Abstract
Over the past fourteen years, ubiquitination has emerged as a centrally important mechanism governing the subcellular trafficking of proteins. Ubiquitination, interaction with sorting factors that contain ubiquitin binding domains, and finally deubiquitination govern the itineraries of cargo proteins that include yeast carboxypeptidase S, the epithelial sodium channel ENaC, and epidermal growth factor receptor. The molecular structures and mechanisms of the paradigmatic HECT and RING domain ubiquitin ligases, JAMM and USP domain deubiquitinating enzymes, and numerous ubiquitin binding domains involved in these pathways, have been worked out in recent years and are described.
Keywords: Lysosome, vacuole, yeast genetics, EGF, EGF receptor, growth factor receptor, epithelial sodium channel, ENaC, yeast genetics, carboxypeptidase S, protein structure, crystal structure, ubiquitin, RING domain, HECT domain, JAMM domain, isopeptidase, ubiquitin ligase, deubiquitinating enzyme, ubiquitin binding domain, ESCRT complex
INTRODUCTION
Ubiquitin is a small protein with a large footprint in biology. The covalent ubiquitination of proteins is one of the most widespread regulatory post-translational modifications of proteins. The C-terminus of ubiquitin is conjugated to target proteins by the action of three enzymes: an ubiquitin activating enzyme (E1), an ubiquitin conjugating enzyme (E2), and an ubiquitin protein ligase (E3) (30, 53, 88, 122). Ubiquitin is normally conjugated to proteins via an isopeptide bond between the C-terminus of ubiquitin and specific Lys residues in the ubiquitinated protein. Ubiquitin may be attached to proteins as one or a few monomers (mono- or multiubiquitination) or as a polyubiquitin chain. Ubiquitin polymers are formed when additional ubiquitin molecules are attached to one of the seven Lys residues on a previously attached ubiquitin.
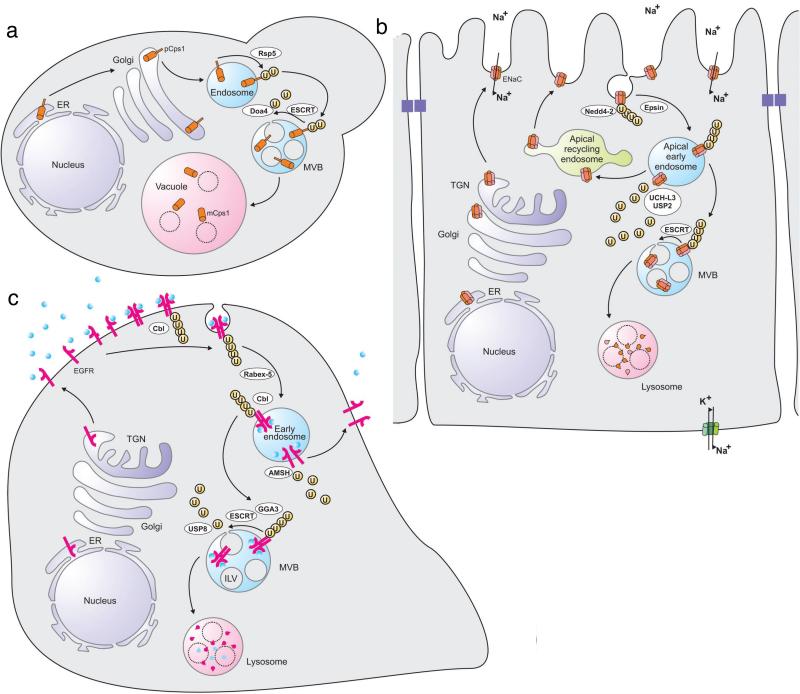
The first demonstration of an involvement of ubiquitin in membrane traffic came with the discovery that ubiquitination of the mating factor receptor Ste2 (32) and the uracil permease Fur4 (26) in budding yeast is required for their efficient endocytosis. Subsequent work has revealed that many (but far from all) membrane proteins in diverse organisms are endocytosed in a ubiquitin-dependent manner, often in response to ligand binding (33, 114). Moreover, even though ubiquitin was first identified as an endocytosis signal, it has turned out to be even more important as a signal for sorting membrane proteins into the intralumenal vesicles (ILVs) of multivesicular bodies (MVBs). The ILVs are essentially vehicles specialized for delivering integral membrane proteins to the lumen of lysosomes for proteolytic processing or degradation, which occurs when MVBs fuse with these degradative organelles (Figure 1a). With very few exceptions, almost all integral membrane proteins that are sorted to the lysosome lumen follow ubiquitin-dependent sorting into MVBs.
