Abstract
Fire blight, caused by the bacterium Erwinia amylovora, is a devastating disease of apple (Malus × domestica). The phytoalexins of apple are biphenyls and dibenzofurans, whose carbon skeleton is formed by biphenyl synthase (BIS), a type III polyketide synthase. In the recently published genome sequence of apple ‘Golden Delicious’, nine BIS genes and four BIS gene fragments were detected. The nine genes fall into four subfamilies, referred to as MdBIS1 to MdBIS4. In a phylogenetic tree, the BIS amino acid sequences from apple and Sorbus aucuparia formed an individual cluster within the clade of the functionally diverse type III polyketide synthases. cDNAs encoding MdBIS1 to MdBIS4 were cloned from fire-blight-infected shoots of apple ‘Holsteiner Cox,’ heterologously expressed in Escherichia coli, and functionally analyzed. Benzoyl-coenzyme A and salicoyl-coenzyme A were the preferred starter substrates. In response to inoculation with E. amylovora, the BIS3 gene was expressed in stems of cv Holsteiner Cox, with highest transcript levels in the transition zone between necrotic and healthy tissues. The transition zone was the accumulation site of biphenyl and dibenzofuran phytoalexins. Leaves contained transcripts for BIS2 but failed to form immunodetectable amounts of BIS protein. In cell cultures of apple ‘Cox Orange,’ expression of the BIS1 to BIS3 genes was observed after the addition of an autoclaved E. amylovora suspension. Using immunofluorescence localization under a confocal laser-scanning microscope, the BIS3 protein in the transition zone of stems was detected in the parenchyma of the bark. Dot-shaped immunofluorescence was confined to the junctions between neighboring cortical parenchyma cells.
The domesticated apple (Malus × domestica Borkh.; Rosaceae) is the main fruit crop in the temperate regions of the world (Luby, 2003; Velasco et al., 2010). Approximately 72 million tons of apples were grown worldwide in 2009 (FAO, 2009). Apples are consumed for their taste, nutritional value, and high content of health-protective constituents, such as polyphenols. The major polyphenol classes are flavanols (catechins and proanthocyanidins), flavonols, dihydrochalcones, hydroxycinnamates, and in red apples anthocyanins (Mayr et al., 1995; Lee et al., 2003; Tsao et al., 2003; Wolfe et al., 2003; Chinnici et al., 2004; Vrhovsek et al., 2004; Mc Ghie et al., 2005). The concentrations of the majority of these constituents are much higher in the apple peel than in the flesh. Compared to other fruits, apples contain highest levels of free phenolic compounds, which may be more available for absorption into the bloodstream (Boyer and Liu, 2004; Manach et al., 2005). Beside phenolics, a considerable number of bioactive triterpenoids are present in apple peels (He and Liu, 2007). Numerous epidemiological studies have linked the consumption of apples with the reduced risk of cancer and cardiovascular disease, which are ranked as the top two leading causes of death in industrialized countries (Boyer and Liu, 2004; Gerhauser, 2008). Apples and apple products exhibit a wide range of biological activities, such as antioxidant effects, cancer chemopreventive potential, and cholesterol-lowering properties (Boyer and Liu, 2004; Gerhauser, 2008).
The progenitor of the cultivated apple is likely to be Malus sieversii, which is widespread in the mountains of Central Asia at elevations between 1,200 and 1,800 m (Luby, 2003; Velasco et al., 2010). Over the centuries, a number of apple cultivars were selected by vegetative propagation of single trees, which coincidentally had more appealing fruit and growth characteristics than other locally known varieties (Gessler and Patocchi, 2007). Few cultivars are the result of an oriented breeding program. Commercial orchards are uniform and commonly consist of only few worldwide distributed cultivars. Although agronomically desirable, the genetic uniformity seriously affects resistance to pests and diseases (Norelli et al., 2003; Gessler and Patocchi, 2007). The most frequent apple pathogens are the fungi Venturia inaequalis, the causal agent of scab, and Podosphaera leucotricha, the causal agent of powdery mildew (Xu and Madden, 2002; Bowen et al., 2011). Among bacteria, the most devastating pathogen is the enterobacterium Erwinia amylovora, which causes fire blight (Thomson, 2000). All three diseases have global economic importance for apple production and trade.
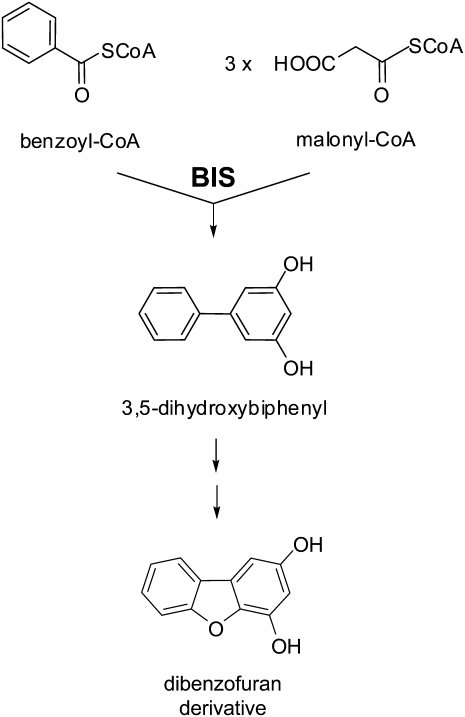
One of the defense strategies of plants against pathogens is formation of phytoalexins. Species of the Rosaceous subtribe Pyrinae (formerly subfamily Maloideae), which includes apple and pear (Pyrus communis), form biphenyls and dibenzofurans as inducible defense compounds (Kokubun and Harborne, 1995). Accumulation of these antimicrobial metabolites as de-novo-formed phytoalexins is confined to the Pyrinae. The biphenyl and dibenzofuran concentrations supposed to be present at localized infection sites inhibit spore germination and hyphal growth (Miyakodo et al., 1985; Hrazdina et al., 1997). Biosynthesis of biphenyls and dibenzofurans is not well understood. Recently, we used cell cultures of Sorbus aucuparia, which is another Pyrinae species, as an in vitro system for studying metabolism of the taxon-specific defense compounds (Hüttner et al., 2010). Three biphenyls and two dibenzofurans accumulated in cultured S. aucuparia cells in response to elicitor treatment. Interestingly, addition of yeast (Saccharomyces cerevisiae) extract and chitosan to the cell cultures resulted in formation of the biphenyls aucuparin and noraucuparin, respectively, as the major phytoalexins. In contrast, treatment of the cell cultures with preparations from either the fire blight bacterium, E. amylovora, or the scab-causing fungus, V. inaequalis, induced accumulation of the dibenzofuran eriobofuran as the major defense compound (Hüttner et al., 2010). The carbon skeleton of the two classes of phytoalexins is formed by biphenyl synthase (BIS), whose activity was first detected in yeast-extract-treated S. aucuparia cell cultures (Liu et al., 2004). A BIS cDNA was cloned and the recombinant enzyme was functionally expressed in Escherichia coli (Liu et al., 2007). BIS transcripts were rapidly and transiently induced by elicitor treatment in S. aucuparia cell cultures. BIS is a type III polyketide synthase (PKS) that catalyzes the iterative condensation of benzoyl-CoA with three molecules of malonyl-CoA to give a linear tetraketide intermediate, which then undergoes intramolecular aldol condensation and loss of the terminal carboxyl group to give 3,5-dihydroxybiphenyl (Fig. 1). Downstream enzymes that metabolize the BIS product have not yet been reported. Thus, the phytoalexin response of the Pyrinae is poorly studied, despite the high economic value of the fruit crops involved.
Figure 1.
Biosynthesis of the biphenyl and dibenzofuran scaffolds.
Very recently, Velasco et al. (2010) have published the genome sequence of the diploid apple variety Golden Delicious. This cultivar was selected for the sequencing project because it is extensively used in fruit production and as parent in apple breeding programs worldwide. The genome had 57,386 predicted genes, the highest number among plant genomes so far sequenced. Analysis of the apple genome was suggestive of a whole-genome duplication event that happened around 50 million years ago in an ancestral nine-chromosome genome. Subsequent loss of a single chromosome, in addition to interchromosomal rearrangements, led to the 17-chromosome karyotype of the cultivated apple (Velasco et al., 2010). The characteristic fruit type has probably evolved after the genome-wide duplication event. Here we report analysis of the apple BIS gene family and differential expression of gene family members in apple ‘Holsteiner Cox’ after inoculation with the fire blight bacterium, E. amylovora.
