure conservation aids similarity based gene prediction. Nucleic Acids Res 32: 776–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak P. (2008) Coexpression, coregulation, and cofunctionality of neighboring genes in eukaryotic genomes. Genomics 91: 243–248 [DOI] [PubMed] [Google Scholar]
- Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, Saw JH, Senin P, Wang W, Ly BV, Lewis KL, et al. (2008) The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452: 991–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. (2010) Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci USA 107: 4623–4628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movahedi S, Van de Peer Y, Vandepoele K. (2011) Comparative network analysis reveals that tissue specificity and gene function are important factors influencing the mode of expression evolution in Arabidopsis and rice. Plant Physiol 156: 1316–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn A. (2010) Gene clusters for secondary metabolic pathways: an emerging theme in plant biology. Plant Physiol 154: 531–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn AE, Field B. (2009) Operons. Cell Mol Life Sci 66: 3755–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, et al. (2007) The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res 35: D883–D887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RD, Charleston MA. (1997) From gene to organismal phylogeny: reconciled trees and the gene tree/species tree problem. Mol Phylogenet Evol 7: 231–240 [DOI] [PubMed] [Google Scholar]
- Palenik B, Grimwood J, Aerts A, Rouzé P, Salamov A, Putnam N, Dupont C, Jorgensen R, Derelle E, Rombauts S, et al. (2007) The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc Natl Acad Sci USA 104: 7705–7710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Prochnik SE, Umen J, Nedelcu AM, Hallmann A, Miller SM, Nishii I, Ferris P, Kuo A, Mitros T, Fritz-Laylin LK, et al. (2010) Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329: 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proost S, Fostier J, De Witte D, Dhoedt B, Demeester P, Van de Peer Y, Vandepoele K. (2012) i-ADHoRe 3.0: fast and sensitive detection of genomic homology in extremely large data sets. Nucleic Acids Res (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proost S, Van Bel M, Sterck L, Billiau K, Van Parys T, Van de Peer Y, Vandepoele K. (2009) PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell 21: 3718–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryszcz LP, Huerta-Cepas J, Gabaldón T. (2011) MetaPhOrs: orthology and paralogy predictions from multiple phylogenetic evidence using a consistency-based confidence score. Nucleic Acids Res 39: e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Rouard M, Guignon V, Aluome C, Laporte MA, Droc G, Walde C, Zmasek CM, Périn C, Conte MG. (2011) GreenPhylDB v2.0: comparative and functional genomics in plants. Nucleic Acids Res 39: D1095–D1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, Sasamoto S, Watanabe A, Ono A, Kawashima K, et al. (2008) Genome structure of the legume, Lotus japonicus. DNA Res 15: 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Schuster SC. (2008) Next-generation sequencing transforms today’s biology. Nat Methods 5: 16–18 [DOI] [PubMed] [Google Scholar]
- Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, Jaiswal P, Mockaitis K, Liston A, Mane SP, et al. (2011) The genome of woodland strawberry (Fragaria vesca). Nat Genet 43: 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachana K, Larsson TA, Powell S, Chen WH, Doerks T, Muller J, Bork P. (2011) Orthology prediction methods: a quality assessment using curated protein families. Bioessays 33: 769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Fawcett JA, Proost S, Sterck L, Vandepoele K. (2009) The flowering world: a tale of duplications. Trends Plant Sci 14: 680–688 [DOI] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, et al. (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 42: 833–839 [DOI] [PubMed] [Google Scholar]
- Vernot B, Stolzer M, Goldman A, Durand D. (2008) Reconciliation with non-binary species trees. J Comput Biol 15: 981–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden AZ, Lee JH, Mock T, Rouzé P, Simmons MP, Aerts AL, Allen AE, Cuvelier ML, Derelle E, Everett MV, et al. (2009) Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324: 268–272 [DOI] [PubMed] [Google Scholar]
- Yi G, Sze SH, Thon MR. (2007) Identifying clusters of functionally related genes in genomes. Bioinformatics 23: 1053–1060 [DOI] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GE, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KF, Gouzy J, Schoof H, et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]
- Zmasek CM, Eddy SR. (2001) A simple algorithm to infer gene duplication and speciation events on a gene tree. Bioinformatics 17: 821–828 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.