Figure 1.
Subcellular itineraries of paradigmatic substrates of ubiquitin-dependent sorting pathways. (a) Yeast carboxypeptidase Cps1 is biosynthesized as a transmembrane precursor form (pCps1), which is transported from the biosynthetic pathway to endosomes. Ubiquitination in endosomes, mediated by Rsp5, provides a signal for recognition by the ESCRT machinery. Before the inclusion within the ILVs of MVBs, pCps1 is deubiquitinated by Doa4. When the MVBs fuse with the vacuole, the ILVs and their content are delivered to the vacuole lumen. Here, the ILV membranes are degraded by lipases, whereas pCps1 is processed into the mature form (mCps1) by specific proteases. (b) The Na+ channel ENaC is delivered to the apical plasma membrane of epithelial cells where it functions to channel Na+ from the extracellular space into the cells. Residency of ENaC in the apical plasma membrane is controlled by the E3 ubiquitin ligase Nedd4-2, which ubiquitinates ENaC and thereby signals its Epsin-dependent endocytosis from clathrin-coated pits and delivery to an apical early endosome. Deubiquitination by UCH-L3 or USP2-45 favours recycling of ENaC to the plasma membrane via the apical recycling endosome. ENaC molecules that remain ubiquitinated are recognized by the ESCRT machinery and sorted into the MVB pathway for lysosomal degradation. (c) When the epidermal growth factor (EGF) binds to its receptor (EGFR) at the plasma membrane, this induces a conformation change of the receptor that promotes dimerization. This triggers tyrosine phosphorylation, which, in addition to initiating signal cascades, promotes receptor ubiquitination by the E3 ubiquitin ligase Cbl. The activated receptor is endocytosed and delivered to an early endosome. If the ligand dissociates in the early endosome, ubiquitination is no more sustained, and the EGFR becomes deubiquitinated by the DUB AMSH. This promotes recycling to the plasma membrane. If still activated in the endosome, the EGFR remains ubiquitinated and is recognized by GGA3 and the ESCRT machinery. This causes its sorting into ILVs following deubiquitination by the DUB USP8. The receptor and its ligand are degraded when the MVB fuses with a lysosome.
Ubiquitin-dependent trafficking of membrane proteins is critical for regulating the numbers of transporters, receptors and adhesion molecules at the plasma membrane, and for correct delivery and activation of certain lysosomal hydrolases, thereby having profound impacts on cell signaling and metabolism. In the first part of this review we will discuss three examples of membrane proteins that follow ubiquitin-dependent trafficking pathways. We will follow the trafficking of these proteins from their biosynthesis to their degradation, and we will highlight the E3 ubiquitin ligases, deubiquitinating enzymes (DUBs) and sorting components that these cargoes encounter during their journey through the cell. In the second part of the review we will zoom in on the molecular details, down to the atomic level, of how membrane proteins are ubiquitinated, deubiquitinated and sorted.
