Abstract
Chemotherapy response rates for advanced colorectal cancer remain disappointingly low, primarily due to drug resistance, so there is an urgent need to improve current treatment strategies. In order to identify novel determinants of resistance to the clinically relevant drugs 5-Fluorouracil (5-FU) and SN38 (the active metabolite of irinotecan), transcriptional profiling experiments were carried out on pre-treatment metastatic colorectal cancer biopsies and HCT116 parental and chemotherapy-resistant cell line models using a disease-specific DNA microarray. To enrich for potential chemo-resistance-determining genes, an unsupervised bioinformatics approach was employed, and 50 genes were selected and then functionally assessed using custom-designed siRNA screens. In the primary siRNA screen, silencing of 21 genes sensitised HCT116 cells to either 5-FU or SN38 treatment. Three genes (RAPGEF2, PTRF and SART1) were selected for further analysis in a panel of 7 CRC cell lines. Silencing SART1 sensitised all 7 cell lines to 5-FU treatment and 4/7 cell lines to SN38 treatment. However, silencing of RAPGEF2 or PTRF had no significant effect on 5-FU or SN38 sensitivity in the wider cell line panel. Further functional analysis of SART1 demonstrated that its silencing induced apoptosis that was caspase 8-dependent. Furthermore, silencing of SART1 led to a down-regulation of the caspase 8 inhibitor, c-FLIP, which we have previously demonstrated is a key determinant of drug resistance in colorectal cancer. This study demonstrates the power of systems biology approaches for identifying novel genes that regulate drug resistance and identifies SART1 as a previously unidentified regulator of c-FLIP and drug-induced activation of caspase 8.
Keywords: Drug resistance, colorectal cancer, systems biology, caspase 8, c-FLIP
Introduction
Resistance to chemotherapeutic drugs is a major problem in the treatment of many cancers. In colorectal cancer (CRC), the long established antimetabolite drug 5-fluorouracil (5-FU), even when used in combination with newer cytotoxic drugs such as oxaliplatin and irinotecan (CPT-11), still produces a response in only 50% of patients with advanced disease (1, 2). Hence, there is a pressing need for research to identify the key, clinically relevant molecular determinants of sensitivity to particular chemotherapy drugs as these may constitute novel predictive biomarkers of drug response and drug resistance. In addition, these key molecular determinants of drug sensitivity may identify novel therapeutic strategies for enhancing the clinical effectiveness of chemotherapy.
Recently, high throughput technologies such as DNA microarrays have been employed to identify panels of markers that predict prognosis (3-8) or response to treatments (9-11) based on the expression profiles of those genes. In this report, we carried out unsupervised analyses of microarray expression profiling data to identify gene lists that segregate advanced CRC patients based on response to 5-FU/CPT-11 therapy. In parallel, we profiled and carried out unsupervised analyses of paired drug sensitive and resistant CRC cell lines to further identify genes associated with drug resistance, Furthermore we have functionally tested whether any of the genes contained within these lists are functionally involved in chemotherapeutic resistance/sensitivity in colorectal cancer cell lines. For expression profiling in this study, we have used a Colorectal Cancer disease specific array (DSA), which contains 61,528 probesets and encodes 52,306 transcripts confirmed as being expressed in CRC and normal colorectal tissue. This array contains transcripts that have not been available for previous expression analysis studies (12). The generation and utility of other DSAs using a similar technical approach have been previously reported (13-15). The power of the current study is that we have used a systems biology approach of transcriptional profiling with functional testing of the identified genes to highlight potential novel drug targets and/or biomarkers, which could be used to potentially improve response rates for advanced CRC.
Materials and Methods
Patient Samples
These have been previously described (16). Briefly, twenty patients with metastatic colorectal cancer were included in the present study. All patients provided written fully informed consent as per IRB guidelines in the University of Southern California and approval was granted from this body. These patients underwent biopsy of colorectal liver metastases prior to commencing irinotecan/5-FU chemotherapy on the IFL schedule: CPT-11 125mg/m2 i.v. over 90 minutes, Leucovorin (LV) 20mg/m2 as i.v. bolus injection immediately prior to 5-FU and 5-FU 500mg/m2 as i.v. bolus injection administered weekly for four weeks and repeated every six weeks.
CT imaging for response evaluation using WHO criteria was performed every 6 weeks. Of these 20 patients, 1 had a complete response to treatment, 10 had a partial response to treatment and 9 had progressive disease on treatment. For the purpose of this study we have further defined ‘responders’ as those patients with either complete or partial response and ‘non-responders’ as those patients with progressive disease. We have specifically excluded patients with stable disease on chemotherapy from the study.
Materials
5-FU and SN38 were purchased from Sigma Chemical Co. (St. Louis, MO) and Abatra Technology Co, LTD (China) respectively.
Cell culture
All CRC cells were grown as previously described (17). Following receipt, cells were grown up, and as soon as surplus cells became available, they were frozen as a seed stock. All cells were passaged for a maximum of 2 months, after which new seed stocks were thawed for experimental use. All cell lines were tested for mycoplasma contamination at least every month. LS174T (2008), SW620 (2008) and RKO (2001) cells were obtained from the American Type Culture Collection [ATCC; authentication by short tandem repeat (STR) profiling/karyotyping/isoenzyme analysis] and maintained in Dulbecco’s modified Eagle’s medium (DMEM). The HCT116 human colon cancer cell line was kindly provided by Prof. Bert Vogelstein (Johns Hopkins University, Baltimore, MD) in 2003 and was grown in McCoy’s 5A medium. LoVo cells were provided for Astra Zeneca in 2003. HCT116, HT29, LoVo and RKO cell lines were validated by STR profiling by LGC Standards (18) in May 2011. The 5-FU- and oxaliplatin-resistant HCT116 sub-lines and were generated in our laboratory as previously described (19). The FLIP overexpressing HCT116 cell lines were previously described (20). All medium was supplemented with 10% dialysed FCS, 50μg/mL penicillin-streptomycin, 2mM L-glutamine, and 1mM sodium pyruvate (all medium and supplements from Invitrogen Life Technologies Corp., Paisley, United Kingdom). All cell lines were maintained at 37°C in a humidified atmosphere containing 5% CO2.
Microarray analysis
Total RNA was extracted from 20 pre-treatment metastatic tumour biopsies from patients with advanced colorectal cancer and also profiled on the Colorectal cancer DSA (Almac Diagnostics, Craigavon, UK). In addition, in vitro analyses were also carried out using the Colorectal cancer DSA. Briefly, HCT116 parental cells were either untreated (0h control) or treated with either 5μM 5-FU or 5nM SN38 for 24 hours as outlined in Fig. 2A (acutely altered genes). Also, untreated 5-FU-resistant and SN38-resistant cells were analyzed to identify those genes that are basally deregulated between parental and resistant cells. Total RNA was isolated from three independent experiments using the RNA STAT-60™ Total RNA isolation reagent (Tel-Test, Inc., Texas, USA) according to the manufacturer’s instructions. For both the clinical and in vitro studies, RNA was sent to Almac Diagnostics (Craigavon, UK) for cDNA synthesis, cRNA synthesis, fragmentation and hybridisation onto the Colorectal cancer DSA. Detailed experimental protocols and raw expression data are available within the ArrayExpress repository (21) (Accession number E-MEXP-1692 (Clinical analysis) and E-MEXP-1691 (in vitro analysis)).