RESULTS
Analysis of the BIS Gene Family in the Genome Sequence of Apple 'Golden Delicious'
The genome sequence of cv Golden Delicious (Velasco et al., 2010) was screened for BIS genes using three BIS cDNAs isolated from elicitor-treated S. aucuparia cell cultures (Liu et al., 2010). These cDNAs and their encoded polypeptides shared 92% to 94% and 92% to 96% identities, respectively. The genome sequence of cv Golden Delicious contained nine predicted genes, whose coding sequences shared >90% nucleotide sequence identity with the three S. aucuparia BIS cDNAs (Table I). Complete coding sequences were present in only five (MDP0000641538, MDP0000208999, MDP0000432621, MDP0000287919, MDP0000302905) of the nine gene sequences (Supplemental Fig. S1). Furthermore, four fragments of BIS genes sharing 80% to 97% identity with the S. aucuparia cDNAs were detected (MDP0000573540, MDP0000280274, MDP0000529287, MDP0000308113). However, the upstream and downstream sequences of all four fragments were unrelated to BIS genes. Some sequences with identities <70% were also found, they are annotated as CHALCONE SYNTHASE (CHS) genes in the database. Based on the alignment of the coding sequences (Supplemental Fig. S1) and the promoter sequences, as far as available (data not shown), the nine BIS genes fall into four subfamilies referred to as MdBIS1 to MdBIS4 (Table I). This grouping was confirmed by reconstruction of the phylogeny of the gene family (Supplemental Fig. S2). MdBIS1 (three copies), MdBIS2 (four copies), and MdBIS3 (one copy) correspond to the BIS1, BIS2, and BIS3 cDNAs, respectively, from S. aucuparia. The coding sequence of MdBIS4 (MDP0000302905) has a 12-bp-longer 3′ end than the other open reading frames (Supplemental Fig. S1). A corresponding cDNA from S. aucuparia has not yet been isolated.
Table I. The BIS gene family of M. domestica.
| BIS Gene in the Genome Sequence of cv Golden Delicious | Chromosome No. | Identity with the Corresponding BIS cDNA from cv Holsteiner Cox |
Subfamily | |
| At the Nucleotide Sequence Level | At the Amino Acid Sequence Level | |||
| MDP0000641583a | 14 | 99% (1,159/1,173) b | 97% (381/391) | BIS1 |
| MDP0000257119ac | 2 | 97% (971/1,004) | 95% (316/334) | |
| MDP0000385168 | Unanchored | 94% (665/710) | 90% (212/236) | |
| MDP0000208899 | 2 | 97% (1,137/1,173) | 98% (383/389) | BIS2 |
| MDP0000432621 | 15 | 96% (1,120/1,167) | 98% (380/389) | |
| MDP0000716308c | 15 | 96% (661/689) | 98% (212/216) | |
| MDP0000168735a | 2 | 96% (692/718) | 97% (238/245) | |
| MDP0000287919 | 15 | 99% (1,162/1,167) | 97% (377/388) | BIS3 |
| MDP0000302905 | 14 | 99% (1,183/1,185) | 99% (393/394) | BIS4 |
Predicted splicing (www.rosaceae.org) is modified (Supplemental Figs. S2– S4).
Identical residues versus open reading frame residues available in the genome sequence.
Promoter region is missing in the genome sequence.
Some of the splicing predictions made for the BIS genes present in the cv Golden Delicious genome sequence (www.rosaceae.org) differ from our observations. (1) In MDP0000641538, two introns comprising the nucleotides 170 to 661 and 947 to 975 were annotated, however, the second intron is coding sequence when compared to the corresponding BIS1 cDNA from apple ‘Holsteiner Cox,’ cloning of which is described below (Supplemental Fig. S3). The same is true for the 3′ segment (580–661) of the first intron. In addition, a deletion is present in the genome sequence behind nucleotide 985. The corrected 1,173-bp coding sequence of MDP0000641538 was aligned with the MdBIS1 cDNA (Table I). (2) The predicted MDP0000257119 coding sequence is in accord with the corresponding MdBIS1 cDNA over a length of 711 bp which, however, constitute only the 3′ segment of the coding sequence (Supplemental Fig. S4). The 293-bp upstream sequence was annotated in the genome sequence as 3′ segment of an intron. Furthermore, the 169-bp 5′ stretch of the cloned cDNA lacks a counterpart in the genome sequence. Finally, a deletion is present behind nucleotide 531 in the coding part of the genome sequence. (3) In MDP0000168735, 786 bp are marked as unclear nucleotides, which are annotated together with another 43 nucleotides (577–620) as an intron (Supplemental Fig. S5). However, the open reading frame of the corresponding cloned MdBIS2 cDNA contains both the 43-nucleotide segment and a stretch of 407 unclear nucleotides as coding sequence.
Analysis of Phylogenetic Relationships between MdBISs and Other Type III PKSs
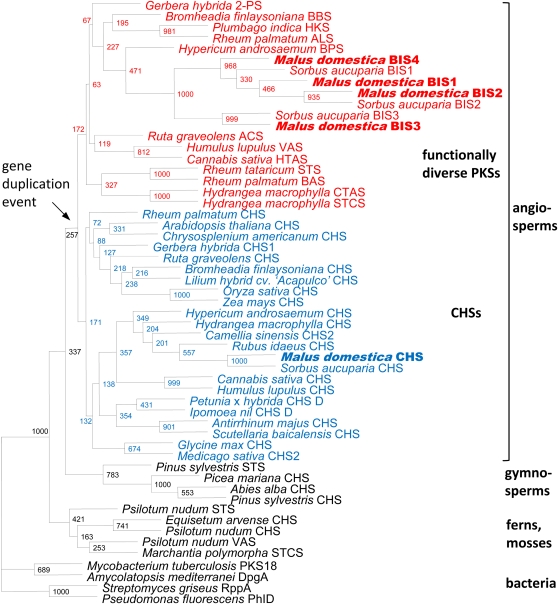
A phylogenetic tree involving MdBIS1 to MdBIS4 and SaBIS1 to SaBIS3 was constructed, as described previously (Beerhues and Liu, 2009). The seven BIS amino acid sequences formed an individual cluster within the clade of the functionally diverse type III PKSs (Fig. 2). BIS1 to BIS3 from apple and S. aucuparia grouped together each. BIS4 has so far only been detected in apple. The amino acid sequence most closely related to the BISs is benzophenone synthase (BPS) from Hypericum androsaemum. This enzyme also uses benzoyl-CoA as a starter substrate (Liu et al., 2003). CHS from apple grouped closely together with CHS from S. aucuparia within the clade of CHSs.
Figure 2.
Neighbor-joining tree of type III PKSs. CHSs and functionally divergent PKSs from angiosperms are highlighted in blue and red, respectively. Bold letters mark BIS1 to BIS4 and CHS from cv Holsteiner Cox. Numbers at the forks are bootstrap values from 1,000 replicates. ACS, Acridone synthase; ALS, aloesone synthase; BAS, benzalacetone synthase; BBS, bibenzyl synthase; CTAS, 4-coumaroyl triacetic acid lactone synthase; DpgA, 3,5-dihydroxyphenylacetate synthase; HKS, hexaketide synthase; HTAS, hexanoyl triacetic acid lactone synthase; PhID, acetylphloroglucinol synthase; 2-PS, 2-pyrone synthase; RppA, 1,3,6,8-tetrahydroxynaphthalene synthase; STS, stilbene synthase; STCS, stilbene carboxylate synthase; VAS, valerophenone synthase.
cDNA Cloning and Functional Analysis of BISs from Fire-Blight-Infected Apple ‘Holsteiner Cox’
Gene-specific primers were derived from the 5′ and 3′ ends of the coding sequences of the BIS1 to BIS4 genes of cv Golden Delicious and used to amplify the open reading frames of the BIS cDNAs from E. amylovora-inoculated shoots of cv Holsteiner Cox. The latter cultivar is used in our laboratories to study the phytoalexin response of apple. The identities of the BIS coding sequences of the two cultivars were 96% to 99% (Table I). The BIS1 cDNA cloned from cv Holsteiner Cox contained an early stop codon at position 211 to 213 and did not yield a functional enzyme protein. In contrast, the BIS2 to BIS4 cDNAs were heterologously expressed in E. coli to give active N-terminally His6-tagged proteins, which were purified by affinity chromatography. The subunit molecular masses of the BISs were approximately 43 kD, as determined by SDS-PAGE (data not shown). In the presence of malonyl-CoA as extender substrate, BIS1 to BIS4 converted benzoyl-CoA, 2-hydroxybenzoyl-CoA (salicoyl-CoA), and 3-hydroxybenzoyl-CoA to 3,5-dihydroxybiphenyl, 4-hydroxycoumarin, and 6-(3′-hydroxyphenyl)-4-hydroxy-2-pyrone, respectively, as observed previously with the S. aucuparia BISs (Liu et al., 2007, 2010). Due to the relevance for biphenyl and dibenzofuran biosynthesis (Fig. 1), kinetic properties were studied with benzoyl-CoA and salicoyl-CoA as starter substrates (Table II). BIS2 to BIS4 exhibited highest affinity for benzoyl-CoA. The turnover rates were slightly higher with salicoyl-CoA. Notably, salicoyl-CoA undergoes only one condensation with malonyl-CoA, however, benzoyl-CoA experiences three. Compared among BIS2 to BIS4, the catalytic efficiencies with benzoyl-CoA and salicoyl-CoA vary by factor 1.6 and 2.2, respectively. The pH and temperature optima of BIS2 to BIS4 were 7.0 to 7.5 and 35°C to 40°C, respectively. Increases in product formation were linear with time up to 20 min and protein concentration up to 10 μg in the standard assay. Benzoyl-CoA concentrations above 15 μm led to increasing inhibition of BIS2 and BIS4 activities, which was not observed with salicoyl-CoA up to 50 μm.