ROLE OF UBIQUITINATION IN THE TRAFFICKING OF YEAST CARBOXYPEPTIDASE S
Carboxypeptidase S (Cps1), a vacuolar hydrolase in the budding yeast Saccharomyces cerevisiae, serves as a prototypic example of a biosynthetic cargo that undergoes ubiquitin-mediated sorting. This protein is synthesized as a type II integral membrane protein that represents a precursor form (pCps1) (111). Its activation requires proteolytic clipping into a mature, soluble form (mCps1) (Figure 1a). This proteolytic activation occurs in the vacuole, the yeast equivalent of mammalian lysosomes, through the activities of the vacuolar proteases Pep4 and Prb1. On its way to the vacuole, Cps1 transits directly from the trans-Golgi network (TGN) to endosomes, where it undergoes sorting into ILVs of MVBs. It is this ILV-associated form that is delivered to the vacuole for efficient processing upon fusion of the MVB with the vacuole membrane (50).
Ubiquitination of Cps1 by Rsp5
Ubiquitination of Cps1 is crucial for its correct trafficking, and Cps1 was actually the first cargo for which ubiquitin was identified as a signal for entry into the MVB pathway (50, 92). Pulse-labeling experiments suggest that pCps1 is ubiquitinated after exiting the Golgi (50). Cps1 is ubiquitinated on Lys8 by the HECT (homologous to E6AP carboxyl terminus)-domain-containing E3 ubiquitin ligase Rsp5 (50, 51, 76). Rsp5 is the only member of the Nedd4 family of HECT domain ligases in budding yeast. Replacement of Lys8 with arginine results in mis-sorting of Cps1 to the limiting (i.e. the membrane delimiting it from the cytosol) membrane of the vacuole (50). In contrast to most known mammalian substrates for HECT E3 ubiquitin ligases, Cps1 does not contain any canonical PPXY motif that serves as a docking site for one of the WW domains of the ubiquitin ligase. Instead, Cps1 recruits Rsp5 through association with an adaptor protein that contains a PPXY motif, the endosomal integral membrane protein Bsd2 (31). In addition, Cps1 contains MVB targeting information within the amino acid sequence PVEKAPR, and direct interaction of this motif with the HECT domain of Rsp5 appears to facilitate its ubiquitination and correct sorting (62).
Mono- vs.polyubiquitination of Cps1
According to SDS-PAGE analyses, the bulk of ubiquitinated Cps1 contains a single ubiquitin moiety, and this has led to the view that a single ubiquitin is sufficient for correct sorting of Cps1 (50). Recent data using a yeast strain that expresses ubiquitin-Lys63Arg, which cannot form Lys63-linked polyubiquitin, as the only ubiquitin species, have challenged this view (61). Cps1 is normally transported from the biosynthetic to the endocytic pathway whereas in the Ub-K63R mutant strain, surprisingly transport into the MVB pathway is compromised. This suggests that Lys63-linked polyubiquitination is required for Cps1 sorting into the MVB pathway, an interpretation that is consistent with the finding that Rsp5 preferentially catalyzes Lys63-linked polyubiquitination of its substrates (55). It should be noted, though, that it is difficult to detect any polyubiquitinated Cps1 by immunobotting – the only higher ubiquitinated form that can be detected convincingly in such experiments is a di-ubiquitinated form (61). It is therefore conceivable that Lys63-linked di-ubiquitination is a sufficient signal for sorting Cps1 into the MVB pathway. The difficulties in detecting polyubiquitinated Cps1 might also be explained by the presence of (unidentified) DUBs that could serve to trim the polyubiquitin chains at high rate. At present one also cannot rule out the possibility that the failure of Cps1 to become correctly sorted in the Ub-Lys63Arg yeast strain might be unrelated to an eventual inhibition of Cps1 polyubiquitination, for instance by preventing (unidentified) polyubiquitination of components of the sorting machinery.