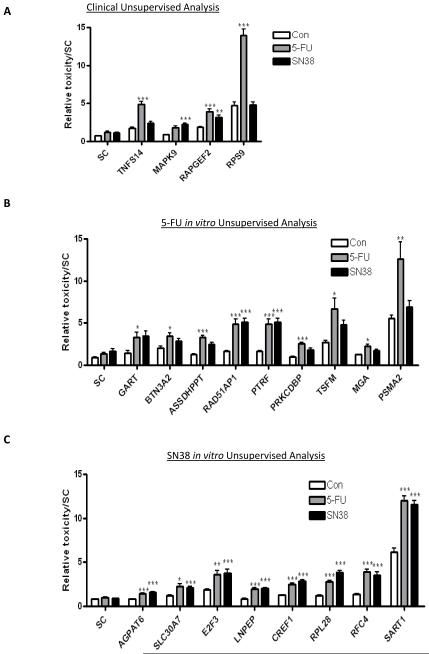
Figure 2.
Results from the initial siRNA screen in the HCT116 cells. The graphs show the relative toxicity, as assessed from MTT and toxilight assays, for all positive genes from the: (A) clinical analysis, (B) the 5-FU in vitro analysis, and (C) the SN38 in vitro analysis. Displayed are those genes that show an interaction between gene silencing and either 5-FU or SN38 treatment as measured by 2-way ANOVA. Synergistic interactions are highlighted as *=p<0.05, **=p<0.01 and ***=p<0.001.
Unsupervised Classification Analysis
Unsupervised classification analysis was carried out using Principal Components Analysis (PCA). All PCA was carried out using the Partek® software (version 6.3, Partek Inc., St. Louis, MO, USA). Briefly, each microarray experiment (5-FU basal, 5-FU inducible, SN38 basal, SN38 inducible and 5-FU/irinotecan clinical) was initially normalised and then underwent minimal flag filtering prior to PCA analysis. Following PCA analysis of each experiment, the results were examined to determine which principal component (PC) lead to the greatest separation between the two treatment groups. The PC that lead to the maximal treatment group separation was then isolated and the top ten positive and top ten negative component loadings were listed for each experiment.
Quantitative reverse transcription-PCR analysis
Total RNA was isolated as described above. Reverse transcription was carried out using 2μg of RNA using a Moloney murine leukemia virus–based reverse transcriptase kit (Invitrogen) according to the manufacturer’s instructions. Quantitative reverse transcription-PCR (RT-PCR) amplification was carried out in a final volume of 10μL containing 5μL of 2xSYBR green master mix (Qiagen), 4μL of primers (2μM), and 1μl of cDNA using an Opticon DNA Engine Thermal Cycler (Bio-Rad Laboratories, Inc., Waltham, MA) using methods previously described (16). All amplifications were primed by pairs of chemically synthesized 18- to 22-mer oligonucleotides designed using the Primer3 primer design software (22).
Statistical Analysis
All t tests and 2-way ANOVAs were calculated using the GraphPad software (Prism4). Specifically, t tests were unpaired, 2-tailed using 95% confidence intervals. 2-way ANOVA was calculated using 95% confidence intervals and a Bonferroni post-hoc test.
siRNA Plate Analysis
All siRNAs were supplied by Qiagen (Crawley, UK). siRNA screening was performed using siRNAs targeting pre-selected genes identified from microarray analysis. All Stars Negative control and All Stars Death control were used as non-targeting (scrambled) and positive controls, respectively. Transfection conditions were optimized using siRNAs with known effect on cell survival (FLIP and XIAP) (23-25). HCT116 cells were reverse-transfected using HiPerFect transfection reagent (Qiagen, Crawley, UK) to a final concentration of 5nM siRNA. After 24h, drug/solvent control was added: 5FU and SN38 (~IC30 doses). 48 hours later, MTT (cell viability) and ToxiLight (cell death) (Promega UK Ltd) assays were performed. The measurement from each well was normalised to the scrambled control. ToxiLight values were divided by MTT values to give a measurement of ‘Relative toxicity’ for each silenced gene. Significance and interaction effects were measured using t-tests, one-way ANOVA and two-way ANOVA to assess statistical significance of changes compared to control siRNA.
Cell viability analysis
Cell viability was determined using MTT ((3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide). For the siRNA plate analysis the analysis was carried out at 72h post transfection. For the combination index (CI) synergy calculation the analysis was carried out at 24h or 48h post transfection and drug treatment. CI values <1, 1, and >1 indicating synergism, additivity, and antagonism, respectively. For synergistic interactions, CI values between 0.8-0.9 indicate slight synergy, 0.6-0.8 indicate moderate synergy, 0.4-0.6 indicate synergy and those <0.4 indicate strong synergy (26).
Western Blot analysis
Western blot analysis was carried out as previously described SART1 (Abcam, Cambridge, UK), FLIP (NF6; Alexis, San Diego, CA), Caspase 8 (12F5; Alexis) and Poly-ADP-Ribose-Polymerase-1 (PARP; eBioscience, San Diego, CA) mouse monoclonal antibodies were used in conjunction with a horseradish peroxidase-conjugated sheep anti-mouse secondary antibody (Amersham, Buckinghamshire, UK). Equal loading was assessed using GAPDH (AbD Serotec, Kidlington, UK).
Flow cytometric analysis
Annexin V/PI analysis was carried out using the EPICS XL Flow Cytometer (Coulter, Miami, FL, USA). Cells were harvested and analyzed according to manufacturer’s instructions (BD Biosciences). Annexin V/PI staining was carried out at 72h post transfection.
Results
Transcriptional profiling of metastatic CRC biopsies and drug resistant cell lines
Using the Colorectal DSA, we performed microarray expression profiling of pre-treatment metastatic CRC patient biopsies (n=20). After appropriate background corrections and normalisations using the Robust Multichip Average (RMA) method (27), expression values from all the 61,528 probes were used for further analysis. In addition, we also carried out an in vitro transcriptional profiling experiment using the same platform. We used a HCT116 colorectal cancer cell line panel made up of parental drug sensitive cells and daughter cell lines with acquired resistance to 5-FU or SN38 (the active metabolite of irinotecan) (19). The transcriptional profiles of the HCT116 parental cells following treatment with either 5μM 5-FU or 5nM SN38 for 24h were examined. In addition, we also compared the basal transcriptional profiles of the HCT116 5-FU-resistant and SN38-resistant daughter cell lines with the parental cell line. All in vitro microarray analyses were validated by quantitative RT-PCR and the results demonstrated a strong overall concordance with the original microarray study (16) (Supplementary Table 1). In order to identify potential targets that may regulate drug sensitivity, we used the unsupervised classification approach of Principal Components Analysis (PCA).