Table II. Steady-state kinetic parameters of BIS2 to BIS4 from cv Holsteiner Cox.
| Isoenzyme | Benzoyl-CoA |
Salicoyl-CoA |
Malonyl-CoA: Km | ||||
| kcat | Km | kcat/Km | kcat | Km | kcat/Km | ||
| min−1 | μm | m−1s−1 | min−1 | μm | m−1s−1 | μm | |
| BIS1 | Not functionala | ||||||
| BIS2 | 0.87 | 3.0 | 4,844 | 1.10 | 4.7 | 4,025 | 14.2 |
| BIS3 | 0.60 | 2.2 | 4,698 | 0.73 | 5.5 | 2,193 | 9.8 |
| BIS4 | 0.50 | 1.2 | 7,288 | 0.70 | 6.3 | 1,856 | 10.0 |
Due to an early stop codon.
Morphological Stages of Fire Blight Spread on Shoots of cv Holsteiner Cox
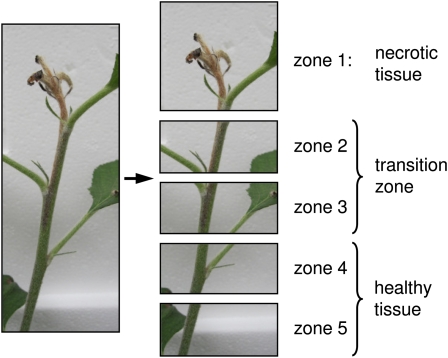
Greenhouse-grown shoots of cv Holsteiner Cox grafted onto M9 rootstocks were inoculated with E. amylovora by cutting the top two leaves with scissors dipped into a bacterial suspension (109 colony-forming units [cfu]/mL). After 4 d, the shoot tip started to turn necrotic. After 6 d, the two youngest leaves that had been cut and the top 2 cm of the stem had died (Fig. 3; zone 1). The next 3-cm segment (zones 2 and 3) looked brownish at the outside but was still green inside. This stem segment was referred to as transition zone because it is the interface between necrotic and healthy tissues (zones 4 and 5). Nine days after inoculation, the transition zone had extended downward by 1 cm and the third leaf from the top showed typical wilting symptoms. In this way, the transition zone gradually advanced down the stem with increasing postinoculation time. For analysis, stems were dissected into the five zones, with the length of the top segment being 2 and 3 cm (6 and 9 d postinoculation, respectively) and that of the following segments 1.5 cm each.
Figure 3.
Dissection of an E. amylovora-inoculated shoot of cv Holsteiner Cox (6 d postinoculation) into five zones for separate analysis.
Differential Expression of BIS Genes in Fire-Blight-Infected Shoots of cv Holsteiner Cox
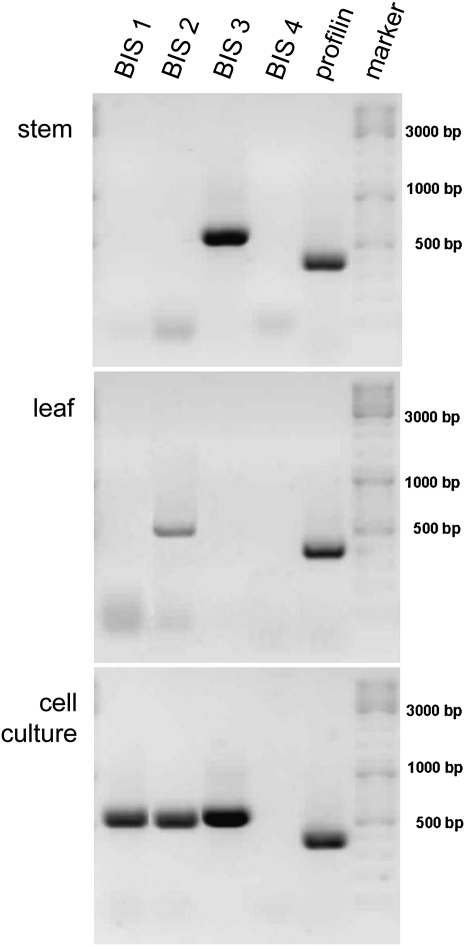
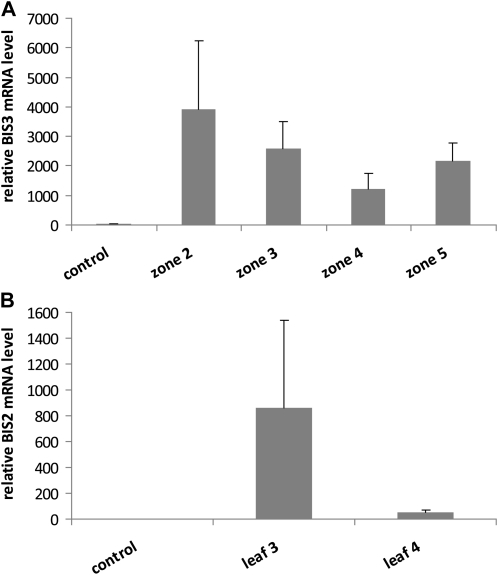
Both E. amylovora-inoculated greenhouse-grown shoots and mock-inoculated control shoots were harvested at various times after inoculation and dissected into five zones (Fig. 3). These samples were studied for expression of the MdBIS1 to MdBIS4 genes using a combination of semiquantitative reverse transcription (RT)-PCR and quantitative real-time PCR. The primer pairs used, which were demonstrated to be gene specific, led to amplification of 560-, 546-, 565-, and 1,185-bp fragments of the BIS1, BIS2, BIS3, and BIS4 coding sequences, respectively. High levels of expression in both stems and leaves were detected 6 d after inoculation of shoots with E. amylovora (Fig. 4). Stems contained only BIS3 transcripts at levels detectable by RT-PCR. Subsequent real-time PCR studies (Fig. 5A) demonstrated that BIS3 expression was highest in the transition zone (zones 2 and 3) and somewhat lower in the yet healthy segment (zones 4 and 5). In zone 2, the BIS3 transcript level was approximately 4,000 times that in the respective stem segment of mock-inoculated control shoots. Extraction of mRNA from necrotic tissue (zone 1) was not feasible.
Figure 4.
RT-PCR amplification of BIS1 to BIS4 transcripts in stems (transition zone) and leaves (first living leaf from the top) of cv Holsteiner Cox 6 d after E. amylovora inoculation, and in cell cultures of cv Cox Orange 9 h after treatment with an autoclaved suspension of E. amylovora. Profilin transcript levels served as control for use of equal RNA template amounts.
Figure 5.
Determination by real-time PCR of relative BIS3 (A) and BIS2 (B) transcript levels in stem segments and leaves, respectively, of cv Holsteiner Cox 6 d after E. amylovora inoculation. mRNA isolation from zone 1 and leaves 1 and 2, which were necrotic, was not feasible. Control mRNA was obtained from stems and leaves of mock-inoculated shoots. sds are indicated (n = 3).
Contrary to stems, leaves of E. amylovora-inoculated shoots expressed the BIS2 gene (Fig. 4). The transcript level in the third leaf from the top was approximately 800 times that in the respective leaf of mock-inoculated control shoots (Fig. 5B). The two youngest leaves were necrotic and the fourth leaf from the top contained only low BIS2 transcript amounts. Mock-inoculated shoots also served as wounding control because their top two leaves were cut with scissors dipped in sterile medium rather than bacterial suspension. By RT-PCR, transcripts for none of the four BISs were detectable, neither in stems nor in leaves. Amplification of profilin transcripts served as control for equal RNA template amounts.