Deubiquitination of Cps1 by the DUB Doa4
Whereas only a very minor fraction of Cps1 is found ubiquitinated in wild-type yeast, the level of (mono-)ubiquitination increases dramatically in yeast strains deleted for the DUB Doa4, indicating that Doa4 is the main DUB for Cps1 (50). Like other MVB cargoes, the vacuolar sorting of Cps1 is dramatically impaired in doa4 mutants (4). The most straightforward explanation to this is that depletion of Doa4 prevents mono-ubiquitin to be recycled and therefore causes a depletion of cellular ubiquitin. However, Cps1 fails to become correctly sorted to the vacuole lumen in doa4 mutants even if the cellular levels of ubiquitin are restored experimentally, suggesting that Doa4 could also have an additional function in Cps1 sorting. Doa4 might deubiquitinate components of the sorting machinery, or cargo deubiquitination itself could promote cargo internalization (80, 95).
ENaC, A SODIUM CHANNEL THAT UNDERGOES UBIQUITIN-DEPENDENT TRAFFICKING
The epithelial Na+ channel ENaC is widely expressed in epithelial tissues such as sweat ducts, salivary glands, colon, airway and kidney nephrons, where it shows a polarized distribution to apical membranes (11). ENaC is a major regulator of salt and water reabsorption, which has been particularly well characterized in the distal nephrons of the kidney.
Subunit structure and channel properties of ENaC
ENaC is composed of three homologous subunits, α, β and γ. These channels form heterotetramers comprised of two α, one α and one γ subunit (24). They have a high selectivity for Na+ over K+ and can be efficiently inhibited by amiloride (Ki ca 100 nM). The α, β and γ subunits have a similar topology with two transmembrane domains, a large extracellular region, and cytoplasmic N- and C-termini (107). Both the β and γ subunits are palmitoylated on specific cytosol-exposed cysteine residues, and this posttranslational modification modulates the gating properties of the channel, possibly by facilitating interactions between the cytoplasmic domains and the plasma membrane (77). ENaC channels are also regulated by channel activating proteases, which cleave extracellular loops and thereby increase Na+ conductance by increasing the channel-open probability (11, 56).
Intracellular trafficking of ENaC
Biosynthesized ENaC molecules transit from the Golgi complex via the TGN to the apical plasma membrane (11). Inactivation of ENaC channels can occur either by the transient alteration of gating properties, or by internalization of the channels. The latter is triggered by ubiquitination, and will be discussed here. Endocytosis of ENaC occurs primarily from clathrin-coated pits and is dependent on the clathrin- and ubiquitin-binding endocytosis adaptor Epsin1 (113). The endocytosed ENaC is delivered to early endosomes, from where a minor fraction is recycled to the TGN, while the rest is either recycled to the plasma membrane or transported to lysosomes, depending on the ubiquitination status of the channel (11)(Figure 1b). Trafficking of ENaC is hormonally controlled, and both the mineralocorticoid aldosterone and the antidiuretic hormone vasopressin cause a rapid redistibution of ENaC from an intracellular pool to the apical plasma membrane to increase Na+ conductance (110). The molecular mechanisms of this hormone-controlled translocation of ENaC remain to be characterized.
Control of ENaC endocytosis and lysosomal trafficking through Nedd4-2-mediated ubiquitination
The E3 ubiquitin ligase Nedd4-2 binds to PPXY motifs in the C-termini of the α and γ subunits (and possibly also the β subunit) of ENaC at the plasma membrane, thereby causing their ubiquitination on N-terminal lysine residues (113). Mutation of these lysines to arginines profoundly inhibits endocytosis from the apical membrane, thereby increasing channel half-time, as well as mutations that prevent Nedd4-2 binding (11). Although recent data indicate that multiple ubiquitin molecules are conjugated to the ENaC subunits, it is not yet clear whether this modification represents poly- or multi-mono-ubiquitin, or a combination of these. What is more certain is that ubiquitinated ENaC subunits are recognized by HRSby the Hrs subunit of the ESCRT-0 complex and that this entry into the ESCRT pathway mediates lysosomal degradation of ubiquitinated ENaC molecules (127). Given the crucial function of Nedd4-2 in the regulation of ENaC trafficking, it is not surprising that ubiquitination of ENaC by this ubiquitin ligase is under strict control by physiological cues. In particular, phosphorylation of Nedd4-2 by the serum and glucocorticoid kinase SGK1 or by the AMP-activated kinase AMPK prevents its interaction with ENaC, thereby increasing the half-life of ENaC (1, 16). Likewise, phosphorylation of the C-terminus of the β-subunit of ENaC by the G-protein receptor-coupled kinase GRK2 causes insensitivity to Nedd4-2-mediated downregulation (18).