Unsupervised classification
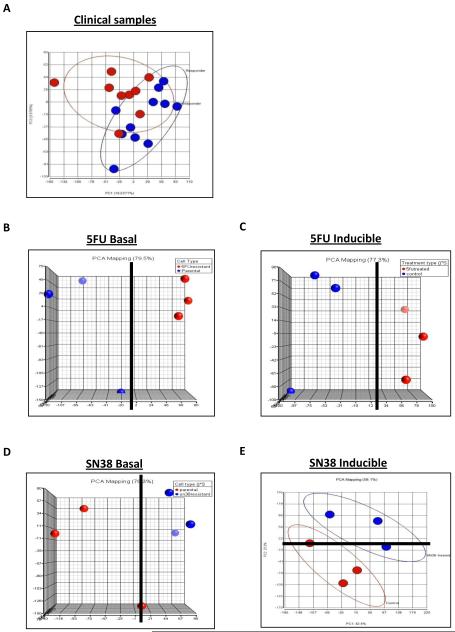
For the unsupervised analysis, five ‘experiments’ were created: clinical (responders v non-responders), 5-FU basal (sensitive v resistant), 5-FU inducible (parental untreated v parental 5-FU treated), SN38 basal (sensitive v resistant) and SN38 inducible (parental untreated v parental treated). Principal Components Analysis (PCA) was used as the unsupervised method, and all data from each of the five experiments was initially flag filtered. For the clinical analysis it was the 3rd Principal Component (PC) (9.8% variability) that gave the best separation based on patient response, although within this there were still a number of mis-classifications (Figure 1A). The top 10 probesets that correlated positively or negatively with PC3 were selected (Supplementary Table 2). For the 5-FU in vitro experiments (basal and inducible), PCA of the flag-filtered data showed a clear separation of sample groups. In both cases, this distinction was evident from the first principal component (Figures 1B and 1C). The top 10 probesets from the extreme positive or negative values of the component loadings that correlated maximally with PC1 were selected (Supplementary Tables 3 and 4). For the SN38 in vitro experiments, the PC that accounted for the maximum variability in the dataset, PC1 (35.8%), was able to differentiate between the basal expression changes observed between parental and SN38-resistant daughter cells, although one of the parental cells was misaligned (Figure 1D). However, in the case of SN38 treatment in the parental cells, separation based on drug treatment was only evident from the second principal component (PC2: 25.6%) of the dataset, with one replicate from each class misaligned (Figure 1E). In each case the top 10 probesets from the extreme positive or negative values of the component loadings that correlated maximally with PC1 (SN38 basal) or PC2 (SN38 inducible) were selected (Supplementary Table 5 and 6). For each of the above described experiments, 5 probesets from the top ten positive and negative component loadings were selected for further functional analysis (Table 1); those that were omitted were either transcribed sequences or in antisense orientation.
Figure 1.
Results from unsupervised classification analysis using principal component analysis (PCA) for (A) clinical data displaying PC1 against PC3, (B) in vitro 5-FU basal data displaying PC1 against PC2, (C) in vitro 5-FU inducible data displaying PC1 against PC2, (D) in vitro SN38 basal data displaying PC1 against PC2 and (E) in vitro SN38 inducible data displaying PC1 against PC2. A solid black line denotes the point of separation in the respective PC. Each sample group is displayed in either red or blue.
Table 1.
The top 50 genes identified from PCA of the clinical, 5-FU in vitro basal, 5-FU in vitro inducible, SN38 in vitro basal and SN38 in vitro inducible transcriptional profiling experiments. Listed is the probe ID, gene symbol and gene name.
| ProbeID | Gene Symbol | Gene Name |
|---|---|---|
| ADXCRIH.1569.C1_s_at | RPS27L | Homo sapiens ribosomal protein S27-like (RPS27L) mRNA. |
| ADXCRAD_BI820604_s_at | MAPK9 | Homo sapiens mitogen-activated protein kinase 9 (MAPK9) transcript |
| ADXCRAG_AF536980_at | PDE4D | Homo sapiens cAMP-specific phosphodiesterase (PDE4D) mRNA 3 untranslatedregion p |
| ADXCRIH.4.C1_at | RPS9 | Homo sapiens ribosomal protein S9 (RPS9) mRNA. |
| ADXCRPDRC.2813.C1_s_at | CYB5D2 | cytochrome b5 domain containing 2 (CYB5D2) mRNA. |
| ADXCRAG_BX640769_x_at | RAPGEF2 | Rap guanine nucleotide exchange factor (GEF) 2 |
| ADXCRAG_AL133646_s_at | GMEB2 | Homo sapiens glucocorticoid modulatory element binding protein 2 |
| ADXCRAG_AL832400_x_at | NLRP1 | NLR family pyrin domain containing 1 |
| ADXCRAG_NM_152345_s_at | ANKRD13B | Homo sapiens ankyrin repeat domain 13B (ANKRD13B) mRNA |
| ADXCRAD_CV571495_x_at | TNFSF14 | Homo sapiens tumor necrosis factor (ligand) superfamily member 14 |
| ADXCRPD.6977.C1_at | GART | Homo sapiens phosphoribosylglycinamide formyltransferase |
| ADXCRIH.2762.C1_s_at | MRPL21 | Homo sapiens mitochondrial ribosomal protein L21 (MRPL21) nuclear |
| ADXCRAD_CN279751_s_at | IRAK1 | Homo sapiens interleukin-1 receptor-associated kinase 1 (IRAK1) |
| ADXCRAG_BC016330_s_at | RAD51AP1 | Homo sapiens RAD51 associated protein 1 (RAD51AP1) mRNA. |
| ADXCRAD_BX415970_x_at | AASDHPPT | aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| ADXCRAD_AL135445_at | BTN3A2 | Homo sapiens butyrophilin subfamily 3 member A2 (BTN3A2) mRNA. |
| ADXCRPD.4326.C1_s_at | OMA1 | Homo sapiens OMA1 homolog zinc metallopeptidase (S. cerevisiae) |
| ADXCRAD_AL545542_x_at | PTRF | Homo sapiens polymerase I and transcript release factor (PTRF) |
| ADXCRAG_NM_032242_s_at | PLXNA1 | Homo sapiens plexin A1 (PLXNA1) mRNA. |
| ADXCRPD.7079.C1_x_at | PRKCDBP | Homo sapiens protein kinase C delta binding protein (PRKCDBP) |
| ADXCRAD_BX100631_s_at | SLC12A8 | Homo sapiens solute carrier family 12 (potassium/chloride |
| ADXCRPD.888.C1_s_at | ASH2L | Homo sapiens ash2 (absent small or homeotic)-like (Drosophila) |
| ADXCRAD_AF155810_s_at | SLC25A14 | Homo sapiens solute carrier family 25 (mitochondrial carrier |
| ADXCRPD.1085.C1_at | TSFM | Homo sapiens Ts translation elongation factor mitochondrial |
| ADXCRIHRC.3317.C1_s_at | PSMA2 | Homo sapiens proteasome (prosome macropain) subunit alpha type 2 |
| ADXCRPD.6687.C1_s_at | CDK3 | Homo sapiens cyclin-dependent kinase 3 (CDK3) gene complete cds. |
| ADXCRAG_XM_031689_s_at | MGA | Homo sapiens MAX gene associated (MGA) mRNA. |
| ADXCRAD_H10318_s_at | BAT4 | Homo sapiens HLA-B associated transcript 4 (BAT4) mRNA. |
| ADXCRPD.3851.C1_s_at | FAM102A | Homo sapiens family with sequence similarity 102 member A |
| ADXCRPD.3229.C1_s_at | TMC4 | Homo sapiens transmembrane channel-like 4 (TMC4) mRNA. |