Treatment of cell cultures of apple ‘Cox Orange’ with an autoclaved suspension of E. amylovora resulted in expression of the BIS1, BIS2, and BIS3 genes (Fig. 4). High transcript levels were found 9 h after the addition of the elicitor. No detectable expression of the BIS genes was found in untreated control cell cultures, in contrast to expression of the profilin gene.
Accumulation of Phytoalexins in Fire-Blight-Infected Shoots of cv Holsteiner Cox
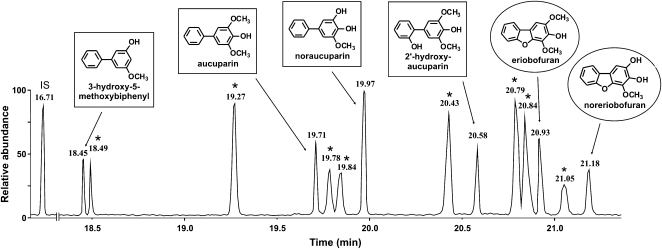
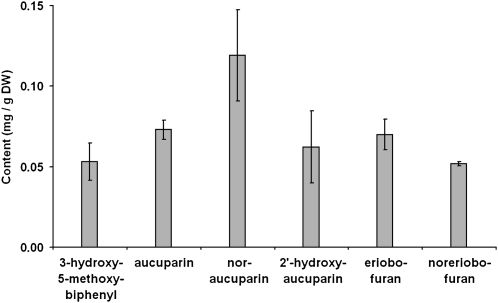
E. amylovora-inoculated shoots were dissected into zones (Fig. 3), which were extracted with methanol (80%). Phytoalexins were identified using a gas chromatography-mass spectrometry (GC-MS) library comprising the previously structure-elucidated compounds from elicitor-treated S. aucuparia cell cultures (Hüttner et al., 2010). Biphenyls and dibenzofurans were detected in the transition zone (Fig. 6). The flanking stem segments above and below this zone lacked detectable amounts of phytoalexins. The extracts from the transition zone contained four biphenyls (3-hydroxy-5-methoxybiphenyl, aucuparin, noraucuparin, 2′-hydroxyaucuparin) and two dibenzofurans (eriobofuran, noreriobofuran). The total phytoalexin content was 0.43 ± 0.08 mg/g dry weight. The concentrations of the individual biphenyl and dibenzofuran phytoalexins are documented in Figure 7. In mock-inoculated shoots, the stem segment corresponding to the transition zone lacked biphenyls and dibenzofurans. Methanolic extracts from leaves of both fire-blight-infected and mock-inoculated shoots were also devoid of phytoalexins.
Figure 6.
GC-MS analysis of biphenyls and dibenzofurans present in the transition zone of fire-blight-infected stems of cv Holsteiner Cox. The phytoalexins were separated as trimethylsilyl derivatives. Stars indicate constitutive compounds also detected in mock-inoculated shoots. IS, Internal standard (4-phenylphenol).
Figure 7.
Concentrations of biphenyls and dibenzofurans in the transition zone of fire-blight-infected stems of cv Holsteiner Cox. Data are mean values ± sd of four independent experiments.
Immunohistochemical Studies
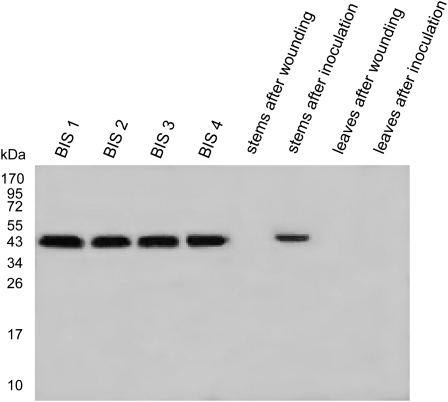
A polyclonal antiserum was raised in rabbits against BIS1 from S. aucuparia. The IgG fractions were isolated from the antiserum and the preimmune serum by affinity chromatography. On a protein blot following SDS-PAGE, the antibodies recognized all four BISs from cv Holsteiner Cox to similar extents (Fig. 8). Preimmune IgG failed to detect any BIS isoenzyme. In protein extracts from stems of cv Holsteiner Cox collected 6 d after E. amylovora inoculation, the presence of BIS protein was readily detectable. A single protein band of approximately 43 kD was immunostained, indicating the exclusive specificity of the antibodies for BIS (Fig. 8). Immunodetection was strongest with protein extracts from zone 2 of the transition zone. An intense immunoreaction was also found 9 d after inoculation, whereas no protein band was stained 2 d postinoculation. Wounding without E. amylovora inoculation failed to induce formation of detectable BIS protein amounts, which was also true for leaves (Fig. 8). However, leaves from E. amylovora-inoculated shoots also lacked immunostainable amounts of BIS protein. This finding was obtained with both leaves harvested at various times after inoculation (2, 4, 6, 9, 12, and 14 d) and leaves present at different developmental stages (first four leaves from the top).
Figure 8.
Immunoblotting after SDS-PAGE of BIS1 to BIS4 (100 ng) and protein extracts (50 μg) from stems and leaves of cv Holsteiner Cox 6 d after E. amylovora inoculation and wounding only.
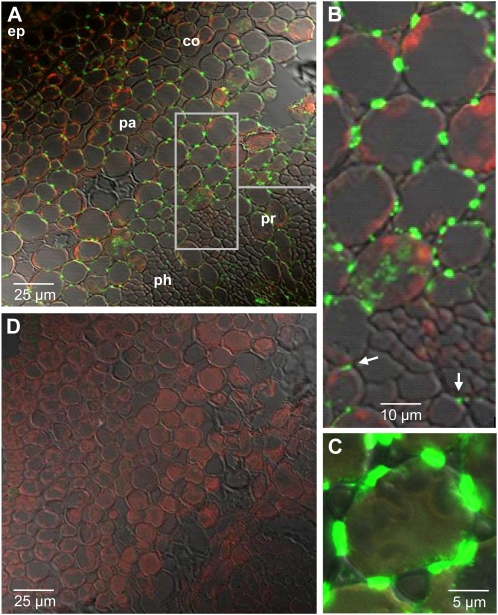
For immunofluorescence localization of BIS protein under a confocal laser-scanning microscope, stems were harvested 6 d after E. amylovora inoculation and cross sections of zone 2 were incubated with the anti-BIS IgG fraction. Bright immunofluorescence was observed in the bark (Fig. 9A). Interestingly, immunofluorescence was dot shaped and restricted to the junctions between neighboring cortical parenchyma cells (Fig. 9, B and C). Fluorescent dots were also observed between ray parenchyma cells and rarely between phloem parenchyma cells (arrows in Fig. 9B). Immunostaining was absent from sections incubated with preimmune IgG (Fig. 9D) or phosphate-buffered saline (PBS) instead of primary antibodies.
Figure 9.
Immunofluorescence localization of BIS protein in cross sections of stems of cv Holsteiner Cox 6 d after E. amylovora inoculation. A and D, Cross sections of the transition zone (zone 2) after incubation with anti-BIS IgG (A) and preimmune IgG (D). B, Detail from A displaying cortex parenchyma and phloem. The arrows indicate fluorescent dots between phloem ray cells and phloem parenchyma cells. C, Detail from A displaying a single cortical parenchyma cell with dot-shaped immunofluorescence at the junctions with the neighboring cells. ep, Epidermis; pa, parenchyma; co, collenchyma; ph, phloem; pr, phloem ray.
DISCUSSION
Apple BIS is encoded by a gene family consisting of four subfamilies, based on the alignment of the promoter and coding sequences from cv Golden Delicious and the analysis of the phylogenetic relationships of nine coding sequences. The ancestral apple genome comprising nine chromosomes underwent a whole-genome duplication event, followed by loss of one chromosome and interchromosomal rearrangements (Velasco et al., 2010). The BIS2 subfamily consists of four gene copies, two of which each are located on the chromosomes 2 and 15 (Table I). Consistently, these two chromosomes contain copies of the same ancient chromosome, IX (Velasco et al., 2010), which obviously contained two BIS genes resulting from a tandem duplication event. The BIS3 subfamily contains a single-copy gene on chromosome 15, the counterpart of which after genome duplication may be one of the incomplete BIS pseudogenes on chromosome 8 (MDP0000280274, MDP0000529287). A possible counterpart of the single-copy gene of the BIS4 subfamily on chromosome 14 has not yet been detected. The BIS1 subfamily comprises two gene copies on chromosomes 2 and 14, which do not stem from a common ancestral chromosome. One of the duplicated counterparts may be the third unanchored gene copy (Table I).