Control of ENaC recycling through deubiquitination
ENaC molecules that remain ubiquitinated in endosome membranes appear to be committed to lysosomal degradation by ESCRT-dependent sorting. Conversely, deubiquitination mediated by the DUBs UCH-L3 and USP2 promote recycling of ENaC to the apical plasma membrane (11). Consistent with this, knockdown of UCH-L3 causes increased ubiquitination of ENaC and a loss of ENaC-mediated Na+ transport (12). Likewise, overexpression of USP2 causes decreased ubiquitination of ENaC and increases its activity at the plasma membrane (22). These findings illustrate the importance of ENaC ubiquitination for its evasion of the endocytic recycling route and targeting to the degradative MVB pathway.
ENaC ubiquitination and genetic disease
Liddle syndrome is an autosomal dominant salt-sensitive hypertension syndrome caused by gain-of-function mutations in the β and γ subunits of ENaC (41, 96). The common theme of these mutations is that they preclude Nedd4-2 binding to ENaC, thereby preventing endocytosis and causing ENaC to reside at the apical plasma membrane for a prolonged time (11). Increased residence time of ENaC in the apical membranes of the distal kidney nephrons results in excessive Na+ reabsorption and ensuing hypertension.
UBIQUITIN-DEPENDENT TRAFFICKING OF EPIDERMAL GROWTH FACTOR RECEPTORS
Epidermal growth factor (EGF) receptors (EGFRs) are expressed in diverse cell types and serve to mediate signalling that results in proliferation, growth, survival and migration (124). The fact that EGFRs are frequently overexpressed or contain gain-of-function mutations in cancers have made them an attractive target for cancer therapy (123). EGFR has six known ligands, of which EGF and transforming growth factor α (TGFα) have been best characterized. Binding of EGF to EGFR has been characterized at the atomic level, and these analyses show that the monomeric EGFR exists in an autoinhibited state which is relieved by ligand binding (23). Ligand-mediated release of autoinhibition results in the exposure of a dimerization interface, and two ligand-bound EGFR monomers will therefore dimerize efficiently. Dimerization of EGFRs has the consequence that the tyrosine kinase domains of two monomers get sufficiently close to the other monomer to cause trans-phosphorylation of multiple cytosolic tyrosine residues. The phosphotyrosine groups recruit a host of important signalling mediators, including the regulatory subunit of class I phoshoinositide 3-kinase, the SH2- and SH3-domain-containing adaptor Grb2, the signal transducer and activator of transcription STAT3, and phospholipase C γ (124). In addition, phosphorylated EGFR recruits a molecule that profoundly controls its downregulation, namely the E3 ubiquitin ligase c-Cbl (64).