| ADXCRPD.12486.C1_at | AGPAT6 | 1-acylglycerol-3-phosphate O-acyltransferase 6 (lysophosphatidic acid acyltransfase |
| ADXCRAD_CB161421_s_at | PIK3AP1 | Homo sapiens phosphoinositide-3-kinase adaptor protein 1 (PIK3AP1) |
| ADXCRPD.5659.C1_s_at | PAPSS1 | Homo sapiens 3-phosphoadenosine 5-phosphosulfate synthase 1 |
| ADXCRPD.9008.C2_s_at | CC2D1A | Homo sapiens coiled-coil and C2 domain containing 1A (CC2D1A) |
| ADXCRIH.3630.C1_s_at | BTN3A2 | Homo sapiens butyrophilin subfamily 3 member A2 (BTN3A2) mRNA. |
| ADXCRAG_BX640630_s_at | SLC30A7 | Homo sapiens solute carrier family 30 (zinc transporter) member 7 |
| ADXCRAD_CA776658_s_at | WDR48 | Homo sapiens WD repeat domain 48 (WDR48) mRNA. |
| ADXCRAG_AL834333_s_at | SUHW3 | Homo sapiens suppressor of hairy wing homolog 3 (Drosophila) |
| ADXCRAG_BC016847_s_at | E2F3 | Homo sapiens E2F transcription factor 3 (E2F3) mRNA. |
| ADXCRAD_AA311408_x_at | LNPEP | Homo sapiens mRNA for leucyl/cystinyl aminopeptidase variant protein. |
| ADXCRPD.8916.C1_at | CGREF1 | Homo sapiens cell growth regulator with EF-hand domain 1 (CGREF1) |
| ADXCRAD_BE870333_at | RPL28 | Homo sapiens ribosomal protein L28 (RPL28) mRNA. |
| ADXCRPD.8036.C1_at | ZNF669 | Homo sapiens zinc finger protein 669 (ZNF669) mRNA. |
| ADXCRSS.Hs#S634877_at | RPL32 | Homo sapiens ribosomal protein L32 (RPL32) transcript variant 2 |
| ADXCRAD_BQ216862_at | WDR68 | Homo sapiens WD repeat domain 68 (WDR68) mRNA. |
| ADXCRAD_BM542286_s_at | RFC4 | Homo sapiens replication factor C (activator 1) 4 37kDa (RFC4) |
| ADXCRAD_CN368539_s_at | DOLPP1 | Homo sapiens dolichyl pyrophosphate phosphatase 1 (DOLPP1) mRNA. |
| ADXCRAD_BM793751_s_at | BMS1 | Homo sapiens BMS1 homolog ribosome assembly protein (yeast) |
| ADXCRAD_CN297084_s_at | MAST2 | Homo sapiens microtubule associated serine/threonine kinase 2 |
| ADXCRPD.5126.C1_s_at | SART1 | Homo sapiens squamous cell carcinoma antigen recognized by T cells |
Functional assessment of unsupervised genes
The clinical and in vitro genes identified from the unsupervised analyses (Table 1) were investigated further by RNAi for their functional relevance in mediating sensitivity to either 5-FU or SN38. For the initial screening process, custom-designed siRNA plates included all genes for which siRNA sequences were available (n=50). In each case, the effect of target gene silencing was examined alone and also in combination with either 5μM 5-FU (IC30(48h)) or 5nM SN38 (IC30(48h)) in HCT116 CRC cells. For each experiment, two assays were carried out, cell viability (MTT assay) and cell death (Toxilight assay). From this the relative toxicity was calculated as the ratio of cell death to cell viability. Cells were transfected with siRNA for 24h prior to 48h treatment with chemotherapy.
Of the genes identified from the clinical PCA analysis, TNFS14, MAPK9, RAPGEF2 and RPS9 silencing resulted in increased relative toxicity when combined with either 5-FU or SN38 compared to either treatment alone (Figure 2A). Silencing of TNFS14 or RPS9 resulted in enhanced toxicity with 5-FU only, while silencing of MAPK9 enhanced the effect of SN38, but not 5-FU. Silencing of the genes identified from the in vitro PCA which were associated with 5-FU response identified a number of chemo-sensitizing genes: GART, BTN3A2, ASSDHPPT, RAD51AP1, PTRF, PRKCDBP, TSFM, MGA and PSMA2 (Figure 2B). The silencing of either RAD51AP1 or PTRF significantly enhanced the relative toxicity of both 5-FU and SN38 treatment, whereas, silencing the remainder of these 5-FU response-associated genes only significantly increased sensitivity to 5-FU treatment. Analysis of the genes identified from the SN38 in vitro PCAs identified 8 positive hits: AGPAT6, SLC30A7, E2F3, LNPEP, CREF1, RPL28, RFC4 and SART1 (Figure 2C). In all cases, gene silencing resulted in supra-additive interactions with both 5-FU and SN38. Validation of gene silencing was confirmed by quantitative RT-PCR, with silencing of target genes ranging from 45%-96% knock-down compared to non-targeting control siRNA (Supplementary Figure 1).
Validation of positive hits in colorectal cancer cell line panel
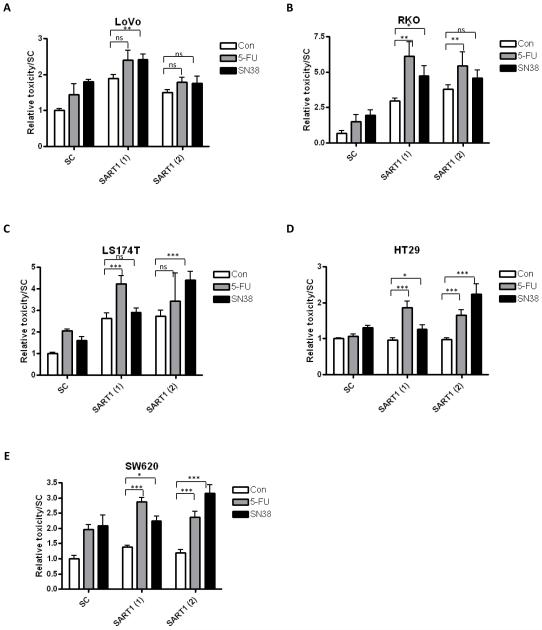
One gene was selected from each of the analysis for testing across a CRC cell line panel. These three genes were selected based on displaying a synergistic interaction with both 5-FU and SN38 when silenced. Therefore, RAPGEF2 was selected from the clinical analysis, PTRF from the 5-FU in vitro analysis and SART1 from the SN38 in vitro analysis. A range of CRC cell lines with varying mutational backgrounds were tested: LoVo, RKO, HT29, SW620 and LS174T. Cell death and cell viability assays were carried out as before, and the relative toxicity measured. In this screen, two siRNA sequences were included. The results showed that silencing either RAPGEF2 or PTRF did not sensitise any cell line, other than HCT116, to either 5-FU or SN38 treatment, therefore the effect of these genes appear to be cell line-dependent (data not shown). However, in the extended cell line panel, silencing of SART1 with either siRNA produced similar results with either additive or synergistic increases in relative toxicity when combined with either chemotherapy treatment (Figure 3A-E).
Figure 3.
Results from the cell line panel siRNA screen. The graphs show the relative toxicity, as assessed from MTT and toxilight assays, for SART1, PTRF and RAPGEF2 for the (A) LoVo, (B) RKO (C) LS174T, (D) HT29, (E) SW620 cell lines. Displayed is the effect of gene silencing alone and in combination with either 5-FU or SN38 treatment. Statistical significance between gene silencing alone (siRNA) and gene silencing in combination with chemotherapy are assessed by an unpaired 2 way t test. statistical significance are highlighted as *=p<0.05, **=p<0.01 and ***=p<0.001. Two siRNA sequences were included for each gene, indicated by (1) and (2).