The high degrees of nucleotide sequence identity between the members of the BIS1 and BIS2 subfamilies led us to select one representative gene each, which, together with the single-copy genes of the BIS3 and BIS4 subfamilies, were referred to as MdBIS1 to MdBIS4. In a phylogenetic tree of type III PKSs, the products of the MdBIS1 to MdBIS4 genes of cv Holsteiner Cox and the SaBIS1 to SaBIS3 genes of S. aucuparia (Liu et al., 2010) grouped together within a cluster that comprised the functionally diverse type III PKSs. As proposed previously (Beerhues and Liu, 2009), an ancient duplication of a common ancestral gene before the speciation of the angiosperms may have caused formation of two clusters, one of which contains the orthologs of CHS, which is the prototype enzyme of the type III PKSs (Schröder, 1999), whereas the second one comprises PKSs that functionally diverge from CHS. The clade of the BIS proteins closely grouped together with BPS, which exhibits similar starter substrate but different product specificities (Liu et al., 2003). The two enzymes form the same tetraketide intermediate that then is converted by BPS to 2,4,6-trihydroxybenzophenone, the precursor of benzophenone and xanthone metabolism. In the Pyrinae, no detection of benzophenone and xanthone derivatives and BPS activity has so far been reported.
The MdBIS1 to MdBIS4 isoenzymes from cv Holsteiner Cox did not appreciably differ in their kinetic properties. Highest affinities (Km) were observed for the starter substrate benzoyl-CoA, which undergoes conversion to 3,5-dihydroxybiphenyl in the presence of malonyl-CoA as extender substrate (Fig. 1). The immediate BIS product, 3,5-dihydroxybiphenyl, is metabolized by downstream enzymes to give aucuparin and noraucuparin, which are the major phytoalexins in S. aucuparia cell cultures after treatment with yeast extract and chitosan, respectively (Hüttner et al., 2010). In contrast, addition of preparations from E. amylovora and V. inaequalis resulted in accumulation of dibenzofurans as the major phytoalexins. Dibenzofurans have been proposed to arise by intramolecular cyclization of a 2′-hydroxylated biphenyl, which in turn may originate from salicoyl-CoA as starter substrate (Hüttner et al., 2010). Consistently, all four MdBIS isoenzymes had slightly higher turnover rates (kcat) with salicoyl-CoA. However, as observed previously with the three SaBIS isoenzymes (Liu et al., 2010), salicoyl-CoA was converted to 4-hydroxycoumarin after a single decarboxylative condensation with malonyl-CoA rather than 2′,3,5-trihydroxybiphenyl after three additions of acetyl units. It will be interesting to study if in E. amylovora-inoculated shoots of cv Holsteiner Cox the starter substrate salicoyl-CoA is available and, if so, what enzymatic product is formed thereof.
Expression of the MdBIS1 to MdBIS4 genes in cv Holsteiner Cox was differentially regulated in response to E. amylovora inoculation. The BIS3 gene was expressed in stems, leading to accumulation of four biphenyls and two dibenzofurans in the transition zone. The BIS2 gene was transcribed in leaves which, however, failed to accumulate immunodetectable amounts of BIS protein, irrespective of the developmental stage studied and the harvest time after inoculation, suggesting that the BIS2 transcripts detected in leaves were not translated. Members of the MdBIS gene family thus appear to be regulated at both the transcriptional and translational levels. The absence of BIS protein from leaves was in accord with the failure to detect biphenyls and dibenzofurans. Consistently, phytoalexins were previously only detected in stems of diseased trees of various Malus species (M. sieversii and Malus sylvestris; Kemp et al., 1985; Kokubun and Harborne, 1995). This finding was also true for the majority of the Pyrinae species examined for phytoalexin accumulation. Leaves of diseased plants commonly lacked phytoalexins, except for leaves of a few species (S. aucuparia, Photinia glabra, Rhaphiolepis umbellata, Eriobotrya japonica; Miyakodo et al., 1985; Watanabe et al., 1990; Widyastuti et al., 1992; Kokubun and Harborne, 1994; Jiang and Xuan, 2006). In cell cultures of apple ‘Cox Orange,’ treatment with an autoclaved E. amylovora suspension induced expression of the BIS1 to BIS3 genes. In contrast, cell cultures of S. aucuparia responded to elicitor treatment with the expression of the BIS1 and BIS3 genes only (Liu et al., 2010).
Biphenyls and dibenzofurans are the inducible defense compounds (phytoalexins) of the Pyrinae (Kokubun and Harborne, 1995). Other classes of polyphenols function as preformed defense compounds (phytoanticipins). A well-known major constituent of apple is phloridzin, a dihydrochalcone glucoside. However, the contribution of phloridzin and its aglucone, phloretin, to pathogen resistance is still under debate (Gosch et al., 2010). Phloretin is structurally and biochemically closely related to chalcones that are formed by CHS and serve as immediate precursors for flavonoids. The involvement of this class of secondary compounds in disease resistance of apple is supported by several lines of evidence (Treutter, 2005, 2006 and refs. therein). Enhanced biosynthesis, especially of 3-deoxycatechins after transient inhibition of flavanone 3-hydroxylase, correlated with improved resistance to pathogens, whereas decreased flavonoid levels after competitive inhibition of Phe ammonia-lyase, which initiates phenylpropanoid metabolism (Hanson and Havir, 1981), enhanced the susceptibility to pathogens. Recently, constitutive expression in cv Holsteiner Cox of the maize (Zea mays) regulatory leaf color (Lc) gene, which encodes a transcription factor, resulted in both increased transcript levels of flavonoid biosynthetic enzymes and increased concentrations of specific flavonoid classes, such as anthocyanins, flavan-3-ols, and proanthocyanidins (Li et al., 2007). These Lc-transgenic apple plants exhibited improved resistance to fire blight and scab (Flachowsky et al., 2010).
Immunofluorescence localization of BIS3 in the transition zone of fire-blight-infected stems of cv Holsteiner Cox revealed the presence of the enzyme protein in the parenchyma of the bark. Consistently, migration of E. amylovora was previously observed in the intercellular spaces of the cortical parenchyma, however, the pathogen can also invade and multiply in mature xylem vessels for long distances down the tree (Billing, 2011 and refs. therein). With young shoots on potted plants, the site of inoculation in relation to the most recently unfolded leaf may be critical in determining whether cortical parenchyma or xylem vessels or both are invaded. Inoculation above the most recently unfolded leaf, as carried out in this study, is likely to favor invasion of the cortical parenchyma, while inoculation at this leaf or older levels is likely to favor invasion of xylem vessels (Billing, 2011 and refs. therein). The latter finding agrees with previous observations that biphenyls and dibenzofurans in field trees, whose mature stem tissue was either naturally infected or artificially inoculated, commonly accumulated in the sapwood, which is that part of the secondary xylem that contains living parenchyma cells (Kokubun and Harborne, 1995).
At the subcellular level, the BIS3 protein was detected at the junctions between cortical parenchyma cells. It is tempting to speculate that the enzyme is associated with plasmodesmata in primary pit fields. Plasmodesmata serve symplastic communication between neighboring cells and provide the potential for the translocation of informational molecules, thereby playing a key role in the coordination of growth and development (Maule, 2008; Lucas et al., 2009). However, they can also be exploited by microbial pathogens as gateways to spread infection from cell to cell (Lee and Lu, 2011). Fire blight is caused by the bacterial pathogen, E. amylovora. Unlike viruses and fungi, bacterial pathogens do not need to cross cell wall boundaries, they mostly limit their habitat to intercellular spaces between cell walls (Lee and Lu, 2011). One of the early defense responses induced by bacterial infection is pathogen-associated molecular pattern-triggered immunity. This cell-wall-based host defense can be suppressed by pathogens that inject effector proteins directly into the plant cells. E. amylovora has the type III secretion system, which delivers effector proteins to the plant cytoplasm and helper proteins to the apoplast (Oh and Beer, 2005). Twelve type III secretion system secreted proteins have so far been identified (Nissinen et al., 2007). In return, plants can activate effector-triggered immunity, a much stronger form of defense, upon recognition of one or more bacterial effectors (Lee and Lu, 2011). Closure of the cellular borders would be vital to block diffusion of any toxic molecules, produced by either the infected cells or the microbe, into the healthy neighboring cells. How BIS3 and biphenyl and dibenzofuran phytoalexins may be involved in these processes remains to be studied.
