Summary
The tad (tight adherence) locus encodes a protein translocation system that produces a novel variant of type IV pili. The pilus assembly protein TadZ (called CpaE in Caulobacter crescentus) is ubiquitous in tad loci, but is absent in other type IV pilus biogenesis systems. The crystal structure of TadZ from E. rectale (ErTadZ), in complex with ATP and Mg2+, was determined to 2.1 Å resolution. ErTadZ contains an atypical ATPase domain with a variant of a deviant Walker-A motif that retains ATP binding capacity while displaying only low intrinsic ATPase activity. The bound ATP plays an important role in dimerization of ErTadZ. The N-terminal atypical receiver domain resembles the canonical receiver domain of response regulators, but has a degenerate, stripped-down “active site”. Homology modeling of the N-terminal atypical receiver domain of CpaE indicates that it has a conserved protein-protein binding surface similar to that of the polar localization module of the social mobility protein FrzS, suggesting a similar function. Our structural results also suggest that TadZ localizes to the pole through the atypical receiver domain during early stage of pili biogenesis, and functions as a hub for recruiting other pili components, thus providing insights into the Tad pilus assembly process.
Keywords: Type IV pili assembly, TadZ, atypical receiver domain, atypical ATPase, localization factor
Introduction
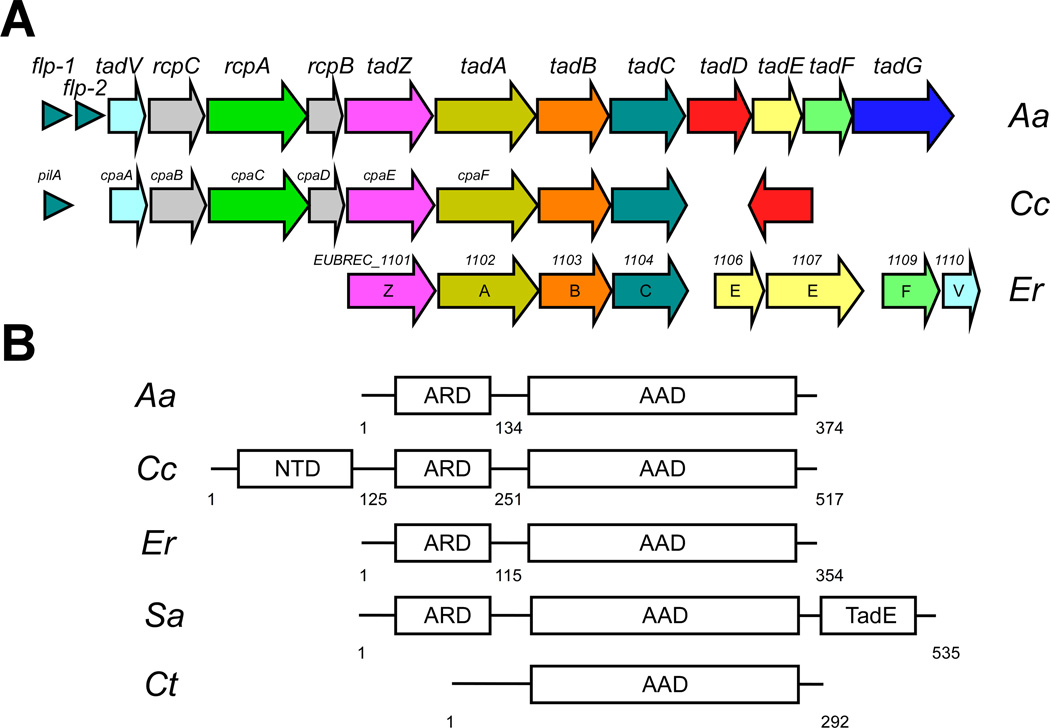
Pili (or fimbriae) consist of filaments of pilin subunits that form hair-like appendages on the surface of many bacteria (Proft et al., 2009). They play important roles in processes such as adhesion to other bacteria especially in the context of bacterial conjugation, bacteriophage absorption, motility, and biofilm formation. The tad (tight adherence) locus is a widespread colonization island that is found in both Gram-negative and Gram-positive bacteria, as well as the archaea (Tomich et al., 2007). In Aggregatibacter actinomycetemcomitans, the tad locus encodes a protein translocation system for the assembly of Flp pili. These pili of A. actinomycetemcomitans mediate strong, nonspecific adherence to solid surfaces (such as teeth), and are important in colonization and pathogenesis (Schreiner et al., 2003; Planet et al., 2003; Kachlany et al., 2001a; Bhattacharjee et al., 2001; Kachlany et al., 2000). Similar loci have also been characterized in Caulobacter crescentus (Skerker et al., 2000; Viollier et al., 2002b; Viollier et al., 2002a), Haemophilus ducreyi (Nika et al., 2002) and Pseudomonas aeruginosa (de Bentzmann et al., 2006). The A. actinomycetemcomitans tad locus consists of a 14 genes flp-1-flp-2-tadV-rcpCAB-tadZABCDEFG (Fig. 1A). The tad operon in C. crescentus consists of at least 7 genes including pilA and cpaABCDEF. In Gram-negative bacterium, the Tad pilus spans the periplasmic region, possibly with the help of proteins TadD, TadE, TadF and TadG, and exits through the secretion channel in the outer membrane formed by the secretin RcpA (CpaC), and associated membrane proteins RcpB and RcpC (Tomich et al., 2007; Chen et al., 2005). The cytoplasmic ATPase TadA (Bhattacharjee et al., 2001) may facilitate the assembly of the pilin subunits into the pilus filament. TadV is a peptidase that cleaves the prepilin subunits prior to assembly into a pilus (and also processes the pseudopilins TadE and TadF). The Tad secretion system is evolutionarily related to the type II secretion systems (T2SSs) (Peabody et al., 2003; Tomich et al., 2007); however, several components are unique (e.g. TadZ, TadE, and TadF) and not found in other T2SSs. Thus, the tad-encoded Flp pili are classified as a subfamily of type IVb pili (Kachlany et al., 2001b). Due to the absence of an outer membrane, the tad loci of Gram-positive bacteria are shorter, usually consisting of tadZABC, and lack the components responsible for outer membrane secretion.
Fig. 1.
Genetic maps of tad loci and the domain architecture of the TadZ proteins. (A) The tad loci for the Gram-negative bacteria A. actinomycetemcomitans (Aa) and C. crescentus (Cc), and for the Gram-positive bacterium E. rectale (Er). The locus number for each E. rectale gene is also shown. (B) Representative domain architectures of TadZs from A. actinomycetemcomitans, C. crescentus, E. rectale, Streptomyces avermitilis (Sa) and Chlorobium tepidum TLS (Ct). NTD represents the N-terminal domain, ARD: the atypical receiver domain, and AAD: the atypical ATPase domain.
The gene tadZ is present in all tad loci, and TadZ is an essential component of the Tad secretion system. Studies of TadZ of A. actinomycetemcomitans (AaTadZ) and the homologous CpaE of C. crescentus (CpaE referred to as CcTadZ hereafter) indicated that they likely function as localization factors (Viollier et al., 2002b; Viollier et al., 2002a; Skerker and Shapiro, 2000; Christen et al., 2010) (B.A. Perez-Cheeks, unpublished, submitted as a companion paper). In C. crescentus, CcTadZ mediates polar positioning of the pilus secretin protein CpaC, as well as the histidine kinase PleC. However, the molecular mechanism of the TadZ function in the pilus assembly process remains unclear. Here, we report the crystal structure of a TadZ protein from E. rectale (ErTadZ) as part of our ongoing structural genomics effort targeting novel protein structures from the human microbiome (Elsliger et al., 2010; Xu et al., 2010b; Xu et al., 2010a). E. rectale is an anaerobic Gram-positive bacterium that is prevalent in the human colon. It is one of the major bacterial producers of butyrate, a short-chain fatty acid that is the preferred energy source for colonocytes, and thus is important for maintaining colon health (Duncan et al., 2008). The ErTadZ structure in complex with ATP and a magnesium ion exhibits a unique molecular architecture combining components from the bacterial cytoskeleton (MinD/ParA/Soj ATPases) and two-component signal transduction (response regulators). However, both domains have lost the canonical active sites. The ATPase domain, with a degenerate Walker-A motif, mediates dimerization in the presence of ATP while exhibiting only low intrinsic ATPase activity. The atypical receiver domain of CcTadZ is structurally similar to the social mobility protein FrzS, which is involved in polar localization to the leading pole (Fraser et al., 2007; Mignot et al., 2007), and indicates a conserved role for these modules in the biogenesis of type IV pili. Thus, we now provide a structural basis for understanding the role of TadZ in the pili assembly process and in polar localization.
Results and discussion
Functional assignment of ErTadZ and sequence features of the TadZ family
Sequence similarities between ErTadZ and CcTadZ or AaTadZ are weak using direct amino–acid sequence comparison (sequence identity <10%). However, the conservation of tad locus genes as a single operon (Tomich et al., 2007) provides reliable evidence for the functional assignment of ErTadZ. The E. rectale genome contains the typical tad operon found in Gram-positive bacteria (Fig. 1A). The tadZ gene is always located immediately upstream of the ATPase gene tadA. In addition to conserved proteins, the E. rectale tad operon also contains two proteins of unknown function (EUBREC_1105 and EUBREC_1108, not shown). A large third protein in this operon (EUBREC_1107, 1082aa) contains a TadE-like region and a Colicin IA-like domain. ErTadZ is related by sequence to proteins in Ruminococcus bromii (gi 291541803), Oribacterium sinus (gi 227872973), Erysipelotrichaceae bacterium (gi 309777432), and Clostridium asparagiforme (gi 225389010) (seq id ~25%, Fig. S1). A search of distant homologs identifies a large number of ATPases, including a few CcTadZ-like proteins, indicating a remote evolutionary relationship between ErTadZ and CcTadZ.