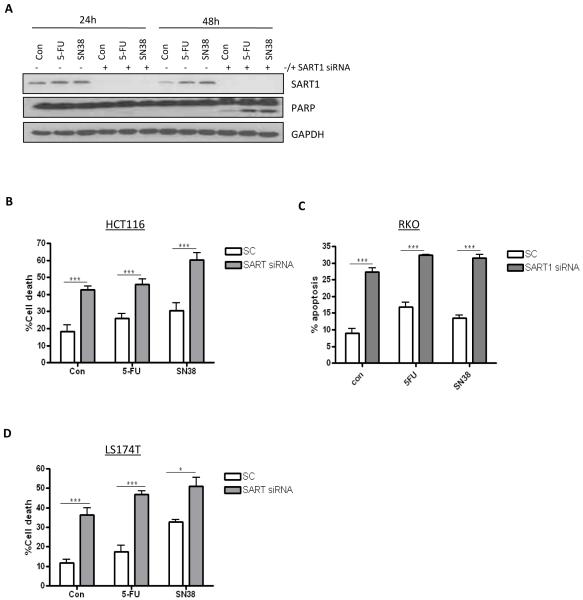
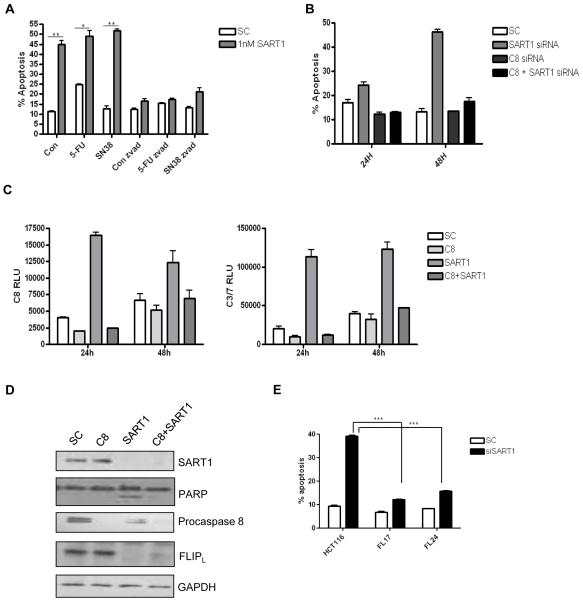
SART1 silencing induces apoptosis in colorectal cancer cells
SART1 protein expression was up-regulated in response to chemotherapy treatment (Figure 4A). Importantly, analysis of PARP cleavage (a hallmark of apoptosis) indicated that SART1 silencing induced enhanced chemotherapy-induced apoptosis after 48h treatment (Figure 4A). To examine the levels of cell death induced following SART1 silencing alone and in combination with chemotherapy, Annexin V/PI staining was carried out in several CRC cell lines. In the HCT116 cell line, SART1 silencing alone resulted in ~43% cell death, and apoptosis was further increased when SART1 silenced cells were co-treated with 5-FU or SN38 (Figure 4B). Similar results were demonstrated with a second siRNA sequence for SART1 (data not shown). Moreover, similar results were obtained in other CRC cell line models, with SART1 silencing inducing apoptosis in LS174T and RKO cell lines (Figures 4C and D).
Figure 4.
(A) Western blot analysis of SART1 expression and PARP cleavage in HCT116 cells transfected with control (−) or SART1 (+) siRNAs and co-treated with 5-FU (IC30(48h)) or SN38 (IC30(48h)) for 24 and 48 h. (B, C and D) Flow cytometric analyses of apoptosis following SART1 silencing alone or in combination with either 5-FU (IC30(48h)) or SN38 (IC30(48h)) in HCT116 (B), RKO (C) and LS147T (D). All cells were reversed transfected with SART1 siRNA for 24h prior to a 48h treatment with chemotherapy.
Silencing of SART1 results in synergistic interactions with either 5-FU or SN38
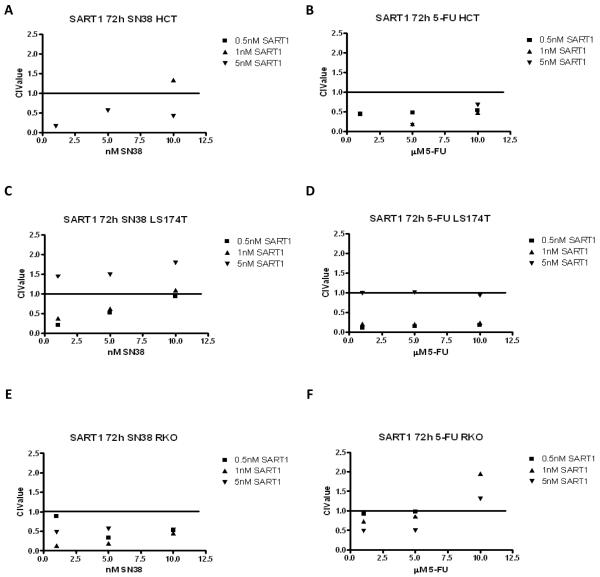
To determine whether silencing of SART1 results in synergistic interactions with chemotherapy, cell viability assays were carried out, and Combination Index (CI) values calculated. Synergistic interactions were observed between chemotherapy and SART1 silencing in the HCT116 (Figures 5A and B), the LS174T (Figures 5C and D) and the RKO (Figures 5E and F) cell lines.
Figure 5.
Cell viability assays were conducted in (A and B) HCT116 (C and D) LS174T, and (E and F) RKO cell lines in response to SART1 siRNA (0.5, 1 and 5nM) and either SN38 (1, 5 and 10nM) or 5-FU (1, 5 and 10μM) for 72 h. To evaluate the interaction between chemotherapy and SART1 silencing, we used the method of Chou and Talalay (26). CI values <1, =1, and >1 indicating synergism, additivity, and antagonism, respectively. For synergistic interactions, CI values between 0.8-0.9 indicate slight synergy, 0.6-0.8 indicate moderate synergy, 0.4-0.6 indicate synergy and those <0.4 indicate strong synergy.
The cell death induced by SART1 silencing is caspase 8- and FLIP-dependent
Previous experiments had demonstrated that SART1 silencing resulted in apoptotic cell death, therefore we investigated whether the mechanism was caspase-dependent, using the pan-caspase inhibitor ZVAD. The cell death observed following SART1 silencing was apoptotic (Annexin V positive) and was completely abrogated following co-treatment with ZVAD (Figure 6A). Analysis of caspase activity following SART1 silencing indicated that caspase 8 was highly activated (Figure 6B). To assess whether SART1 siRNA-induced apoptosis was caspase 8-dpendent, caspase 8-specific siRNA was co-transfected with SART1 siRNA. Apoptotic cell death induced following SART1 silencing was completely abrogated following caspase 8 co-silencing at both 24 and 48h (Figure 6B). Caspase 8 activity assays demonstrated that caspase 8 activity was inhibited following caspase 8 silencing (Figure 6C). In addition, the increased caspase 3/7 activity that was observed following SART1 silencing at 24 and 48h was also completely abrogated following caspase 8 co-silencing (Figure 6C). Furthermore, Western blotting demonstrated that PARP cleavage following SART1 silencing was prevented by caspase 8 co-silencing (Figure 6D). The decreased expression of procaspase 8 in the SART1 silenced cells (Figure 6D, lane 3) is indicative of procaspase 8 activation. Notably, the expression of the endogenous caspase 8 inhibitor c-FLIPL was observed to be down-regulated following SART1 silencing. Importantly, c-FLIPL down-regulation following SART1 silencing was not a downstream effect of apoptosis induction as it was also observed in samples in which caspase 8 was co-silenced. Moreover, SART1 siRNA-induced apoptosis was attenuated in HCT116 cells stably overexpressing FLIPL (Figure 6E). These results suggest that down-regulation of c-FLIPL following SART1 silencing is an upstream effect that leads to caspase 8-dependent apoptosis and enhanced drug-induced apoptosis.