CONCLUSION
Although fire blight can lead to devastating economic losses in apple production, formation of phytoalexins in this fruit tree is poorly investigated. Biosynthesis of biphenyl and dibenzofuran phytoalexins is confined to the Pyrinae including apple, with BIS being the key enzyme. The recently published genome sequence of apple ‘Golden Delicious’ provides insight into the complexity of the BIS gene family. Members of this gene family are differentially expressed in response to fire blight infection and their expression is regulated at the transcriptional and translational levels. However, much effort has still to be directed at elucidating the detailed control of this large gene family. Targeting of the BIS3 protein to the junctions between neighboring cortical parenchyma cells raises interesting questions as to the precise function of this protein and its immediate and distant products in disease resistance. BIS genes may provide a new group of candidates for designing disease control strategies and improving the natural resistance of economically valuable apple cultivars to E. amylovora and other pathogens.
MATERIALS AND METHODS
Plant Material and Inoculation with Erwinia amylovora
Approximately 40-cm-high shoots of apple (Malus × domestica) ‘Holsteiner Cox’ grafted onto M9 rootstocks were grown in a greenhouse. The E. amylovora strain Ea222(pfdC1Z′-gfp) (Flachowsky et al., 2008) was grown for 48 h on glycerine bouillon agar (Garrett and Crosse, 1963), and the suspension was adjusted to 109 cfu/mL. For inoculation, scissors were dipped into the suspension and used to cut the top two leaves of actively growing shoots of cv Holsteiner Cox. For mock inoculation, the bacterial suspension was replaced with sterile medium. At different times after inoculation, the first four leaves from the top were collected and either the top 8 (6 d postinoculation) or the top 9 cm of the stem (9 d postinoculation) were dissected into 1.5-cm segments, except for the 2- and 3-cm-long top segments after 6 and 9 d, respectively. The samples were frozen in liquid nitrogen and stored at −80°C.
Callus of apple ‘Cox Orange’ was purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen. Cell suspension cultures were initiated in liquid Linsmaier-Skoog medium (Linsmaier and Skoog, 1965) at 25°C in the dark on an orbital shaker at 100 rpm. Cultured cells were transferred into 50 mL fresh medium every 10 d. Five-day-old cell cultures were treated with 2 mL of a suspension of Ea222(pfdC1Z′-gfp) (109 cfu/mL). At various times after the onset of elicitation, cells were harvested and stored at −80°C.
Cloning of BIS cDNAs
Total RNA was isolated from leaves and stems of cv Holsteiner Cox (100 mg each) using the RNeasy plant mini kit (Qiagen). Aliquots of the RNA pools (1 μg) were reverse transcribed at 42°C using RevertAid H Minus M-MuLV reverse transcriptase (Fermentas). The genome sequence of apple ‘Golden Delicious’ served as template to derive 5′ and 3′ primers for the amplification of the coding sequences of the MdBIS1 to MdBIS4 genes (Table III). PCR with proofreading DNA polymerase (Pfu; Fermentas) consisted of denaturation at 95°C (3 min), 30 cycles at 95°C (30 s), 56°C (40 s), and 72°C (2 min), followed by a 10-min final extension. Using the restriction sites NheI and KpnI, the amplified sequences were cloned to the pRSET B vector (Invitrogen) and sequenced on both strands.
Table III. Primers.
| Labela | Sequenceb |
| For amplification of the coding sequences of MdBIS1 to MdBIS4 of cv Holsteiner Cox | |
| MdBIS1 ORF forw | 5′-GATCGCTAGCATGGCGCCTTCGGTTAAGAATCATG-3′ |
| MdBIS1 ORF rev | 5′-AGTCGGTACCTTAGTATGTAATAGATTCACTACGCAGTATGAC-3′ |
| MdBIS2 ORF forw | 5′-GATCGCTAGCATGGCGCCCTTGGTTAAGGATCAA-3′ |
| MdBIS2 ORF rev | 5′-AGTCGGTACCTTAGCATGTAATAGATTCACTACGCAGCAC-3′ |
| MdBIS3 ORF forw | 5′-GATCGCTAGCATGGCGCCTTTGGTTAAGAATGAGC-3′ |
| MdBIS3 ORF rev | 5′-AGTCGGTACCTTAGCATCTAATAGATTCACTACGCAGCAC-3′ |
| MdBIS4 ORF forw | 5′-GATCGCTAGCATGGCGCCTTTGGTTAAGAATCAAGTAG-3′ |
| MdBIS4 ORF rev | 5′-AGTCGGTACCTCATGCAAGTTTTTCGCATGCAATAGATTCAC-3′ |
| For expression analysis by RT-PCR | |
| MdBIS1 RT-PCR forw | 5′-TCCACCCAATGTCTACTACCAAAAAGAC-3′ |
| MdBIS1 RT-PCR rev | 5′-CCTGGCCCACCAGTATGTCAAGG-3′ |
| MdBIS2 RT-PCR forw | 5′-TACCAAGAGGACTATCCTGATTTCTTG-3′ |
| MdBIS2 RT-PCR rev | 5′-AGCTTGCCCCACTAGTATGTCTAG G-3′ |
| MdBIS3 RT-PCR forw | 5′-AAATCCACCCAACGTCTACC-3′ |
| MdBIS3 RT-PCR rev | 5′-AGCTTGACCCACCAATATGTC-3′ |
| MdBIS4 RT-PCR forw | 5′-ATGGCGCCTTTGGTTAAGAATCAAGTAG-3′ |
| MdBIS4 RT-PCR rev | 5′-TCATGCAAGTTTTTCGCATGCAATAGATTCAC-3′ |
| Profilin forw | 5′-ACGACCACCTGATGTGCG-3′ |
| Profilin rev | 5′-AGAGACCCTGCTCGATAAGG-3′ |
| For expression analysis by quantitative real-time PCR | |
| MdBIS2 Q-PCR forw | 5′-CCATCAAATGTCTACTACCAAGAG-3′ |
| MdBIS2 Q-PCR rev | 5′-AGCTTTGCCTTCTTCAATCGACTTC-3′ |
| MdBIS3 Q-PCR forw | 5′-CGCCTTTGGTTAAGAATGAGCCTC-3′ |
| MdBIS3 Q-PCR rev | 5′-CCTCAATGTTACAAATGTCTTGGCGC-3′ |
| β-Actin forw | 5′-GTGAGGCTCTATTCCAACCATC-3′ |
| β-Actin rev | 5′-GGAACACAAATTGGGCAAGTAT-3′ |
forw, Forward; rev, reverse.
Restriction sites are underlined.
Heterologous Expression, Affinity Purification, and Functional Analysis
Expression in Escherichia coli, isolation of N-terminally His6-tagged proteins, and determination of enzyme activity and kinetic parameters were carried out as described previously (Liu et al., 2007), except that enzyme assays containing radiolabeled malonyl-CoA were omitted. Analysis of the enzymatic products by HPLC and GC-MS was carried out according to Liu et al. (2010). Authentic reference compounds were either synthesized (Liu et al., 2004) or purchased from Sigma.
Expression Analysis by RT-PCR
Total RNA (1 μg) was reverse transcribed at 42°C using RevertAid H Minus M-MuLV reverse transcriptase (Fermentas). Core fragments of the MdBIS1 to MdBIS4 cDNAs were amplified with gene-specific primer pairs (Table III). Amplification of profilin transcripts served as control for equal RNA template amounts, as used previously for normalization (Pike et al., 2005). PCR was carried out using Taq DNA polymerase (Peqlab). The annealing temperature descended from 60°C to 55°C over 10 cycles, followed by 20 cycles with descending temperature from 55°C to 50°C. The extension temperature was 72°C (2 min).
Expression Analysis by Quantitative Real-Time PCR
Quantitative RT-PCR was performed on an iCycler iQ real-time PCR detection system (Bio-Rad Laboratories) using the iQ SYBR green supermix (Bio-Rad Laboratories). Gene-specific amplification was evaluated by melt curve analysis and agarose gel electrophoresis. Amplification and correlation efficiencies of each PCR reaction were determined using serial dilutions of cDNA from inoculated cell cultures. The PCR efficiency was used to transform the Ct values into raw data for relative quantification. The Gene Expression Macro Version 1.1 (Bio-Rad Laboratories) was employed for determination of PCR efficiency and calculation of the mRNA transcript levels. Expression of the MdBIS genes was evaluated using the primers listed in Table III. All samples were normalized on the basis of β-actin mRNA as internal control sample. Scaling of the BIS transcript levels was performed in relation to the mRNA expression level of the reference gene. Scaling of the expression of the BIS genes was performed in relation to the respective mRNA expression levels in mock-inoculated tissues, which were set to be one.
Phylogenetic Reconstruction
A neighbor-joining tree involving the BISs and CHS from cv Holsteiner Cox was constructed as described previously (Liu et al., 2007).