We investigated the domains of the TadZ family of proteins using separate profiles for the receiver domain (PF00072, RD thereafter) and the NifH/FrxC family ATPase Pfam domain (PF00142) (Bateman et al., 2004). Of the top 250 hits of CcTadZ homologs identified by a BLAST search, 242 and 233 proteins were found to contain an N-terminal RD or a C-terminal ATPase domain respectively, suggesting similar domain architecture, as subsequently revealed by the ErTadZ structure (see below). Further examination of the RD of these TadZ homologs indicate that they have lost critical residues corresponding to the phosphorylation site of canonical RDs (Bourret, 2010). The C-terminal ATPase domain of TadZ contains a sequence motif ([KR]GGxGs[ST]) that is a variant of the deviant Walker-A motif (KGGxGK[ST]) with the invariant lysine (underlined) replaced by a residue with a small side-chain (s) (see below).
Thus, ErTadZ is a member of the diverse TadZ family, and part of the E. rectale tad gene cluster. Most members of this family have the same two-domain architecture as ErTadZ and AaTadZ (Fig. 1B). CcTadZ contains an additional ~120aa proline-rich region, which is expected to be only partially ordered. TadZ from Streptomyces avermitilis is fused to a TadE-like tail, while the N-terminal RD is absent in TadZ from Chlorobium tepidum.
Structure determination and model quality
Full-length ErTadZ contains 354 residues with a molecular weight of 39.8 kDa and a calculated pI of 4.9. The crystal structure of ErTadZ was determined by the multiple-wavelength anomalous diffraction (MAD) phasing method using the high-throughput structural genomics pipeline implemented at the Joint Center for Structural Genomics (JCSG, http://www.jcsg.org) (Lesley et al., 2002; Elsliger et al., 2010). The selenomethionine (SeMet) derivative of ErTadZ was expressed in E. coli with an N-terminal His-tag and was purified by metal affinity chromatography. Three-wavelength MAD data were collected to 2.1 Å resolution at Stanford Synchrotron Radiation Lightsource (SSRL) beamline BL11-1. The data were indexed and processed in monoclinic space group C2 with unit cell dimensions of a=121.5 Å, b=79.5 Å, c=55.0 Å and β=96.2°. The asymmetric unit contains one protein molecule, with a solvent content of 60.3% (Vm=3.1). The final model was refined to an Rcryst of 19.0% and an Rfree of 21.8%. TLS parameters were refined with each domain defined as a rigid body unit (residues 1–114,115–354). The electron density map showed clear density for the full-length protein. The model of ErTadZ displays good geometry with an all-atom clash score of 7.1, and the Ramachandran plot produced by MolProbity (Davis et al., 2004) shows that all residues are in allowed regions, with 98.3 % in favored regions. The final model of ErTadZ contains all residues of the full-length protein, one ATP, one magnesium ion (Mg2+), one chloride ion, one sulfate ion, two glycerol molecules, two polyethylene glycol fragments and 140 water molecules. The side-chains of nine residues (Met1, Lys12, Glu13, Arg17, Lys46, Arg49, Arg189, Arg300 and Lys312) on the protein surface, and the purification tag are disordered and are absent from the final model. The data processing and refinement statistics are summarized in Table 1.
Table 1.
Data collection and refinement statistics
| Data collection | λ1 MADSe | λ2 MADSe | λ3 MADSe |
| Wavelength (Å) | 0.9791 | 0.9184 | 0.9785 |
| Resolution range (Å) | 29.2-2.10 | 29.2-2.10 | 29.2-2.10 |
| No. observations | 98,489 | 98,386 | 98,604 |
| No. unique reflections | 30,457 | 30,416 | 30,493 |
| Completeness (%) | 99.8 (100)a | 99.8 (100) | 99.9 (100) |
| Mean I/σ (I) | 10.4 (1.9)a | 9.8 (1.7) | 9.7 (1.7) |
| Rsym on I | 0.078 (0.56)a | 0.085(0.63) | 0.087 (0.61) |
| Model and refinement statistics | |||
| Data set used in refinement | λ2 MADSe | ||
| No. reflections (total) | 30,416 | ||
| No. reflections (test) | 1,535 | ||
| Completeness (% total) | 99.8 | ||
| Cutoff criteria | |F|>0 | ||
| Rcryst | 0.190 | ||
| Rfree | 0.218 | ||
| Restraints (RMS observed) | |||
| Bond angle (°) | 1.38 | ||
| Bond length (Å) | 0.015 | ||
| Molprobity Scores | |||
| All-atom clash score | 7.11 | ||
| Ramachandran plot favored (allowed, %) | 98.3 (100) | ||
| Average isotropic B-value (Å2) | 47.6 (33.6/27.1)b | ||
| ESU based on Rfree (Å) | 0.15 | ||
| Protein residues / atoms | 354 / 2773 | ||
| Ligands | 1 ATP, 1 Mg2+ |
Highest resolution shell (2.15-2.10 Å) in parentheses.
This value represents the total B that includes TLS and residual B components. The B-values of ATP and Mg2+ are listed in parenthesis.
ESU = Estimated Standard Uncertainty in coordinates.
Rsym = Σ|Ii−<Ii>| / Σ|Ii|, where Ii is the scaled intensity of the ith measurement and <Ii> is the mean intensity for that reflection.
Rcryst = Σ| |Fobs|−|Fcalc| | / Σ|Fobs|, where Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively.
Rfree = as for Rcryst, but for 5.0% of the total reflections chosen at random and omitted from refinement.
Overall structural description
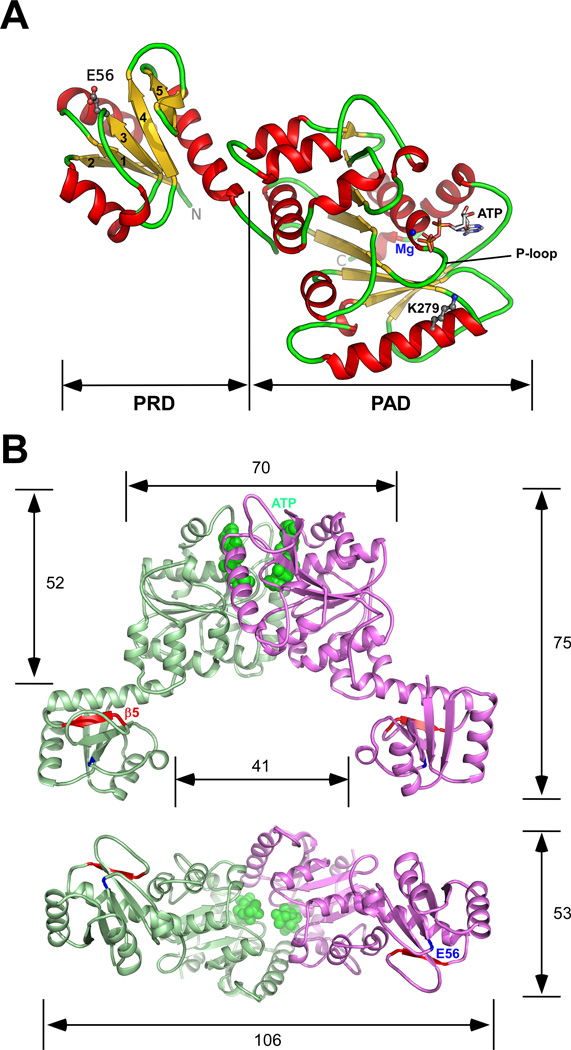
The structure of ErTadZ consists of two domains (Fig. 2A). The N-terminal domain (residues 1–114) is structurally similar to the RD of response regulators. The C-terminal domain (residues 125–354) shows structural homology to a class of ATPases that are involved in diverse activities such as cell division (MinD), DNA segregation (ParA/Soj), and nitrogen fixation (NifH). An ATP molecule and a magnesium ion are located in the canonical ATP binding site. We refer to these domains as the atypical receiver domain (ARD) and the atypical ATPase domain (AAD), since they have lost the catalytic residues of canonical domains (see below).
Fig. 2.
The crystal structure of TadZ from E. rectale (ErTadZ). (A) The monomer of ErTadZ consists of two domains: the atypical receiver domain (ARD) and the atypical ATPase domain (AAD). Glu56 and Lys279 are shown as ball-and-sticks; ATP is shown as sticks, and Mg2+ as a blue sphere. (B) ErTadZ dimer and molecular dimensions (in Å) with ATP in green spheres. Glu56 and strand β5, corresponding to the canonical phosphorylation site and output surface of a canonical RD, are highlighted in blue and red respectively.
The ARD is connected to the AAD through a short linker (residues 115–124). This linker and the C-terminal end of the last helix of the ARD (residues 108–114) interact with the backside of the AAD (distal to the ATP binding site) and together bury 1384 Å2 of total surface area. ErTadZ dimerizes via the AAD domain through the crystallographic two-fold axis, forming a V-shaped molecule with molecular dimensions of ~106 Å × 75 Å × 53 Å (Fig. 2B). The ARDs are located as appendages emanating from the central dimer core with a median intermolecular distance of ~74 Å.