Figure 6.
(A). Flow cytometric analysis of apoptosis in HCT116 cells following treatment with control (SC) or SART1 siRNA (1nM) alone or in combination with chemotherapy (IC30(48h))) for 72h in the presence and absence of the pan-caspase inhibitor, ZVAD (10μM). (B). Flow cytometric analysis of apoptosis in HCT116 cells following transfection with SC or SART1 siRNA (5nM) alone or in combination caspase 8 siRNA (10nM). (C) Caspase-Glo® assay measuring caspase 8 activity or caspase 3/7 activity in HCT116 cells following transfection with SC or SART1 siRNA (5nM) alone or in combination caspase 8 siRNA (10nM). (D). Western blot analysis of SART1, procaspase 8 and FLIPL expression following transfection with SC or SART1 siRNA (5nM) alone or in combination caspase 8 siRNA (10nM). All experiments are representative of three independent experiments. (E). Flow cytometric analysis of apoptosis in parental and c-FLIPL overexpressing cells (FL17 and FL24) following transfection with SC or SART1 siRNA (5nM) for 24 hours. Statistical significance was assessed by using an unpaired two-tailed t test with *** = p<0.001, ** = p<0.01 and * = P<0.05.
Discussion
In the current study we utilised a systems biology approach incorporating transcriptional profiling, bioinformatics and functional analyses to identify key mediators of 5-FU and SN38 sensitivity in colorectal cancer that may represent novel drug targets and/or biomarkers. Following minimal filtering, unsupervised analysis was carried out on both the clinical and in vitro genelists using PCA. The reason for using both clinical and in vitro data was to try and identify potentially clinically relevant targets that could also be validated within an in vitro system. In addition, PCA was chosen as the classification approach as it represents a completely unbiased approach. Furthermore, due to the fact that the clinical sample size used was small, a supervised method approach may have significantly over estimated the predictive ability of the identified genes. Using the unsupervised approach, PCA identified a total of 100 genes, 50 of which were taken forward for functional testing using a custom-designed siRNA screen. The siRNA screen incorporated multiple siRNA sequences across a panel of several CRC cell lines and identified squamous cell carcinoma antigen recognized by T cells or SART1 as a potential mediator of 5-FU and SN38 sensitivity in this disease setting.
We functionally tested 50 of the 100 transcripts by siRNA screening and found that a significant number of these did play a role in mediating chemotherapy response in the initial cell line model system. Many of the 50 untested transcripts represented hypothetical proteins, and some were antisense RNAs, which made testing their functional significance more difficult. However, while most of the remaining untested transcripts have not been previously implicated in either colorectal cancer progression or chemotherapy response, some potentially may have a role. Such genes include CSNK2A2 (5-FU in vitro-related) and CCNDBP1, TNFAIP8 and GDI2 (clinically-related). CCNDBP1 negatively regulates cell cycle progression through the inhibition of the cyclinD1/CDK4 complex (28, 29). CCNDBP1 is located on chromosome 15q15, which is associated with loss of heterozygosity (LOH) in many tumour types including colon (28, 29). It is down-regulated in tumours compared to matched normal and therefore has been associated with tumour suppression (28). TNFAIP8 has been shown to regulate apoptosis in thymocytes (30). Interestingly, TNFAIP8 can block caspase-mediated apoptosis, contains a death effector domain (DED) and has been hypothesized to be a novel member of the FLIP family (31). GDI2 has been identified in a number of proteomic studies as a potential biomarker for pancreatic and ovarian cancer (32, 33). Indeed, GDI2 has been identified as a marker for chemo-resistance in ovarian cancer (32). CSNK2A2 is known to be over-expressed in cancers, including CRC. It has been associated with disease progression in CRC via a proteomic approach (34). Of interest, CSNK2A2 protects colon cancer cells from TRAIL-induced apoptosis (35). The pro-survival and inhibition of apoptosis that is associated with CSNK2A2 is mediated through its ability to induce survivin expression via the Wnt pathway (36). Although these targets were not assessed for functional significance, they may still represent potential biomarkers or therapeutic targets for inhibiting drug resistance in this disease setting.
SART1 is a biscistronic gene that encodes two proteins, one of which is 800 amino acids long and contains a leucine zipper and the other that is 259 amino acids long that does not contain the leucine zipper. The SART1(800) protein is located in the nucleus of the majority of proliferating cells, while the SART1(259) protein is located in the cytosol of epithelial cancers. In this study, we focussed on SART1(800), the main function of which is thought to be in the recruitment of the tri-snRNP to the pre-spliceosome (37). Therefore, SART1 may be critical for pre-mRNA splicing. SART1(800) protein is expressed in 100% of CRC cell lines, 55% of CRC tissue and 0% of non-tumour tissue (38). Hypoxia inducible factor or HAF is the murine homolog of SART1, and several studies have demonstrated that HAF binds to and regulates the activity of the EPO receptor, VEGF and HIF-1α (39, 40). HAF binds and regulates HIF-1α activity independent of oxygen levels (40). SART1 has also been identified as a novel SUMO1 and SUMO2 target protein (41-43).
SART1 has been associated with the development and progression of head and neck squamous cell carcinoma. Again, in this disease setting, there was a much higher tumour expression compared to normal (44). Kittler et al. carried out an siRNA screen to identify genes that were essential for cell division in breast cancer. They identified SART1 as a gene essential for cell division in breast cancer, with SART1 depletion displaying similar defects to either CENPE or KIF11 depletion. These defects following SART1 depletion may indicate that SART1 plays a direct role in cell division or may have an indirect effect caused by defective pre-mRNA splicing (45). Following this study, a second study was carried out by Olson et al., which examined whether the genes identified from the former study were associated with breast cancer risk. The results demonstrated that four single nucleotide polymorphisms (SNPs) were present in SART1, and two were associated with increased risk of breast cancer, while the other two were associated with a decreased risk of breast cancer development (46). The authors concluded that genetic variation in SART1 may be associated with breast cancer development.