Extraction and Analysis of Phytoalexins
Freeze-dried stem segments and leaves (1 g) were extracted three times with 10 mL of methanol (80%). 4-Phenylphenol (0.1 mg) was added as an internal standard for relative quantification. The combined extract was concentrated in vacuo at 40°C to remove methanol. The residual aqueous solution was extracted three times with 15 mL of ethyl acetate. The combined organic phase was dried over anhydrous sodium sulfate, filtered, and concentrated to dryness in vacuo. The residue was dissolved in 1 mL of ethyl acetate, filtered through a 0.2-μm syringe filter, and analyzed by GC-MS, as described previously (Hüttner et al., 2010). Identification of biphenyls and dibenzofurans was based on a GC-MS library comprising the previously structure-elucidated phytoalexins from elicitor-treated Sorbus aucuparia cell cultures (Hüttner et al., 2010).
Preparation of Antiserum and Isolation of IgG
Polyclonal antibodies were raised against heterologously expressed and affinity-purified SaBIS1 in rabbits (Seqlab). The IgG fractions of the antiserum and the preimmune serum were isolated according to the manufacturer’s instructions by affinity chromatography on a HiTrap protein A HP column (1 mL; GE Healthcare) coupled to a Biologic-FPLC system (Bio-Rad Laboratories). The IgG fractions were stored at −20°C in PBS after buffer exchange on a PD-10 column (GE Healthcare).
Protein-Blot Analysis
Protein extracts were prepared from stems and leaves of cv Holsteiner Cox collected at various times (0–14 d) after E. amylovora inoculation. Protein concentrations were determined by the method of Bradford (1976). The protein extracts (50 μg) and the affinity-purified MdBIS1 to MdBIS4 proteins (100 ng) were separated on 10% SDS-polyacrylamide gels and electrophoretically transferred (Bio-Rad; 100 V, 350 mA) to polyvinylidene difluoride membranes (Immobilon P; Millipore). The membranes were blocked at 4°C overnight with 5% (w/v) skim milk powder in TBS plus Tween 20 (TBST; 50 mm Tris, pH 7.5, 150 mm NaCl, 0.05% [v/v] Tween 20) and incubated with either the anti-BIS IgG fraction (1:10,000 [v/v] dilution with TBST containing 5% [w/v] skim milk powder) or the preimmune IgG fraction (1:10,000 [v/v]) for 1 h at room temperature with gentle agitation. After washing three times with TBST (7 min each), the membranes were incubated with peroxidase-conjugated AffiniPure goat anti-rabbit IgG (Jackson Immuno Research; 1:3,300 [v/v] in TBST containing 5% [w/v] skim milk powder) at room temperature for 45 min with gentle agitation. Finally, the membranes were washed with TBST (3 × 7 min), rinsed with PBS buffer, and processed using the ECL western-blotting detection system according to the manufacturer’s instructions (Lumi-LightPlus western blotting substrate; Roche).
Immunohistochemical Analysis
Using the resin technique, plant tissue was analyzed under a confocal laser-scanning microscope. Transition zones of E. amylovora-inoculated shoots of cv Holsteiner Cox were harvested 6 d postinoculation, cut into small pieces (about 0.3 × 0.8 cm), and fixed immediately on ice for 2 h under reduced pressure (vacuum 0.3 mbar) in fixative solution (2% [w/v] formaldehyde, 0.1% [v/v] glutaraldehyde, 0.1% Triton X-100 [w/v] in 0.1 m phosphate buffer, pH 7.2). The samples were washed twice for 10 min with PBS buffer. Water was gradually removed by 30%, 50%, 70%, and 90% aqueous ethanol and finally 3 × 100% ethanol. Each step of incubation took 30 min at room temperature. After dehydration, the plant tissue was embedded in Technovit 7100 (Heraeus-Kulzer) according to the manufacturer’s instructions. Fixed tissues were cut into thin segments (3.5 μm) using a microtome (HM 355 S; Microm). The sections were transferred onto diagnostic microscope slides (Teflon; Roth) using a drop of distilled water and left to dry. The thin sections were incubated in 50 mm ammonium chloride solution at 37°C for 15 min, washed with water, incubated in 50 mm glycin solution at 37°C for 15 min, and washed again with water. The sections were then blocked at 37°C for 30 min with bovine serum albumin (BSA)-blocking solution (10% [w/v] BSA and 0.1% [w/v] fish gelatin in PBS buffer, pH 7.2), washed once with water and 3 × 10 min with PBS buffer. Thereafter, the sections were incubated with either anti-BIS IgG oder preimmune IgG (1:25 to 1:100 dilution) with 1% BSA in PBS buffer at 37°C for 1 h. They were washed once with water and 3 × 10 min with PBS buffer. Finally, the sections were incubated in the dark with fluorescence dye-conjugated goat anti-rabbit antibody as secondary antibody (Alexa Fluor 488 goat anti-rabbit IgG [H + L]; Molecular Invitrogen) diluted 1:100 with 1% (w/v) BSA in PBS buffer at 37°C for 1 h. After washing 3 × 15 min with PBS buffer and then with water, sections were left in the dark at 37°C until dryness. Mounting medium (Citifluor; Agar Scientific) was used to adhere the coverslip to the tissue section. The tissues were viewed using a laser-scanning microscope (Zeiss LSM 510META). The scan mode was channel scan. The imaging of whole-mount specimens was recorded using a digital camera (Axioskop 2; Zeiss) and the software Axio Vision 3.0 (Zeiss).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers MdBIS1, JQ390521; MdBIS2, JQ390522; MdBIS3, JQ390523; and MdBIS4, JQ390524.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of BIS coding sequences.
Supplemental Figure S2. Phylogenetic tree of BIS coding sequences.
Supplemental Figure S3. Structural analysis of the BIS1.1 gene.
Supplemental Figure S4. Structural analysis of the BIS1.2 gene.
Supplemental Figure S5. Structural analysis of the BIS2.4 gene.
Supplementary Material
References
- Beerhues L, Liu B. (2009) Biosynthesis of biphenyls and benzophenones—evolution of benzoic acid-specific type III polyketide synthases in plants. Phytochemistry 70: 1719–1727 [DOI] [PubMed] [Google Scholar]
- Billing E. (2011) Fire blight: why do views on host invasion by Erwinia amylovora differ? Plant Pathol 60: 178–189 [Google Scholar]
- Bowen JK, Mesarich CH, Bus VGM, Beresford RM, Plummer KM, Templeton MD. (2011) Venturia inaequalis: the causal agent of apple scab. Mol Plant Pathol 12: 105–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer J, Liu RH. (2004) Apple phytochemicals and their health benefits. Nutr J 3: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Chinnici F, Bendini A, Gaiani A, Riponi C. (2004) Radical scavenging activities of peels and pulps from cv. Golden Delicious apples as related to their phenolic composition. J Agric Food Chem 52: 4684–4689 [DOI] [PubMed] [Google Scholar]
- FAO (2009) Food and Agriculture Organization of the United Nations. FAOSTAT Home Page. http://faostat.fao.org/site/567/default.aspx#ancor (September 8, 2011)
- Flachowsky H, Richter K, Kim WS, Geider K, Hanke MV. (2008) Transgenic expression of a viral EPS-depolymerase is potentially useful to induce fire blight resistance in apple. Ann Appl Biol 153: 345–355 [Google Scholar]
- Flachowsky H, Szankowski I, Fischer TC, Richter K, Peil A, Höfer M, Dörschel C, Schmoock S, Gau AE, Halbwirth H, et al. (2010) Transgenic apple plants overexpressing the Lc gene of maize show an altered growth habit and increased resistance to apple scab and fire blight. Planta 231: 623–635 [DOI] [PubMed] [Google Scholar]
- Garrett CME, Crosse JE. (1963) Observations on lysogeny in the plant pathogens Pseudomonas morsprunorum and Ps. syringae. J Appl Bacteriol 26: 27–34 [Google Scholar]
- Gerhauser C. (2008) Cancer chemopreventive potential of apples, apple juice, and apple components. Planta Med 74: 1608–1624 [DOI] [PubMed] [Google Scholar]