A novel subfamily of atypical ATPases
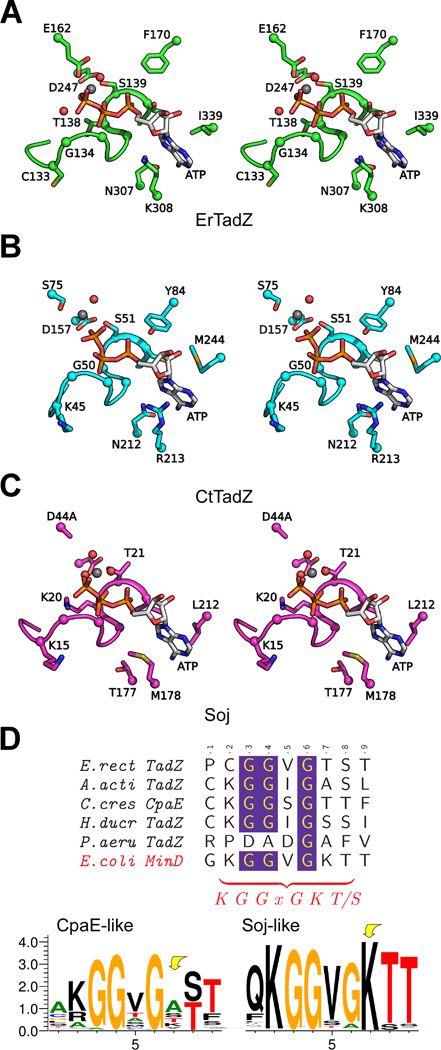
The AAD domain of ErTadZ is structurally most similar to a truncated CT0433 (residues 36–277) of the Gram-negative green sulfur bacterium C. tepidum with an rmsd of 3.0 Å for 221 equivalent Cα atoms and 18% sequence identity (PDB code 3ea0, the Midwest Structural Genomics Center, unpublished, Dali Z=19.8) (Fig. S2). CT0433 is located in a conserved tad locus and, thus, is also a TadZ homolog (referred to as CtTadZ hereafter). AAD of ErTadZ is also highly similar to MinD of Pyrococcus furiosus (PDB code 1g3r, Z=19.4) (Hayashi et al., 2001) and the chromosome segregation protein Soj from Thermus thermophilus (PDB code 2bek, Z=17.9) (Leonard et al., 2005). Compared to the AAD of ErTadZ, the AAD of CtTadZ is phylogenetically closer to CcTadZ, AaTadZ, MinD and Soj with higher sequence and structural similarities [e.g. Z=26.5 for 1g3r (MinD) and Z=24.3 for 2bek (Soj)]. However, ErTadZ does not possess a C-terminal “amphipathic helix” present in MinD. AaTadZ, on the other hand, has a slightly longer sequence that could accommodate a C-terminal “amphipathic helix” (Fig. S2).
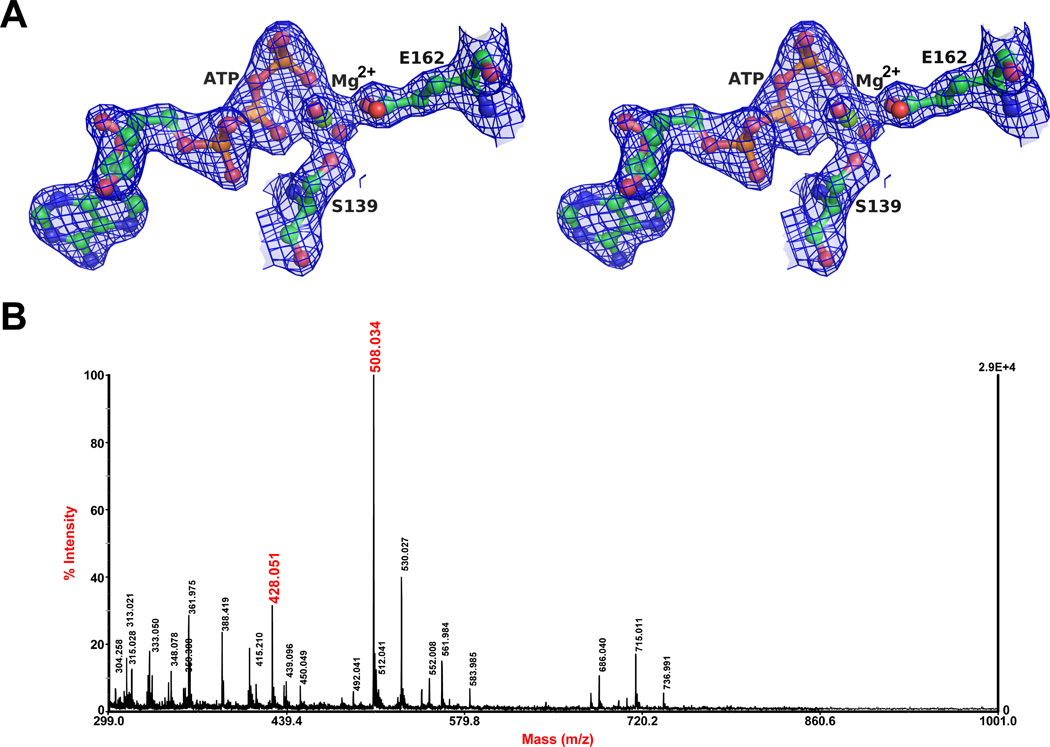
ErTadZ contains a bound ATP molecule, supported by well-defined electron density maps and detection of ATP in the purified ErTadZ sample, used for crystallization, by MALDI mass spectrometry (Fig. 3). A magnesium ion interacts with β- and γ- phosphates of ATP, Ser139 Oγ1, Glu162 Oε2 and two waters with a near perfect hexacoordinate geometry (Fig. 4A). ATP and Mg2+ were not added during ErTadZ purification or crystallization stages and, thus, were co-purified from the expression host. ATP and Mg2+ were also detected in the crystal structure of CtTadZ; however, the location of the magnesium ion and the γ-phosphate of ATP are altered (Fig. 4B). The conformation and location of the ATP and Mg2+ in ErTadZ are essentially identical to those of the Soj ATPase mutant D44A structure (PDB code 2bek) (Leonard et al., 2005) (Fig. 4C). MinD/Soj-like ATPases contain a deviant Walker-A (P-loop) motif (xKGGxGK[ST], compared to the canonical GxxGxGK[ST]) with two lysines, one near the carboxyl end of the motif that is common to all Walker-A motifs, and a second at the beginning of the motif that is unique to this subgroup of ATPases (Koonin, 1993; Lutkenhaus et al., 2003). The Walker-A motif of ErTadZ is further degraded (132PCGGVGTS139) with the loss of both lysines (Fig. 4D). The P-loop of CtTadZ contains an alanine at the second lysine position, but has the first conserved lysine.
Fig. 3.
ATP and Mg2+ in the active site of AAD. (A) The σA-weighted 2Fo-Fc density around the bound ATP is contoured at 1.0 σ. ATP and protein side chains of the final refined model are shown ball-and-sticks. Mg2+ and water molecules are shown as green and red spheres, respectively. (B) MALDI MS spectrum showing the presence of ATP (m/z 508.03 Da) and ADP (m/z 428.05 Da) in the ErTadZ protein sample used for structure determination.
Fig. 4.
Stereoview of the ATP binding sites of (A) ErTadZ, (B) CtTadZ, and (C) the Soj D44A mutant. Magnesium ions and water molecules are shown as gray and red spheres, respectively. (D) The P-loop regions (Walker-A motifs) of the TadZ family of proteins lack the normally conserved second lysine. (Top panel) Sequence alignment of the P-loop regions of several characterized TadZ homologs and E. coli MinD. The consensus sequence of the deviant Walker-A motif is shown at the bottom. (Bottom panel) Sequence conservation pattern of the P-loop in TadZ homologs (490 sequences) and Soj homologs (1631 sequences) as a sequence logo representation, where the overall height of the stack indicates the sequence conservation, and the height of the symbols indicate the relative frequency of each amino acid at that position. Sequences of each set were created from top hits of BLAST (cutoff Expect<1.E-10) using either the AAD domain of CcTadZ or Soj as probes, respectively.
The loss of the second lysine in the Walker-A motif is also observed in other TadZ homologs (sequence motif s[KR]GGsGs[ST], where s are residues with small side chains A/S/T/V, Fig. 4D), suggesting that this is a common property of the TadZ family. Nevertheless, these P-loop regions containing the degraded Walker A- motif are still highly conserved (Figs. S1 and S2), indicating that the TadZ proteins likely have retained the ATP binding capability, as supported by the ErTadZ and CtTadZ structures.