The current study has demonstrated that SART1 silencing sensitises CRC cells to 5-FU or SN38 treatment via apoptosis induction. The mechanism of cell death is caspase-dependent, specifically caspase 8-dependent. In addition, the results demonstrate that SART1 silencing leads to c-FLIPL down-regulation. Our previous studies have demonstrated that c-FLIPL is a key regulator of chemotherapy-induced cell death in CRC and other tumour types (20, 23-25, 47). The down-regulation of c-FLIPL was not a consequence of cell death as we also carried out experiments in which caspase 8 and SART1 were co-silenced and demonstrated that the cells did not undergo apoptosis, however, c-FLIPL was still down-regulated. Moreover, apoptosis induced by SART1 silencing was attenuated in c-FLIPL overexpressing cells. These results suggest that SART1 expression is important for continued c-FLIPL expression. c-FLIP regulation is highly complex and involves transcriptional and post-transcriptional regulation by a number of key signal transduction pathways, such as NF*B, JNK, c-myc, c-fos and PKC (48). Preliminary data suggest that the effects of SART1 on c-FLIP expression are post-transcriptional (data not shown): the mechanistic basis of SART1’s regulation of c-FLIP is the subject of ongoing studies. The current study has demonstrated that silencing of SART1 enhanced chemotherapy-induced cell death via c-FLIP down-regulation. Thus, c-FLIP and/or SART1 may represent predictive biomarkers of response to chemotherapy in CRC.
In conclusion, the current study has used a systems biology approach to identify a number of novel regulators of chemo-resistance in colorectal cancer, most notably SART1, which regulates expression of the critical apoptosis-regulating and drug resistance-associated protein c-FLIP. Future studies will assess the clinical relevance of SART1 expression as a prognostic and predictive biomarker in colorectal cancer.
Supplementary Material
Acknowledgements
We would like to thank Ms Cathy Fenning for technical support.
Financial support from Cancer Research UK (C212/A7402); Cancer Research UK Bobby Moore Fellowship, Research and Development Office Northern Ireland, Department of Health, Social Services and Public Safety (RRG/3261/05, RRG 6.42) and Staff Training and Development Unit (STDU), Queen’s University Belfast.
Footnotes
Conflict of interest: P. Johnston is employed by and has an ownership interest in Almac Diagnostics.
References
- 1.Douillard JY, Cunningham D, Roth AD, Navarro M, James RD, Karasek P, et al. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: a multicentre randomised trial. Lancet. 2000;355:1041–1047. doi: 10.1016/s0140-6736(00)02034-1. [DOI] [PubMed] [Google Scholar]
- 2.Giacchetti S, Perpoint B, Zidani R, Le Bail N, Faggiuolo R, Focan C, et al. Phase III multicenter randomized trial of oxaliplatin added to chronomodulated fluorouracil-leucovorin as first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2000;18:136–147. doi: 10.1200/JCO.2000.18.1.136. [DOI] [PubMed] [Google Scholar]
- 3.Barrier A, Boelle PY, Lemoine A, Tse C, Brault D, Chiappini F, et al. Gene expression profiling of nonneoplastic mucosa may predict clinical outcome of colon cancer patients. Dis Colon Rectum. 2005;48:2238–2248. doi: 10.1007/s10350-005-0175-9. [DOI] [PubMed] [Google Scholar]
- 4.Barrier A, Boelle PY, Roser F, Gregg J, Tse C, Brault D, et al. Stage II colon cancer prognosis prediction by tumor gene expression profiling. J Clin Oncol. 2006;24:4685–4691. doi: 10.1200/JCO.2005.05.0229. [DOI] [PubMed] [Google Scholar]
- 5.Barrier A, Lemoine A, Boelle PY, Tse C, Brault D, Chiappini F, et al. Colon cancer prognosis prediction by gene expression profiling. Oncogene. 2005;24:6155–6164. doi: 10.1038/sj.onc.1208984. [DOI] [PubMed] [Google Scholar]
- 6.Barrier A, Roser F, Boelle PY, Franc B, Tse C, Brault D, et al. Prognosis of stage II colon cancer by non-neoplastic mucosa gene expression profiling. Oncogene. 2006 doi: 10.1038/sj.onc.1210060. [DOI] [PubMed] [Google Scholar]
- 7.Eschrich S, Yang I, Bloom G, Kwong KY, Boulware D, Cantor A, et al. Molecular staging for survival prediction of colorectal cancer patients. J Clin Oncol. 2005;23:3526–3535. doi: 10.1200/JCO.2005.00.695. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Jatkoe T, Zhang Y, Mutch MG, Talantov D, Jiang J, et al. Gene expression profiles and molecular markers to predict recurrence of Dukes’ B colon cancer. J Clin Oncol. 2004;22:1564–1571. doi: 10.1200/JCO.2004.08.186. [DOI] [PubMed] [Google Scholar]
- 9.Del Rio M, Molina F, Bascoul-Mollevi C, Copois V, Bibeau F, Chalbos P, et al. Gene expression signature in advanced colorectal cancer patients select drugs and response for the use of leucovorin, fluorouracil, and irinotecan. J Clin Oncol. 2007;25:773–780. doi: 10.1200/JCO.2006.07.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DS, Nobel AB, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 11.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 12.Allen WL, Jithesh PV, Oliver GR, Proutski I, Longley DB, Lenz HJ, et al. The colorectal cancer disease-specific transcriptome may facilitate the discovery of more biologically and clinically relevant information. BMC Cancer. 2010;10:687. doi: 10.1186/1471-2407-10-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grigoriadis A, Oliver GR, Tanney A, Kendrick H, Smalley MJ, Jat P, et al. Identification of differentially expressed sense and antisense transcript pairs in breast epithelial tissues. BMC Genomics. 2009;10:324. doi: 10.1186/1471-2164-10-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanney A, Oliver GR, Farztdinov V, Kennedy RD, Mulligan JM, Fulton CE, et al. Generation of a non-small cell lung cancer transcriptome microarray. BMC Med Genomics. 2008;1:20. doi: 10.1186/1755-8794-1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosey AM, Gorski JJ, Murray MM, Quinn JE, Chung WY, Stewart GE, et al. Molecular basis for estrogen receptor alpha deficiency in BRCA1-linked breast cancer. J Natl Cancer Inst. 2007;99:1683–1694. doi: 10.1093/jnci/djm207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allen WL, Coyle VM, Jithesh PV, Proutski I, Stevenson L, Fenning C, et al. Clinical determinants of response to irinotecan-based therapy derived from cell line models. Clin Cancer Res. 2008;14:6647–6655. doi: 10.1158/1078-0432.CCR-08-0452. [DOI] [PubMed] [Google Scholar]
- 17.Kyula JN, Van Schaeybroeck S, Doherty J, Fenning CS, Longley DB, Johnston PG. Chemotherapy-induced activation of ADAM-17: a novel mechanism of drug resistance in colorectal cancer. Clin Cancer Res. 2010;16:3378–3389. doi: 10.1158/1078-0432.CCR-10-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. http://www.lgcstandards.com/home/home_en.aspx.