Ubiquitination of EGFR by Ube2D1-4 and c-Cbl
Ubiquitination of activated EGFR takes place at the plasma membrane as a rapid response to ligand binding and continues during endocytosis and early-endosomal trafficking, balanced by deubiquitination (70, 117) (Figure 1c). The E2 ubiquitin-conjugating enzymes Ube2D1-4 are involved, as is the E3 ubiquitin ligase c-Cbl (64, 117). c-Cbl belongs to a small subfamily of RING-domain-containing E3 ubiquitin ligases that also comprises two other ubiquitin ligases capable of ubiquitinating EGFR, Cbl-b and Cbl-c. c-Cbl can bind to the human EGFR in two ways, either directly to phosphorylated tyrosine-1045, or indirectly via binding to Grb2, which in turn binds to phosphorylated tyrosines -1068 and -1086 of the receptor (70). Ubiquitination of the EGFR is readily detectable at EGF concentrations as low as 1 ng/ml (52) but is considerably higher above a threshold EGF concentration of about 10 ng/ml (104). The type of EGFR ubiquitination following EGF stimulation has been determined both by indirect methods and by mass spectrometry (29, 39). The latter analyses have identified 6 major ubiquitination sites in EGFR, all located within the tyrosine kinase domain. More than half of the ubiquitin molecules appended to the EGFR are in the form of poly-ubiquitin (mostly Lys63-linked), whereas the remaining ubiquitination is represented by multi-mono-ubiquitin (39).
Endocytosis of EGFRs
EGFR endocytosis has been characterized since the seventies (13). These receptors can be endocytosed via several alternative pathways, possibly depending on the extent of ubiquitination (105), but internalization via clathrin-coated pits is quantitatively most important (28). Efficient endocytosis of EGFRs requires their tyrosine kinase activity (108), whereas ubiquitination facilitates but is not strictly required for EGFR endocytosis (28). A recent study has suggested that at least four partially redundant mechanisms contribute to EGFR endocytosis through clathrin-coated pits. These include ubiquitination of the tyrosine kinase domain, interaction with the clathrin adaptor AP-2, three C-terminal lysine residues outside the tyrosine kinase domain (which are subject to acetylation), and interaction with Grb2 (28). Consistent with a role for ubiquitination in EGFR endocytosis, knock-down of the ubiquitin-binding endocytosis adaptors Epsin and EPS15 causes a partial inhibition of internalization (52, 105, 112). On the other hand, an EGFR mutant in which all major ubiquitination sites are mutated is internalized at close to normal rate (38). The internalization of this mutant is nevertheless sensitive to Cbl depletion, suggesting that Cbl may have additional roles in endocytosis than mediating receptor ubiquitination. Indeed, Cbl has multiple interaction partners, including the actin- and ubiquitin binding protein CIN85, which in turn interacts with Endophilin to facilitate clathrin-mediated endocytosis of EGFRs (109).
Ubiquitin-dependent activation of GTPase cascades downstream of EGFR
In addition to the well-characterized phosphorylation-dependent activation of the small GTPase RAS downstream of activated EGFR, resulting in activation of the MAP kinase pathway, there is also evidence for a ubiquitin-dependent activation of small GTPase cascades downstream of ligand –bound EGFR. The unconventional E3 ubiquitin ligase Rabex-5 has been reported to bind to EGF-activated EGFR through its two ubiquitin-binding domains (84). Even though the functional implications of this interaction still have to be worked out, it is worth noting that Rabex-5 was originally identified as a guanine nucleotide exchange factor (GEF) for the small GTPase RAB5, which controls endosomal membrane fusion (36, 83). Moreover, Rabex-5 has also been identified as a downstream effector of a structurally related endosomal GTPase, RAB22 (128). Interestingly, EGFRs are endocytosed in Rabex-5-deficient cells but escape degradation in lysosomes (128). This opens the possibility that one of the consequences of EGFR ubiquitination may be, via recruitment of Rabex-5, to activate RAB5/RAB22-dependent EGFR trafficking through the early-endocytic pathway.