ATPase activity of ErTadZ
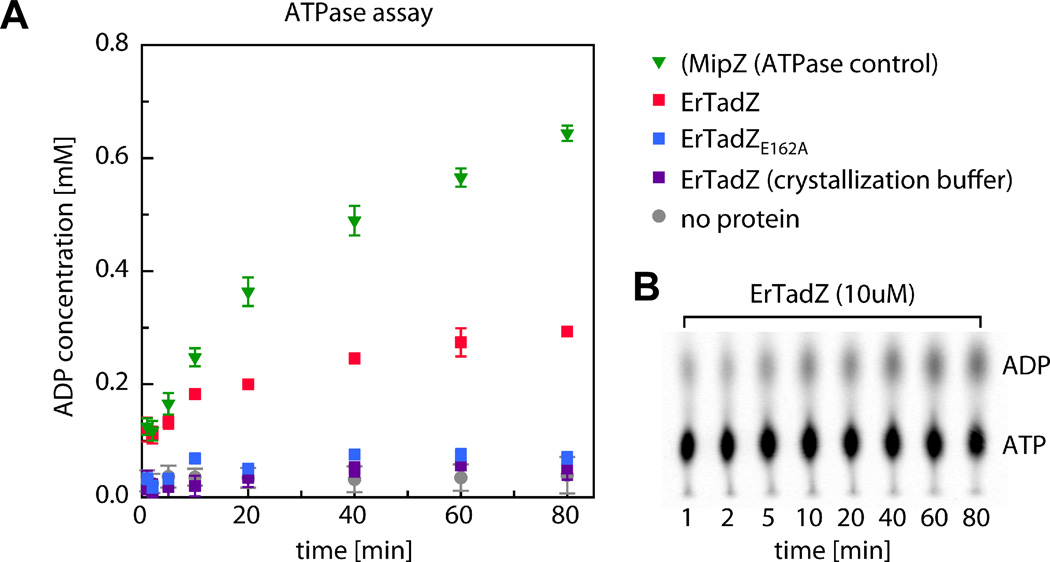
To probe whether the ATP binding site has catalytic activity, we analyzed the nucleotide composition of the ErTadZ sample used for crystallization by MALDI mass spectrometry. In addition to ATP (m/z 508.03), smaller amounts of ADP (m/z 428.5) were identified in the ErTadZ sample (Fig. 3B, see Experimental procedures), suggesting that ATP bound to ErTadZ is either slowly hydrolyzed or ADP co-purified with ErTadZ. Further biochemical analysis using radiolabeled ATP and nucleotide separation by TLC-chromatography revealed that ErTadZ exhibits a low intrinsic ATP hydrolysis rate under physiological buffer conditions, but lost ATPase activity under the buffer conditions used for crystallization (Fig. 5). In addition, we found that an ErTadZE162A mutation that substitutes the Mg2+-chelating glutamate residue in the ATP binding pocket with an alanine also lost ATPase activity. These results suggest that ErTadZ exhibits a low intrinsic ATPase activity.
Fig. 5.
ATPase activity of ErTadZ. (A) ATPase activities were assayed using [33P] α-labeled ATP (1mM). Under physiological buffer conditions, ErTadZ (red) shows a moderate ATP hydrolysis rate as compared to the previously reported ATPase MipZ (green), a distant homolog of ErTadZ and a spatial regulator of cell division in C. crescentus (Thanbichler et al., 2006). An ErTadZE162A point mutation (blue) that substitutes the Mg2+ chelating glutamine residue in the ATP binding pocket with an alanine residue abrogates ATPase activity under physiological buffer conditions. Similarly, the ATPase activity of wild type ErTadZ is lost under buffer conditions used for crystallization (purple). (B) The formation of ADP was quantified by PEI-cellulose TLC as shown for the wild type ErTadZ sample under physiological buffer conditions.
It is generally believed the second lysine in the Walker-A motif is required for ATP hydrolysis. For example, mutations of the second conserved lysine in MinD cause it to lose activity (de Boer et al., 1991; Hayashi et al., 2001; Hu et al., 2002). Our results indicate that ErTadZ has a low intrinsic ATPase activity at 35°C at physiological relevant buffer conditions, despite the absence of this highly conserved lysine. It remains possible, but highly speculative at present, that another protein can further stimulate the ATPase activity of ErTadZ, in vivo. Thus, since the crystallization buffer completely abolishes the ATPase activity, the crystal structure may represent a snapshot of ErTadZ locked in the ATP-bound state.
A conserved dimer mediated by ATP
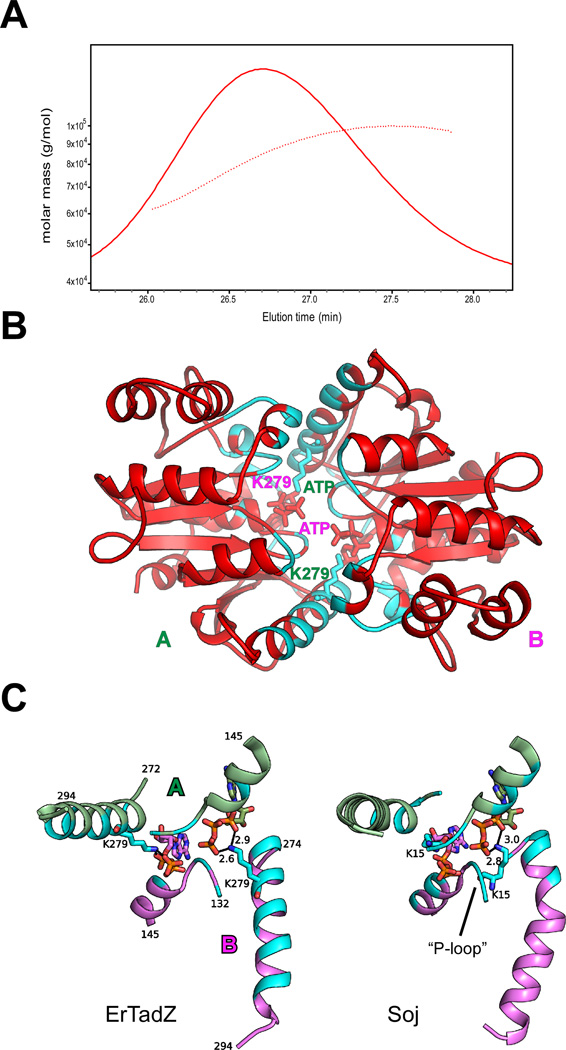
Liquid chromatography–mass spectrometry (LCMS) indicated that the molecular weight of an ErTadZ monomer was 42.46 kDa, which would correspond to the SeMet protein containing the purification tag. Analytical size exclusion gave an estimation of molecular weight the oligomer as 72.1 kDa (equivalent to 1.7 monomers, Fig. S3). A more accurate molar mass, estimated from Static Light Scattering (SLS) averaged across the majority of the peak, was 85.95 kDa as calculated by the ASTRA software (Wyatt Technology), with an oligomer number of 2.02, indicating that a dimer is the dominant species in solution (Fig. 6A).
Fig. 6.
A conserved dimer formed by the AAD domains. (A) Profile of the SEC/SLS experiment displaying the refractive index signal (SEC, solid trace) against elution time (minutes) for ErTadZ (red). The dotted trace represents the molar mass (g/mol) calculated according to SLS. BSA (bovine serum albumin, 66.4 kDa) was used as the standard. (B) AAD dimer of ErTadZ with protomers labeled A and B. Lys279 and ATP are shown as sticks. Residues near the domain interface of ErTadZ highlighted in cyan. (C) A close-up view of ATP-mediated interactions between the two protomers of ErTadZ A and B (A, pale green; B, magenta). ATP plays a conserved role in both dimerization of ErTadZ (left) and Soj (right). Residues near the domain interface of ErTadZ highlighted in cyan. Hydrogen bonds are shown in dashed lines and their distance labeled in Å.
The dimer interface involves a relatively flat surface encompassing the bound ATP (Fig. 6B) and buries a surface area of ~2630 Å2 per monomer. The dimerization brings the ATPs and the glycine-rich P-loops of the two protomers together. The two equivalent helices (residues 138–152) following the P-loop align collinearly in a head-to-head arrangement, with the phosphoryl groups of the ATP molecules sandwiched in between. Similar dimeric assemblies are also conserved in CtTadZ and Soj (Fig. S4). In ErTadZ and CtTadZ, a helix (residues 274–293 in ErTadZ; Fig 6B) makes more significant contribution to the dimer interface, compared to Soj (Fig. 6C).
The first conserved lysine of the deviant Walker-A motif promotes dimer formation upon ATP binding, through interactions with the phosphoryl groups of the neighboring protomer (Leonard et al., 2005). Interestingly, Lys279 in ErTadZ, located on an α-helix adjacent to the P-loop, interacts with the β- and γ-phosphate of the neighbor ATP in the same manner, thus fulfilling a similar role as the first lysine in Walker-A motif (Fig. 6C). ATP plays an important role in the dimerization of ErTadZ by contributing 40% of the interaction surface. The ATP-mediated dimerization in CtTadZ is more similar to canonical ATPases as it interacts with ATP through the first conserved lysine of the deviant Walker-A motif. Moreover, this residue is also highly conserved (or substituted by an arginine) in other TadZ proteins (Fig. 4D). Thus, it is likely that TadZ proteins have preserved the function of utilizing ATP as a “molecular switch” in promoting dimer formation (Leonard et al., 2005).