- 19.Boyer J, McLean E, Aroori S, Wilson P, McCulla A, Carey P, et al. Characterization of p53 wild-type and null isogenic colorectal cancer cell lines resistant to 5-fluorouracil, oxaliplatin, and irinotecan. Clin Cancer Res. 2004;10:2158–2167. doi: 10.1158/1078-0432.ccr-03-0362. [DOI] [PubMed] [Google Scholar]
- 20.Longley DB, Wilson TR, McEwan M, Allen WL, McDermott U, Galligan L, et al. c-FLIP inhibits chemotherapy-induced colorectal cancer cell death. Oncogene. 2006;25:838–848. doi: 10.1038/sj.onc.1209122. [DOI] [PubMed] [Google Scholar]
- 21. http://www.ebi.ac.uk/arrayexpress/
- 22. http://frodo.wi.mit.edu/primer3/
- 23.Wilson TR, McEwan M, McLaughlin K, Le Clorennec C, Allen WL, Fennell DA, et al. Combined inhibition of FLIP and XIAP induces Bax-independent apoptosis in type II colorectal cancer cells. Oncogene. 2009;28:63–72. doi: 10.1038/onc.2008.366. [DOI] [PubMed] [Google Scholar]
- 24.Wilson TR, McLaughlin KM, McEwan M, Sakai H, Rogers KM, Redmond KM, et al. c-FLIP: a key regulator of colorectal cancer cell death. Cancer Res. 2007;67:5754–5762. doi: 10.1158/0008-5472.CAN-06-3585. [DOI] [PubMed] [Google Scholar]
- 25.Wilson TR, Redmond KM, McLaughlin KM, Crawford N, Gately K, O’Byrne K, et al. Procaspase 8 overexpression in non-small-cell lung cancer promotes apoptosis induced by FLIP silencing. Cell Death Differ. 2009 doi: 10.1038/cdd.2009.76. [DOI] [PubMed] [Google Scholar]
- 26.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 27.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 28.Ma W, Stafford LJ, Li D, Luo J, Li X, Ning G, et al. GCIP/CCNDBP1, a helix-loop-helix protein, suppresses tumorigenesis. J Cell Biochem. 2007;100:1376–1386. doi: 10.1002/jcb.21140. [DOI] [PubMed] [Google Scholar]
- 29.Xia C, Bao Z, Tabassam F, Ma W, Qiu M, Hua S, et al. GCIP, a novel human grap2 and cyclin D interacting protein, regulates E2F-mediated transcriptional activity. J Biol Chem. 2000;275:20942–20948. doi: 10.1074/jbc.M002598200. [DOI] [PubMed] [Google Scholar]
- 30.Woodward MJ, de Boer J, Heidorn S, Hubank M, Kioussis D, Williams O, et al. Tnfaip8 is an essential gene for the regulation of glucocorticoid-mediated apoptosis of thymocytes. Cell Death Differ. 17:316–323. doi: 10.1038/cdd.2009.125. [DOI] [PubMed] [Google Scholar]
- 31.Kumar D, Whiteside TL, Kasid U. Identification of a novel tumor necrosis factor-alpha-inducible gene, SCC-S2, containing the consensus sequence of a death effector domain of fas-associated death domain-like interleukin- 1beta-converting enzyme-inhibitory protein. J Biol Chem. 2000;275:2973–2978. doi: 10.1074/jbc.275.4.2973. [DOI] [PubMed] [Google Scholar]
- 32.Lee DH, Chung K, Song JA, Kim TH, Kang H, Huh JH, et al. Proteomic identification of paclitaxel-resistance associated hnRNP A2 and GDI 2 proteins in human ovarian cancer cells. J Proteome Res. 9:5668–5676. doi: 10.1021/pr100478u. [DOI] [PubMed] [Google Scholar]
- 33.Sun ZL, Zhu Y, Wang FQ, Chen R, Peng T, Fan ZN, et al. Serum proteomic-based analysis of pancreatic carcinoma for the identification of potential cancer biomarkers. Biochim Biophys Acta. 2007;1774:764–771. doi: 10.1016/j.bbapap.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 34.Nibbe RK, Markowitz S, Myeroff L, Ewing R, Chance MR. Discovery and scoring of protein interaction subnetworks discriminative of late stage human colon cancer. Mol Cell Proteomics. 2009;8:827–845. doi: 10.1074/mcp.M800428-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Izeradjene K, Douglas L, Delaney A, Houghton JA. Casein kinase II (CK2) enhances death-inducing signaling complex (DISC) activity in TRAIL-induced apoptosis in human colon carcinoma cell lines. Oncogene. 2005;24:2050–2058. doi: 10.1038/sj.onc.1208397. [DOI] [PubMed] [Google Scholar]
- 36.Tapia JC, Torres VA, Rodriguez DA, Leyton L, Quest AF. Casein kinase 2 (CK2) increases survivin expression via enhanced beta-catenin-T cell factor/lymphoid enhancer binding factor-dependent transcription. Proc Natl Acad Sci U S A. 2006;103:15079–15084. doi: 10.1073/pnas.0606845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makarova OV, Makarov EM, Luhrmann R. The 65 and 110 kDa SR-related proteins of the U4/U6.U5 tri-snRNP are essential for the assembly of mature spliceosomes. Embo J. 2001;20:2553–2563. doi: 10.1093/emboj/20.10.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasatomi T, Suefuji Y, Matsunaga K, Yamana H, Miyagi Y, Araki Y, et al. Expression of tumor rejection antigens in colorectal carcinomas. Cancer. 2002;94:1636–1641. doi: 10.1002/cncr.10421. [DOI] [PubMed] [Google Scholar]
- 39.Gupta M, Mungai PT, Goldwasser E. A new transacting factor that modulates hypoxia-induced expression of the erythropoietin gene. Blood. 2000;96:491–497. [PubMed] [Google Scholar]
- 40.Koh MY, Darnay BG, Powis G. Hypoxia-associated factor, a novel E3-ubiquitin ligase, binds and ubiquitinates hypoxia-inducible factor 1alpha, leading to its oxygen-independent degradation. Mol Cell Biol. 2008;28:7081–7095. doi: 10.1128/MCB.00773-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schimmel J, Balog CI, Deelder AM, Drijfhout JW, Hensbergen PJ, Vertegaal AC. Positively charged amino acids flanking a sumoylation consensus tetramer on the 110kDa tri-snRNP component SART1 enhance sumoylation efficiency. J Proteomics. 73:1523–1534. doi: 10.1016/j.jprot.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 42.Vertegaal AC, Andersen JS, Ogg SC, Hay RT, Mann M, Lamond AI. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol Cell Proteomics. 2006;5:2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Vertegaal AC, Ogg SC, Jaffray E, Rodriguez MS, Hay RT, Andersen JS, et al. A proteomic study of SUMO-2 target proteins. J Biol Chem. 2004;279:33791–33798. doi: 10.1074/jbc.M404201200. [DOI] [PubMed] [Google Scholar]
- 44.Cromer A, Carles A, Millon R, Ganguli G, Chalmel F, Lemaire F, et al. Identification of genes associated with tumorigenesis and metastatic potential of hypopharyngeal cancer by microarray analysis. Oncogene. 2004;23:2484–2498. doi: 10.1038/sj.onc.1207345. [DOI] [PubMed] [Google Scholar]
- 45.Kittler R, Putz G, Pelletier L, Poser I, Heninger AK, Drechsel D, et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- 46.Olson JE, Wang X, Goode EL, Pankratz VS, Fredericksen ZS, Vierkant RA, et al. Variation in genes required for normal mitosis and risk of breast cancer. Breast Cancer Res Treat. 119:423–430. doi: 10.1007/s10549-009-0386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rogers KM, Thomas M, Galligan L, Wilson TR, Allen WL, Sakai H, et al. Cellular FLICE-inhibitory protein regulates chemotherapy-induced apoptosis in breast cancer cells. Mol Cancer Ther. 2007;6:1544–1551. doi: 10.1158/1535-7163.MCT-06-0673. [DOI] [PubMed] [Google Scholar]
- 48.Shirley S, Micheau O. Targeting c-FLIP in cancer. Cancer Lett. doi: 10.1016/j.canlet.2010.10.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.