- Gessler C, Patocchi A. (2007) Recombinant DNA technology in apple. Adv Biochem Engin Biotechnol 107: 113–132 [DOI] [PubMed] [Google Scholar]
- Gosch C, Halbwirth H, Stich K. (2010) Phloridzin: biosynthesis, distribution and physiological relevance in plants. Phytochemistry 71: 838–843 [DOI] [PubMed] [Google Scholar]
- Hanson KR, Havir EA. (1981) Phenylalanine ammonia-lyase. Stumpf PK, Conn EE, , The biochemistry of Plants, Vol 7. Academic, London, pp 577–625 [Google Scholar]
- He X, Liu RH. (2007) Triterpenoids isolated from apple peels have potent antiproliferative activity and may be partially responsible for apple’s anticancer activity. J Agric Food Chem 55: 4366–4370 [DOI] [PubMed] [Google Scholar]
- Hrazdina G, Borejsza-Wysocki W, Lester C. (1997) Phytoalexin production in an apple cultivar resistant to Venturia inaequalis. Phytopathology 87: 868–876 [DOI] [PubMed] [Google Scholar]
- Hüttner C, Beuerle T, Scharnhop H, Ernst L, Beerhues L. (2010) Differential effect of elicitors on biphenyl and dibenzofuran formation in Sorbus aucuparia cell cultures. J Agric Food Chem 58: 11977–11984 [DOI] [PubMed] [Google Scholar]
- Jiang LL, Xuan LJ. (2006) A new biphenyl glycoside from the leaves of Eriobotrya japonica. Chin Chem Lett 17: 35–37 [Google Scholar]
- Kemp MS, Holloway PJ, Burden RS. (1985) 3β-19α-Dihydroxy-2-oxours-12-en-28-oic acid: a pentacyclic triterpene induced in the wood of Malus pumila Mill. infected with Chondrostereum purpureum (Pers. ex Fr.) Pouzar. and a constituent of the cuticular wax of apple fruits. J Chem Res M 1848–1876 [Google Scholar]
- Kokubun T, Harborne JB. (1994) A survey of phytoalexin induction in leaves of the Rosaceae by copper ions. Z Naturforsch C: J Biosci 49: 628–634 [Google Scholar]
- Kokubun T, Harborne JB. (1995) Phytoalexin induction in the sapwood of plants of the Maloideae (Rosaceae): biphenyls or dibenzofurans. Phytochemistry 40: 1649–1654 [Google Scholar]
- Lee JY, Lu H. (2011) Plasmodesmata: the battleground against intruders. Trends Plant Sci 16: 201–210 [DOI] [PubMed] [Google Scholar]
- Lee KW, Kim YJ, Kim DO, Lee HJ, Lee CY. (2003) Major phenolics in apple and their contribution to the total antioxidant capacity. J Agric Food Chem 51: 6516–6520 [DOI] [PubMed] [Google Scholar]
- Li H, Flachowsky H, Fischer TC, Hanke MV, Forkmann G, Treutter D, Schwab W, Hoffmann T, Szankowski I. (2007) Maize Lc transcription factor enhances biosynthesis of anthocyanins, distinct proanthocyanidins and phenylpropanoids in apple (Malus domestica Borkh.). Planta 226: 1243–1254 [DOI] [PubMed] [Google Scholar]
- Linsmaier EM, Skoog F. (1965) Organic growth factor requirements of tobacco tissue cultures. Physiol Plant 18: 100–127 [Google Scholar]
- Liu B, Beuerle T, Klundt T, Beerhues L. (2004) Biphenyl synthase from yeast-extract-treated cell cultures of Sorbus aucuparia. Planta 218: 492–496 [DOI] [PubMed] [Google Scholar]
- Liu B, Falkenstein-Paul H, Schmidt W, Beerhues L. (2003) Benzophenone synthase and chalcone synthase from Hypericum androsaemum cell cultures: cDNA cloning, functional expression, and site-directed mutagenesis of two polyketide synthases. Plant J 34: 847–855 [DOI] [PubMed] [Google Scholar]
- Liu B, Raeth T, Beuerle T, Beerhues L. (2007) Biphenyl synthase, a novel type III polyketide synthase. Planta 225: 1495–1503 [DOI] [PubMed] [Google Scholar]
- Liu B, Raeth T, Beuerle T, Beerhues L. (2010) A novel 4-hydroxycoumarin biosynthetic pathway. Plant Mol Biol 72: 17–25 [DOI] [PubMed] [Google Scholar]
- Luby JJ. (2003) Taxonomic classification and brief history. Ferree DC, Warrington IJ, , Apples: Botany, Production and Uses. CAB International, Wallingford, UK, pp 1–14 [Google Scholar]
- Lucas WJ, Ham BK, Kim JY. (2009) Plasmodesmata—bridging the gap between neighboring plant cells. Trends Cell Biol 19: 495–503 [DOI] [PubMed] [Google Scholar]
- Manach C, Williamson G, Morand C, Scalbert A, Rémésy C. (2005) Bioavailability and bioefficacy of polyphenols in humans. I. Review of 97 bioavailability studies. Am J Clin Nutr () 81: 230S–242S [DOI] [PubMed] [Google Scholar]
- Maule AJ. (2008) Plasmodesmata: structure, function and biogenesis. Curr Opin Plant Biol 11: 680–686 [DOI] [PubMed] [Google Scholar]
- Mayr U, Treutter D, Santos-Buelga C, Bauer H, Feucht W. (1995) Developmental changes in the phenol concentrations of ‘golden delicious’ apple fruits and leaves. Phytochemistry 38: 1151–1155 [DOI] [PubMed] [Google Scholar]
- McGhie TK, Hunt M, Barnett LE. (2005) Cultivar and growing region determine the antioxidant polyphenolic concentration and composition of apples grown in New Zealand. J Agric Food Chem 53: 3065–3070 [DOI] [PubMed] [Google Scholar]
- Miyakodo M, Watanabe K, Ohno N, Nonaka F, Morita A. (1985) Isolation and structural determination of eriobofuran, a new dibenzofuran phytoalexin from leaves of loquat, Eriobotrya japonica L. J Pest Sci 10: 101–106 [Google Scholar]
- Nissinen RM, Ytterberg AJ, Bogdanove AJ, VAN Wijk KJ, Beer SV. (2007) Analyses of the secretomes of Erwinia amylovora and selected hrp mutants reveal novel type III secreted proteins and an effect of HrpJ on extracellular harpin levels. Mol Plant Pathol 8: 55–67 [DOI] [PubMed] [Google Scholar]
- Norelli JL, Jones AL, Aldwinckle HS. (2003) Fire blight management in the twenty-first century: using new technologies that enhance host resistance in apple. Plant Dis 87: 756–765 [DOI] [PubMed] [Google Scholar]
- Oh CS, Beer SV. (2005) Molecular genetics of Erwinia amylovora involved in the development of fire blight. FEMS Microbiol Lett 253: 185–192 [DOI] [PubMed] [Google Scholar]
- Pike SM, Zhang XC, Gassmann W. (2005) Electrophysiological characterization of the Arabidopsis avrRpt2-specific hypersensitive response in the absence of other bacterial signals. Plant Physiol 138: 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder J. (1999) The chalcone/stilbene synthase-type family of condensing enzymes. Sankawa U, , Comprehensive Natural Products Chemistry, Vol 1. Elsevier Science, Amsterdam, pp 749–771 [Google Scholar]
- Thomson SV. (2000) Epidemiology of fire blight. Vanneste JL, , Fire Blight: The Disease and Its Causative Agent, Erwinia amylovora. CAB International, Wallingford, UK, pp 9–36 [Google Scholar]
- Treutter D. (2005) Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biol (Stuttg) 7: 581–591 [DOI] [PubMed] [Google Scholar]
- Treutter D. (2006) Significance of flavonoids in plant resistance: a review. Environ Chem Lett 4: 147–157 [Google Scholar]
- Tsao R, Yang R, Young JC, Zhu H. (2003) Polyphenolic profiles in eight apple cultivars using high-performance liquid chromatography (HPLC). J Agric Food Chem 51: 6347–6353 [DOI] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, et al. (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 42: 833–839 [DOI] [PubMed] [Google Scholar]
- Vrhovsek U, Rigo A, Tonon D, Mattivi F. (2004) Quantitation of polyphenols in different apple varieties. J Agric Food Chem 52: 6532–6538 [DOI] [PubMed] [Google Scholar]
- Watanabe K, Widyastuti SM, Nonaka F. (1990) Two biphenyl compounds from Rhaphiolepsis umbellata as its phytoalexin. Agric Biol Chem 54: 1861–1862 [Google Scholar]
- Widyastuti SM, Nonaka F, Watanabe K, Sako N, Tanaka K. (1992) Isolation and characterization of two aucuparin-related phytoalexins from Photinia glabra Maxim. Ann Phytopath Soc Japan 58: 228–233 [Google Scholar]
- Wolfe K, Wu X, Liu RH. (2003) Antioxidant activity of apple peels. J Agric Food Chem 51: 609–614 [DOI] [PubMed] [Google Scholar]
- Xu X, Madden LV. (2002) Incidence and density relationships of powdery mildew on apple. Phytopathology 92: 1005–1014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.