Comparison of ErTadZ-ARD to CheY and other ARDs
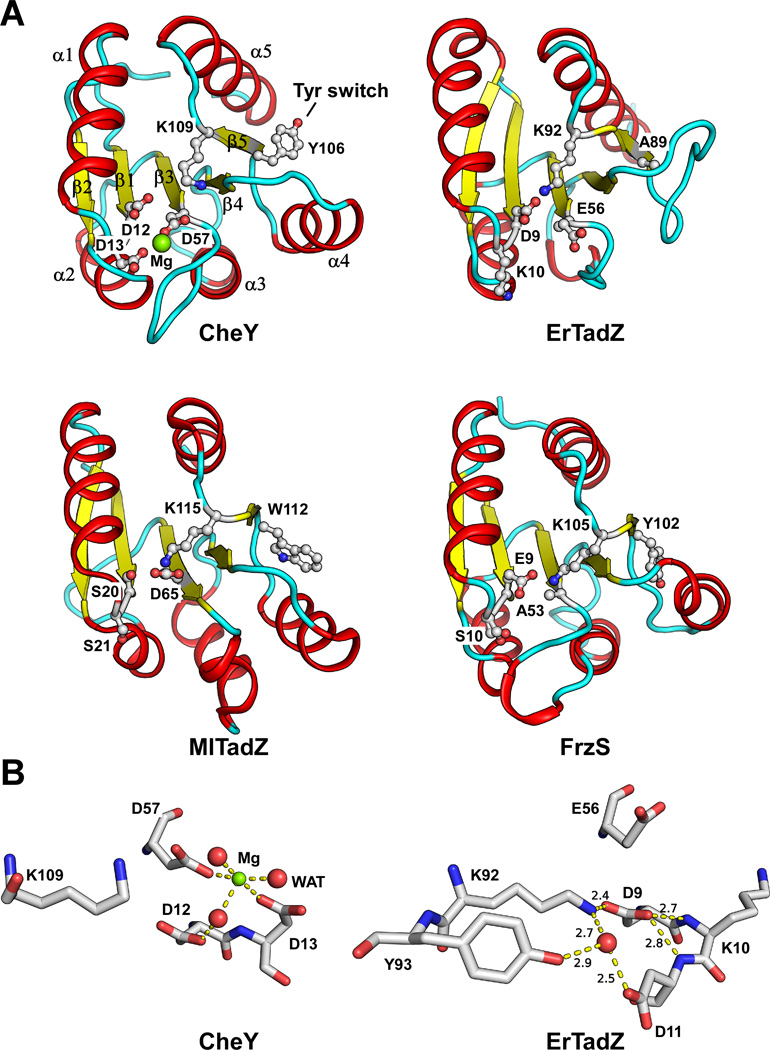
ARD is structurally similar to the RD of response regulators, such as the prototypical chemotaxis protein CheY (Stock et al., 1993). CheY functions as a molecule switch controlled by the phosphorylation state on Asp57. Several additional residues are conserved near the phosphorylation site, including Asp12, Asp13 and Lys109, which are needed either for Mg2+ coordination or for phosphorylation-induced conformational changes. CheY consists of a (βα)5 fold forming a three-layer sandwich with a hydrophobic β-sheet core protected on both sides by amphipathic helices. The structures of CheY and ARDs (from ErTadZ), a recently solved TadZ homolog from Mesorhizobium loti (MlTadZ, PDB code 3snk, JCSG, unpublished), and Myxococcus xanthus social motility protein FrzS (Fraser et al., 2007), are shown in Fig. 7A. The overall fold of the ARD of ErTadZ is similar to CheY, except that helix α3 is substituted by a 310 helix and α4 is absent. The “active site” of ErTadZ still partially resembles the chemotaxis protein CheY with several conserved residues, such as Asp9 and Lys92, suggesting its evolutionary origin from an RD. A glutamate (Glu56) replaces the aspartate at the site of phosphorylation, and is coupled with another change (D to K mutation) at one of the residues (Lys10), which chelates Mg2+ in canonical RDs. As a result of these changes, the environment in the “active site” of ErTadZ differs from that of CheY (Fig. 7B). In ErTadZ, the conserved Lys92 interacts directly with Asp9. MlTadZ and FrzS also have degraded “active sites”, but both preserve the interaction between the conserved lysine and an acidic residue in the “active site”. Thus, we conclude that ARD of ErTadZ has lost its ability to bind Mg2+ or to undergo phosphorylation due to the dramatic changes in the canonical active site. It is also interesting to note that the Asp to Glu substitution at the phosphorylation site mimics phosphorylation-based activation in canonical response regulators (Klose et al., 1993).
Fig. 7.
Structural comparisons of CheY and ARDs. (A) Structures of the ARD of ErTadZ (PDB code 3fkq), MlTadZ (PDB code 3snk) and FrzS (PDB code 2gkg) compared to the prototypical RD CheY (PDB code 2che). All molecules are shown in similar (superimposed) orientations. Key residues of CheY and equivalent residues of other proteins are shown as ball-and-sticks. The bound Mg2+ in CheY is shown as a green sphere. (B) Close-up views of the active site of CheY and the equivalent region in ErTadZ. Hydrogen (and magnesium coordination) bonds are shown as dashed lines and their distances are labeled (Å). Water molecules are shown as red spheres.
ARDs have been characterized in a number of proteins involved in different activities, such as bacteria CikA (Zhang et al., 2006; Gao et al., 2007), KaiA (Ye et al., 2004; Williams et al., 2002), plant pseudo-response regulators that control circadian rhythms (Mizuno et al., 2005), FrzS (Fraser et al., 2007), and Vibrio cholerae transcription regulator VpsT (Krasteva et al., 2010). Despite the structural similarities to canonical RDs, the sequence similarities among ARDs are very limited (Fig. S5). The activity of ARDs is phosphorylation-independent because they lack the invariant phospho-accepting Asp or other residues essential for phosphorylation (Makino et al., 2000; Fraser et al., 2007; Williams et al., 2002).
Phosphorylation of an RD induces conformational changes that are mapped to a region defined by α4β5α5 ("output surface"), which triggers the switching of a conserved tyrosine or phenylalanine from a buried to an exposed conformation (Tyr106 in CheY) (commonly referred as “Tyr/Phe switch”) (Bourret, 2010). ARDs seem to have retained the ability to form protein-protein interactions using a similar interface, but in a phosphorylation-independent manner, as demonstrated in FrzS (Fraser et al., 2007). The same region may undergo structure modification with the attachment of additional structural elements, such as an additional helix in VpsT that allows the binding of c-di-GMP (Krasteva et al., 2010). The corresponding region of ErTadZ is not structurally conserved (α5 is replaced by a loop), compared to CheY and other ARDs. Furthermore, the ARD of ErTadZ does not contain the “Tyr/Phe switch” (Ala89). Nonetheless, a conserved surface among close homologs of ErTadZ (Asp9, Asp11, Tyr14, Leu18, Lys92, Tyr93, and Gln94) near the “active site” suggests that this site may be functionally important (Fig. S1C). Thus, ErTadZ contains a more divergent ARD, whereas sequence analysis indicated that ARDs of AaTadZ and CcTadZ still contain the “Tyr/Phe switch” (Fig. S5).
Homology modeling of CcTadZ and identification of a potential protein docking site
Evidence from both structure and sequence of the TadZ family clearly suggest that they have evolved from fusion of RD and MinD-like ATPase domains, coupled with the loss of catalytic residues of their ancestors. The absence of canonical active sites in these proteins indicates that they are likely involved in mediating protein-protein interactions, consistent with their roles in pili assembly that involves the formation of multi-protein complexes. In order to better understand how the two domains of TadZ are utilized in the pili assembly process, we attempted to identify the potential protein docking sites on TadZ by identifying conserved residues on the protein surface. Clustering of conserved residues on the protein surface is often indicative of functionally important sites.
Sequences of the TadZ family are highly divergent, particularly in the ARD region. Most members of this family are more closely related to the last two domains of CcTadZ, compared to AaTadZ or ErTadZ. Consequently, we built a homology model for the more representative CcTadZ. The sequence of the ARD domain of CcTadZ (residues 125–251) was submitted to I-Tasser server (Zhang, 2008), and the top scoring model was selected for subsequent rounds of manually adjustments and energy minimized to reduce clashes. The resulting model is consistent with the properties of the RD architecture, which consists of conserved hydrophobic β-stands and amphipathic helices.
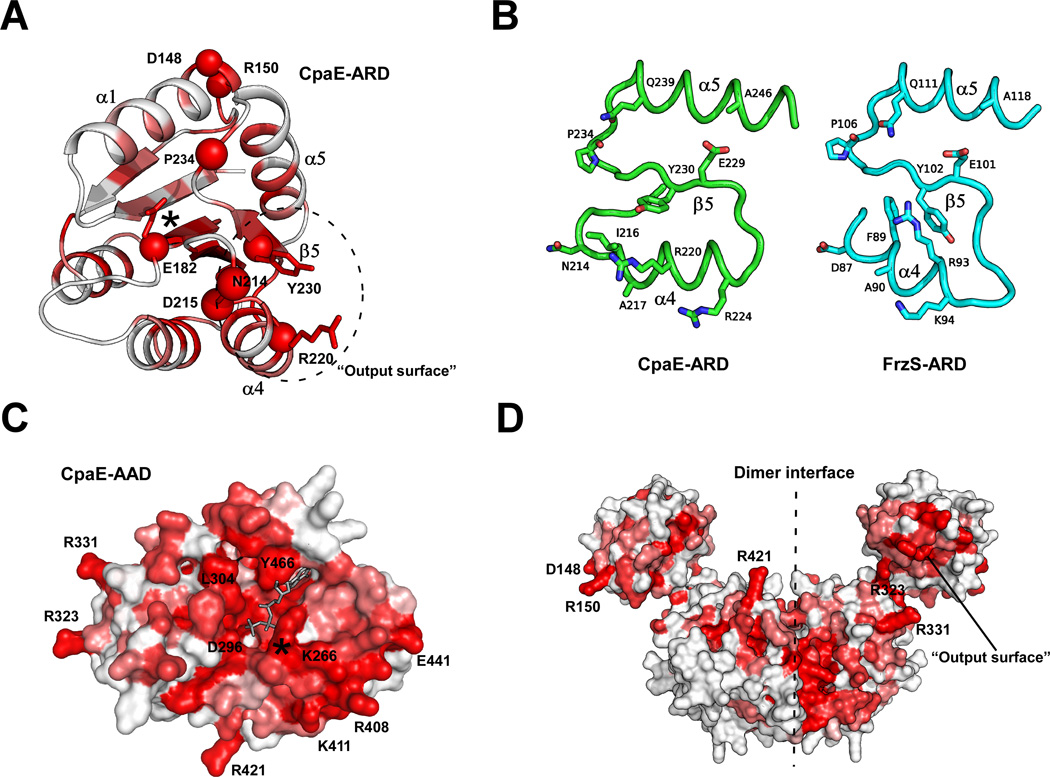
The ARD of CcTadZ is expected to adopt an (βα)5 fold as for canonical RDs (Fig. S5). Similar to ErTadZ, the “phosphorylation site” of CcTadZ is also occupied by a glutamate (Glu182), indicating that it is also an ARD (Fig. 8A). More interestingly, the “output surface” of the CcTadZ ARD is highly conserved, featuring an arginine-tyrosine pair (Arg220 and Tyr230). Many of the residues on the “output surface” of CcTadZ are also conserved in the ARD of FrzS (Fig. 8B).
Fig. 8.
Homology modeling of CcTadZ (CpaE) and mapping of conserved residues. (A) Homology model of the CcTadZ ARD domain colored by sequence conservation in a gradient from the non-conserved (white) to the most conserved (red). The Cα atoms of the most conserved residues are shown in red spheres. (B) Comparison of the “output surface” of the ARD domains of CcTadZ and FrzS. (C) Surface representation of the homology model of the AAD of CcTadZ colored by sequence conservation (color scheme same as A). ATP is shown as grey sticks. The P-loop region is denoted by a star. (D) A CcTadZ dimer model based on the ErTadZ dimer (color scheme same as A).
A model for the AAD of CcTadZ was also constructed in a similar fashion (Fig. 8C). Models of AAD and ARD of CcTadZ were then superimposed on the respective domains of the full length ErTadZ to generate a two-domain model for CcTadZ (residues 125–517, Fig. 8D). Conserved residues on the surface of AAD are all located on or near the dimer interface, indicating the functional importance of this interface (Fig. 8C). Several conserved arginines (Arg323, Arg331, and Arg421) are close to the domain interface between AAD and ARD.
Implications for polar localization
Type IV pili are generally localized to a single pole of the cell (Wall et al., 1999). It is not well understood how this cell polarity of type IV pili is established. A model was proposed based on studies of C. crescentus (Lawler et al., 2006; Huitema et al., 2006; Lam et al., 2006), which divides asymmetrically to produce two daughter cells that are functionally and morphologically different: a swarmer cell containing a flagellum and pili at one pole and a stalked cell with a stalk at the opposite pole. TipN localizes to the middle of the predivisional cell and is inherited by both daughter cells at the new poles after cell division, and then serves as a “molecular beacon” for establishing the new pole. In the stalked cell, which can further divide, TipN serves as a hub for proper localization of the developmental regulator PleC, the polar organelle development protein PodJ (Viollier et al., 2002b), and the pilus assembly protein CcTadZ to the new pole (where new pili will be generated in the swarmer daughter cell). CcTadZ is localized to one pole in the swarmer cell. The accompanying paper by Perez-Cheeks et al. shows polar localization of TadZ in A. actinomycetemcomitans (B.A. Perez-Cheeks, unpublished, submitted as a companion paper). Thus, TadZ likely plays a critical localization role in pili biogenesis. The structure of TadZ from E. rectale reveals a novel protein architecture consisting of an atypical receiver domain (ARD) and an atypical ATPase domain (AAD), thus representing an interesting example of gene fusion.
ARDs were previously found to be involved in mediating polar protein localization. The polar localization of the social mobility protein FrzS and the cyanobacterial circadian clock kinase CikA requires ARDs (Fraser et al., 2007; Zhang et al., 2006). ARD of FimX of P. aeruginosa, a phosphodiesterase that governs twitching motility, is also essential in its localization to a single cell pole (Kazmierczak et al., 2006). These ARDs were likely evolved from canonical polar targeting RDs. Several characterized canonical RDs are capable of localizing to the pole. For example, M. xanthus RomR localizes bipolar asymmetrically with a large cluster at the lagging pole using its RD as a localization module (Leonardy et al., 2007). C. crescentus CpdR has a phosphorylation-dependent, localization pattern and plays an important role in cell cycle control (Iniesta et al., 2006). Two other C. crescentus response regulators, DivK and PleD, also localize to the cell poles, which is regulated by the phosphorylation states of their RDs (Chan et al., 2004; Lam et al., 2003).
Since ARDs are evolved from RDs, it has been speculated that ARD may interact with histidine kinase (HK) by mimicking the His-Asp phosphor-transfer complex. Analysis of the clustering of highly conserved surface residues reveals two potential protein-protein docking sites on the ARD of TadZ, which map to the canonical active site and the “output surface” respectively. The putative binding surface of ErTadZ-ARD is near Lys92/Tyr93 (Fig. S1), corresponding to a location that is directly involved in the interaction between RD and HK in a two-component system, suggesting that ErTadZ may have retained the ability to bind an HK. Recognition of an HK by the ARD in TadZ raises the possibility of regulating pili biogenesis by a two-component signaling system. Computational analysis of CcTadZ and its close homologs suggests another protein-protein docking site that overlaps with the “output surface” of the canonical RD (see previous section), which is absent in ErTadZ. This site is close to, but does not overlap with, the conserved site in ErTadZ. More interestingly, we showed that the ARD of CcTadZ has an “output surface” similar to that of FrzS (Fig. 8B). This finding is surprising since no functional relationship between CcTadZ and FrzS was expected, even though both are involved in polar localization and are functionally related to type IV pili. FrzS is essential for the social mobility of M. xanthus, which is powered by type IV pili that consist of an N-terminal ARD and a coil-coiled tail, which is believed to interact with an ATPase motor (Ward et al., 2000; Mauriello et al., 2010). The architecture of an ARD associated with an ATPase is similar in FrzS and TadZ. Therefore, the structural analysis suggests that the ARD of TadZ likely functions as a localization module.
Based on the significant structural similarities, the AAD of TadZ may have functional similarities to MinD. The dimerization of MinD allows its stable binding to the membrane via a C-terminal amphipathic helix and the subsequent recruitment of MinC and MinE (Zhou et al., 2005). It is feasible that the AAD may also bind membrane in a similar manner once TadZ is localized to the pole, if a similar amphipathic helix exists. In contrast to MinD that oscillates from pole to pole at the expense of ATP, TadZ is localized to a single pole.
Thus, we propose that TadZ localizes to the leading pole via the ARD at an early stage in pili biosynthesis. ARD likely recognizes a landmark that has previously been established at the pole, such as a component of the TipN-PodJ-PleC multi-protein complex in C. crescentus. The AAD, or other regions of ARD, may then serve as a docking portal for recruiting/assembling other pili biosynthesis components. Interacting partners of TadZ are currently unknown. The first likely candidates are the TadA ATPase motor and inner membrane proteins, TadB and TadC, since they always appear together in genomes. Pseudo-pilins (TadE and TadF), which are generally believed to locate at the base of type IV pili, are also possible binding partners. In a few bacteria, such as Streptomyces avermitilis, TadZ is fused to a C-terminal TadE tail (Fig. 1B).
Figurski and coworkers have undertaken an analysis of the functional roles of AaTadZ and its domains (B.A. Perez-Cheeks, et al., unpublished, submitted as a companion paper). Remarkably, they find that AaTadZ is also localized to the leading pole despite significant sequence divergence, indicating a conserved role for TadZ. Both ARD and AAD of AaTadZ are required for the localization to the pole since both the N-terminal and the C-terminal deletion mutants of AaTadZ failed to localize. They show that the AAD of TadZ indeed shares functional similarity with MinD by interacting with the membrane through the C-terminal amphipathic helix. Thus, our structural studies have revealed new potential regulatory functions mediated by the ErTadZ family proteins.
Experimental procedures
Protein production and crystallization
Clones were generated using the Polymerase Incomplete Primer Extension (PIPE) cloning method (Klock et al., 2008). The gene encoding ErTadZ was amplified by polymerase chain reaction (PCR) from E. rectale genomic DNA using PfuTurbo DNA polymerase (Stratagene) and I-PIPE (Insert) primers (forward primer ErTadZfw, reverse primer ErTadZrv, Table S1) that included sequences for the predicted 5' and 3' ends. The expression vector, pSpeedET, which encodes an amino-terminal tobacco etch virus (TEV) protease-cleavable expression and purification tag (MGSDKIHHHHHHENLYFQ/G), was PCR amplified with V-PIPE (Vector) primers (forward primer: 5’-taacgcgacttaattaactcgtttaaacggtctccagc-3’, reverse primer: 5’-gccctggaagtacaggttttcgtgatgatgatgatgatg-3’). V-PIPE and I-PIPE PCR products were mixed to anneal the amplified DNA fragments together. Escherichia coli GeneHogs (Invitrogen) competent cells were transformed with the I-PIPE / V-PIPE mixture and dispensed on selective LB-agar plates. The cloning junctions were confirmed by DNA sequencing. Expression was performed in a selenomethionine-containing medium at 37°C with suppression of normal methionine synthesis. At the end of fermentation, lysozyme was added to the culture to a final concentration of 250 µg/ml, and the cells were harvested and frozen. After one freeze/thaw cycle, the cells were homogenized in lysis buffer [50 mM HEPES pH 8.0, 50 mM NaCl, 10mM imidazole, 1 mM Tris(2-carboxyethyl)phosphine-HCl (TCEP)] and passed through a Microfluidizer (Microfluidics). The lysate was clarified by centrifugation at 32,500 g for 30min and loaded onto a nickel-chelating resin (GE Healthcare) pre-equilibrated with lysis buffer, the resin washed with wash buffer [50 mM HEPES pH 8.0, 300 mM NaCl, 40 mM imidazole, 10% (v/v) glycerol, 1 mM TCEP], and the protein eluted with elution buffer [20 mM HEPES pH 8.0, 300 mM imidazole, 10% (v/v) glycerol, 1 mM TCEP]. The eluate was buffer exchanged with HEPES crystallization buffer [20 mM HEPES pH 8.0, 200 mM NaCl, 40 mM imidazole, 1mM TCEP] and concentrated to 18.1 mg/ml using centrifugal ultrafiltration (Millipore). ErTadZ was crystallized by mixing 200nl protein with 200 nl crystallization solution over a 50 µl reservoir volume using the nanodroplet vapor diffusion method (Santarsiero et al., 2002) with standard JCSG crystallization protocols (Lesley et al., 2002). The crystallization reagent that produced the ErTadZ crystal used for structure solution consisted of 2.1 M ammonium sulfate, 2.25% polyethylene glycol 400, 15.0% glycerol, and 0.1 M HEPES pH 7.57. The crystal used for structure determination was harvested after 36 days at 293K. MPD was added to the crystal as a cryoprotectant to a final concentration of 15% (v/v). Initial screening for diffraction was carried out using the Stanford Automated Mounting (SAM) system (Cohen et al., 2002) and an X-ray microsource installed on a SSRL beamline (Menlo Park, CA). The crystal was indexed in monoclinic space group C2.
Data collection, structure solution, and refinement
MAD data were collected at wavelengths corresponding to the inflection, high energy remote, and peak of a selenium MAD experiment at 100 K using Mar CCD 325 detector (Rayonix) at SSRL beamline 11-1. Data processing and structure solution were carried out using an automated structure solution protocols developed at the JCSG. In summary, the MAD data were integrated and reduced using MOSFLM (Leslie, 1992) and then scaled with the program SCALA (Evans, 2006). Location of selenium sites, initial phasing, and identification of the space group were carried out using SHELXD (Sheldrick, 2008). Phase refinement and model building were performed using autoSHARP (Bricogne et al., 2003) and wARP (Cohen et al., 2004). The above process produced excellent density maps and resulted in an initial model that was ~96% complete. Further model completion and refinement were performed manually with COOT (Emsley et al., 2004) and REFMAC5 (Murshudov et al., 1997). The refinement included experimental phase restraints in the form of Hendrickson-Lattman coefficients and TLS refinement with 2 TLS groups (residues 1–114, 115–354). Data and refinement statistics are summarized in Table 1. Analysis of the stereochemical quality of the model was accomplished using MolProbity (Davis et al., 2004). Molecular graphics were prepared with PyMOL (DeLano Scientific). Sequence logo was produced by Weblogo (Crooks et al., 2004).
Gel filtration analysis
The oligomeric state of ErTadZ in solution was determined using a 1 × 30 cm2 Superdex 200 column (GE Healthcare) coupled with miniDAWN static light scattering (SEC/SLS) and Optilab differential refractive index detectors (Wyatt Technology). The mobile phase consisted of 20 mM Tris pH 8.0, 150 mM sodium chloride, and 0.02% (w/v) sodium azide. The molecular weight was calculated using ASTRA 5.1.5 software (Wyatt Technology).
Nucleotide detection by MALDI Mass Spectrometry
A protein sample of 100µM ErTadZ in 20mM HEPES buffer pH 8.0, 200mM NaCl, 5mM β-mercaptoethanol was heat inactivated at 94°C for 5 min and centrifuged at 10,000 × g for 5 min. Nucleotide pools in the supernatant were analyzed on a MALDI-MS instrument (Voyager-DE, Applied Biosystems).
Site-directed mutagenesis of ErTadZ
Primers ErTadZE162Afw, pSpeedETfw, ErTadZE162Arv, and pSpeedETrv (Table S1) were used to PCR-amplify DNA sequences covering the first 508bp and the last 606bp of the ErTadZ ORF from the pSpeedET::ErTadZ expression vector. PCR products were spliced together in a subsequent SOE-PCR step using the outside primers pSpeedETfw and pSpeedETrv. The resulting SOE-PCR product was cloned into the PacI and AgeI sites of pSpeedET::ErTadZ to produce the pSpeedET::ErTadZE162A construct. The engineered mutation in the ErTadZE162A allele that changed Glu162 (GAA) into Ala (GCA) was confirmed by sequencing.
ATPase assay
Hexahistidine-tagged ErTadZ (10µM) or the ATPase MipZ (10µM) was preincubated in 20mM HEPES buffer pH 8.0, 200mM NaCl, 10mM MgCl2, 5mM β-mercaptoethanol. To assess the ATPase turnover rate of ErTadZ under crystallization conditions, a sample of ErTadZ (10um) was preincubated in crystallization buffer (0.1 M HEPES pH 7.57, 2.1 M ammonium sulfate, 15.0% glycerol) supplemented with 10mM MgCl2. Reactions were supplemented with 1mM ATP containing 30mCi of α-labeled [33P] ATP (3000 Ci/mmol, Perkin Elmer) and incubated at 35°C. Aliquots were taken at regular time intervals and heat-inactivated at 94°C for 5 min. Samples were spotted onto polyethyleneimine-cellulose thin-layer chromatography plates (Merck, Darmstadt) and nucleotide were separated using a mobile phase consisting of 0.5M KH2PO4 pH 3.4. Radioactive ATP and ADP species were detected on a Storage PhosphorScreen (Amersham Bioscience) and quantified using imageJ software v1.43.
Supplementary Material
Acknowledgements
We thank the members of the JCSG high-throughput structural biology pipeline for their contribution to this work. The JCSG is supported by the NIH, National Institute of General Medical Sciences, Protein Structure Initiative grants U54 GM094586 and GM074898. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource (SSRL), SLAC National Accelerator Laboratory. The SSRL is a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health (National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences). The genome of E. rectale was a gift of Dr. Jeffrey Gordon, Washington University in St. Louis, School of Medicine. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
Accession code
Atomic coordinates and experimental structure factors for ErTadZ at 2.1 Å resolution have been deposited in the PDB code under accession code 3fkq.
Supporting information
Additional supporting information may be found in the online version of this article.
References
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee MK, Kachlany SC, Fine DH, Figurski DH. Nonspecific adherence and fibril biogenesis by Actinobacillus actinomycetemcomitans: TadA protein is an ATPase. J Bacteriol. 2001;183:5927–5936. doi: 10.1128/JB.183.20.5927-5936.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret RB. Receiver domain structure and function in response regulator proteins. Curr Opin Microbiol. 2010;13:142–149. doi: 10.1016/j.mib.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci USA. 2004;101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Viollier PH, Shapiro L. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol Microbiol. 2005;55:1085–1103. doi: 10.1111/j.1365-2958.2004.04443.x. [DOI] [PubMed] [Google Scholar]
- Christen B, Fero MJ, Hillson NJ, Bowman G, Hong SH, Shapiro L, McAdams HH. High-throughput identification of protein localization dependency networks. Proc Natl Acad Sci USA. 2010;107:4681–4686. doi: 10.1073/pnas.1000846107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AE, Ellis PJ, Miller MD, Deacon AM, Phizackerley RP. An automated system to mount cryo-cooled protein crystals on a synchrotron beamline, using compact samples cassettes and a small-scale robot. J Appl Crystallogr. 2002;35:720–726. doi: 10.1107/s0021889802016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SX, Morris RJ, Fernandez FJ, Ben Jelloul M, Kakaris M, Parthasarathy V, et al. Towards complete validated models in the next generation of ARP/wARP. Acta Cryst D. 2004;60:2222–2229. doi: 10.1107/S0907444904027556. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bentzmann S, Aurouze M, Ball G, Filloux A. FppA, a novel Pseudomonas aeruginosa prepilin peptidase involved in assembly of type IVb pili. J Bacteriol. 2006;188:4851–4860. doi: 10.1128/JB.00345-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer PA, Crossley RE, Hand AR, Rothfield LI. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J. 1991;10:4371–4380. doi: 10.1002/j.1460-2075.1991.tb05015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Flint HJ. Proposal of a neotype strain (A1-86) for Eubacterium rectale. Request for an opinion. Int J Syst Evol Microbiol. 2008;58:1735–1736. doi: 10.1099/ijs.0.2008/004580-0. [DOI] [PubMed] [Google Scholar]
- Elsliger M-A, Deacon AM, Godzik A, Lesley SA, Wooley J, Wuthrich K, Wilson IA. The JCSG high-throughput structural biology pipeline. Acta Cryst F. 2010;66:1137–1142. doi: 10.1107/S1744309110038212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Cryst D. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Fraser JS, Merlie JP, Jr, Echols N, Weisfield SR, Mignot T, Wemmer DE, et al. An atypical receiver domain controls the dynamic polar localization of the Myxococcus xanthus social motility protein FrzS. Mol Microbiol. 2007;65:319–332. doi: 10.1111/j.1365-2958.2007.05785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T, Zhang X, Ivleva NB, Golden SS, LiWang A. NMR structure of the pseudo-receiver domain of CikA. Protein Sci. 2007;16:465–475. doi: 10.1110/ps.062532007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi I, Oyama T, Morikawa K. Structural and functional studies of MinD ATPase: implications for the molecular recognition of the bacterial cell division apparatus. EMBO J. 2001;20:1819–1828. doi: 10.1093/emboj/20.8.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Gogol EP, Lutkenhaus J. Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. Proc Natl Acad Sci USA. 2002;99:6761–6766. doi: 10.1073/pnas.102059099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH. Bacterial birth scar proteins mark future flagellum assembly site. Cell. 2006;124:1025–1037. doi: 10.1016/j.cell.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc Natl Acad Sci USA. 2006;103:10935–10940. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachlany SC, Planet PJ, Bhattacharjee MK, Kollia E, DeSalle R, Fine DH, Figurski DH. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J Bacteriol. 2000;182:6169–6176. doi: 10.1128/jb.182.21.6169-6176.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachlany SC, Planet PJ, DeSalle R, Fine DH, Figurski DH. Genes for tight adherence of Actinobacillus actinomycetemcomitans: from plaque to plague to pond scum. Trends Microbiol. 2001a;9:429–437. doi: 10.1016/s0966-842x(01)02161-8. [DOI] [PubMed] [Google Scholar]
- Kachlany SC, Planet PJ, Desalle R, Fine DH, Figurski DH, Kaplan JB. flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans. Mol Microbiol. 2001b;40:542–554. doi: 10.1046/j.1365-2958.2001.02422.x. [DOI] [PubMed] [Google Scholar]
- Kazmierczak BI, Lebron MB, Murray TS. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol Microbiol. 2006;60:1026–1043. doi: 10.1111/j.1365-2958.2006.05156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klock HE, Koesema EJ, Knuth MW, Lesley SA. Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins. 2008;71:982–994. doi: 10.1002/prot.21786. [DOI] [PubMed] [Google Scholar]
- Klose KE, Weiss DS, Kustu S. Glutamate at the site of phosphorylation of nitrogen-regulatory protein NTRC mimics aspartyl-phosphate and activates the protein. J Mol Biol. 1993;232:67–78. doi: 10.1006/jmbi.1993.1370. [DOI] [PubMed] [Google Scholar]
- Koonin EV. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J Mol Biol. 1993;229:1165–1174. doi: 10.1006/jmbi.1993.1115. [DOI] [PubMed] [Google Scholar]
- Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H, Matroule JY, Jacobs-Wagner C. The asymmetric spatial distribution of bacterial signal transduction proteins coordinates cell cycle events. Dev Cell. 2003;5:149–159. doi: 10.1016/s1534-5807(03)00191-6. [DOI] [PubMed] [Google Scholar]
- Lam H, Schofield WB, Jacobs-Wagner C. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell. 2006;124:1011–1023. doi: 10.1016/j.cell.2005.12.040. [DOI] [PubMed] [Google Scholar]
- Lawler ML, Brun YV. A molecular beacon defines bacterial cell asymmetry. Cell. 2006;124:891–893. doi: 10.1016/j.cell.2006.02.027. [DOI] [PubMed] [Google Scholar]
- Leonard TA, Butler PJ, Lowe J. Bacterial chromosome segregation: structure and DNA binding of the Soj dimer--a conserved biological switch. EMBO J. 2005;24:270–282. doi: 10.1038/sj.emboj.7600530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardy S, Freymark G, Hebener S, Ellehauge E, Sogaard-Andersen L. Coupling of protein localization and cell movements by a dynamically localized response regulator in Myxococcus xanthus. EMBO J. 2007;26:4433–4444. doi: 10.1038/sj.emboj.7601877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesley SA, Kuhn P, Godzik A, Deacon AM, Mathews I, Kreusch A, et al. Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc Natl Acad Sci USA. 2002;99:11664–11669. doi: 10.1073/pnas.142413399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4+ESF-EAMCB Newsletter on Protein Crystallography. 1992;(No. 26) [Google Scholar]
- Lutkenhaus J, Sundaramoorthy M. MinD and role of the deviant Walker A motif, dimerization and membrane binding in oscillation. Mol Microbiol. 2003;48:295–303. doi: 10.1046/j.1365-2958.2003.03427.x. [DOI] [PubMed] [Google Scholar]
- Makino S, Kiba T, Imamura A, Hanaki N, Nakamura A, Suzuki T, et al. Genes encoding pseudo-response regulators: insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol. 2000;41:791–803. doi: 10.1093/pcp/41.6.791. [DOI] [PubMed] [Google Scholar]
- Mauriello EM, Mouhamar F, Nan B, Ducret A, Dai D, Zusman DR, Mignot T. Bacterial motility complexes require the actin-like protein, MreB and the Ras homologue, MglA. EMBO J. 2010;29:315–326. doi: 10.1038/emboj.2009.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignot T, Merlie JP, Jr, Zusman DR. Two localization motifs mediate polar residence of FrzS during cell movement and reversals of Myxococcus xanthus. Mol Microbiol. 2007;65:363–372. doi: 10.1111/j.1365-2958.2007.05789.x. [DOI] [PubMed] [Google Scholar]
- Mizuno T, Nakamichi N. Pseudo-Response Regulators (PRRs) or True Oscillator Components (TOCs) Plant Cell Physiol. 2005;46:677–685. doi: 10.1093/pcp/pci087. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nika JR, Latimer JL, Ward CK, Blick RJ, Wagner NJ, Cope LD, et al. Haemophilus ducreyi requires the flp gene cluster for microcolony formation in vitro. Infect Immun. 2002;70:2965–2975. doi: 10.1128/IAI.70.6.2965-2975.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody CR, Chung YJ, Yen MR, Vidal-Ingigliardi D, Pugsley AP, Saier MH., Jr Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology. 2003;149:3051–3072. doi: 10.1099/mic.0.26364-0. [DOI] [PubMed] [Google Scholar]
- Planet PJ, Kachlany SC, Fine DH, DeSalle R, Figurski DH. The Widespread Colonization Island of Actinobacillus actinomycetemcomitans. Nat Genet. 2003;34:193–198. doi: 10.1038/ng1154. [DOI] [PubMed] [Google Scholar]
- Proft T, Baker EN. Pili in Gram-negative and Gram-positive bacteria - structure, assembly and their role in disease. Cell Mol Life Sci. 2009;66:613–635. doi: 10.1007/s00018-008-8477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarsiero BD, Yegian DT, Lee CC, Spraggon G, Gu J, Scheibe D, et al. An approach to rapid protein crystallization using nanodroplets. J Appl Cryst. 2002;35:278–281. [Google Scholar]
- Schreiner HC, Sinatra K, Kaplan JB, Furgang D, Kachlany SC, Planet PJ, et al. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc Natl Acad Sci USA. 2003;100:7295–7300. doi: 10.1073/pnas.1237223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM. A short history of SHELX. Acta Cryst A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Skerker JM, Shapiro L. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 2000;19:3223–3234. doi: 10.1093/emboj/19.13.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM, Martinez-Hackert E, Rasmussen BF, West AH, Stock JB, Ringe D, Petsko GA. Structure of the Mg(2+)-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis. Biochemistry. 1993;32:13375–13380. doi: 10.1021/bi00212a001. [DOI] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- Tomich M, Planet PJ, Figurski DH. The tad locus: postcards from the widespread colonization island. Nat Rev Microbiol. 2007;5:363–375. doi: 10.1038/nrmicro1636. [DOI] [PubMed] [Google Scholar]
- Viollier PH, Sternheim N, Shapiro L. A dynamically localized histidine kinase controls the asymmetric distribution of polar pili proteins. EMBO J. 2002a;21:4420–4428. doi: 10.1093/emboj/cdf454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Sternheim N, Shapiro L. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc Natl Acad Sci USA. 2002b;99:13831–13836. doi: 10.1073/pnas.182411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall D, Kaiser D. Type IV pili and cell motility. Mol Microbiol. 1999;32:1–10. doi: 10.1046/j.1365-2958.1999.01339.x. [DOI] [PubMed] [Google Scholar]
- Ward MJ, Lew H, Zusman DR. Social motility in Myxococcus xanthus requires FrzS, a protein with an extensive coiled-coil domain. Mol Microbiol. 2000;37:1357–1371. doi: 10.1046/j.1365-2958.2000.02079.x. [DOI] [PubMed] [Google Scholar]
- Williams SB, Vakonakis I, Golden SS, LiWang AC. Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: a potential clock input mechanism. Proc Natl Acad Sci USA. 2002;99:15357–15362. doi: 10.1073/pnas.232517099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Abdubek P, Astakhova T, Axelrod HL, Bakolitsa C, Cai X, et al. A conserved fold for fimbrial components revealed by the crystal structure of a putative fimbrial assembly protein (BT1062) from Bacteroides thetaiotaomicron at 2.2 Å resolution. Acta Cryst F. 2010a;66:1281–1286. doi: 10.1107/S1744309110006548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Abdubek P, Astakhova T, Axelrod HL, Bakolitsa C, Cai X, et al. Structure of a membrane-attack complex/perforin (MACPF) family protein from the human gut symbiont Bacteroides thetaiotaomicron. Acta Cryst F. 2010b;66:1297–1305. doi: 10.1107/S1744309110023055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S, Vakonakis I, Ioerger TR, LiWang AC, Sacchettini JC. Crystal structure of circadian clock protein KaiA from Synechococcus elongatus. J Biol Chem. 2004;279:20511–20518. doi: 10.1074/jbc.M400077200. [DOI] [PubMed] [Google Scholar]
- Zhang X, Dong G, Golden SS. The pseudo-receiver domain of CikA regulates the cyanobacterial circadian input pathway. Mol Microbiol. 2006;60:658–668. doi: 10.1111/j.1365-2958.2006.05138.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Schulze R, Cox S, Saez C, Hu Z, Lutkenhaus J. Analysis of MinD mutations reveals residues required for MinE stimulation of the MinD ATPase and residues required for MinC interaction. J Bacteriol. 2005;187:629–638. doi: 10.1128/JB.187.2.629-638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.