Endosomal sorting of EGFRs
Endocytosed EGFRs have two alternative fates – either recycling to the plasma membrane, or transport to lysosomes for degradation by hydrolases such as Cathepsin L (25, 35) (Figure 1c). Electron microscopy of EGFRs in cells stimulated with EGF has contributed strongly to the paradigm of lysosomal sorting via the MVB pathway since such studies were the first to demonstrate the ligand-dependent trafficking of a receptor into the ILVs of MVBs (74). Somewhat surprisingly, different ligands cause different trafficking of EGFRs, as best documented in the case of EGF versus TGFα. While stimulation with high concentrations of EGF mainly causes endocytosed EGFRs to be degraded in lysosomes, TGFα causes recycling of the bulk of endocytosed receptors (70). This paradox can be explained by the differential affinities of the two ligands for the receptors at the mildly low pH (5.5-6.0) found in early endosomes. EGF remains bound to EGFR at this pH, resulting in sustained receptor activation, whereas TGFα dissociates (70). The latter leads to a rapid decrease in EGFR ubiquitination and recycling instead of lysosomal sorting. These findings argue that there is a continuous ubiquitination and deubiquitination of EGFRs along the endocytic pathway, and that sustained ubiquitination requires receptor activation through ligand binding. Consistent with this, knock-down of c-Cbl and Cbl-b strongly inhibits ligand-induced degradation of EGFR while having less effect on internalization (39, 63, 85). There is strong evidence that degradative endosomal sorting of ubiquitinated EGFRs is mediated by the ESCRT machinery. EGFRs can be co-immunoprecipitated with Hrs after EGF stimulation (105, 117), and knockdown of various subunits of ESCRT-0, -I, -II or –III strongly inhibits EGF-mediated lysosomal degradation of EGFRs (6, 7, 9, 71, 91). An involvement of GGA3 in degradation of endocytosed EGF has also been reported, possibly reflecting alternative entries into the ESCRT pathway, with a GGA3-containing complex playing the role of an alternative ESCRT-0 (90). In addition, an isoform of EPS15 that associates with Hrs, EPS15B, is required for efficient lysosomal degradation of endocytosed EGFRs (99). The function of this protein in EGFR trafficking is not known, but the fact that it contains two C-terminal UIMs suggests that it may contribute to increase the avidity of ESCRT-0 for ubiquitinated receptors. In line with this, biochemical evidence suggests that poly-ubiquitinated EGFRs interact preferentially with ESCRT-0 and are selectively targeted for degradation (117).
Deubiquitination of EGFRs
Ubiquitinated EGFRs are substrate for the DUBs, AMSH and USP8/UBPY (72, 75). These DUBs share the same binding site on the ESCRT-0 subunit STAM through a non-canonical SH3 domain-binding motif, PX(V/I)(D/N)RXXKP (14). Both DUBs also contain microtubule interaction and transport (MIT) domains that enable them to interact with ESCRT-III. The MIT domains of these DUBs have different but overlapping interactions with ESCRT-III subunits, described in more detail below. AMSH has substrate specificity towards Lys63-linked polyubiquitin chains and has been proposed to oppose E3 ubiquitin ligase activity on the EGFR through deubiquitination at an early stage of the endosomal sorting process, thereby promoting EGFR recycling (118). USP8, on the other hand, has no preference for any particular type of polyubiquitin chains. This DUB is most similar to yeast Doa4 and might fulfill an equivalent role by deubiquitinating EGFRs prior to their inclusion into ILVs, in order to ensure recycling of ubiquitin (2, 98). However, like AMSH, USP8 has also been implicated at an earlier stage, countering the entry of EGFR into the ESCRT pathway (8). The competitive binding of AMSH and USP8 to ESCRT subunits, and the fact that they associate with both ESCRT-0 and –III, suggests that these DUBs may regulate cargo deubiquitination in complex ways.
STRUCTURE AND FUNCTION OF UBIQUITIN AND UBIQUITIN CHAINS
Having described the cellular itineraries of three archetypal ubiquitin-dependent cargoes, we will now consider the mechanisms behind these trafficking pathways in structural detail. Fortunately, ubiquitin is widespread in biology, and many of the relevant molecular mechanisms are conserved between trafficking and other pathways. Thus the field of trafficking benefits from a number of basic mechanistic and structural studies that were motivated in many cases by the role of ubiquitin in non-trafficking processes.
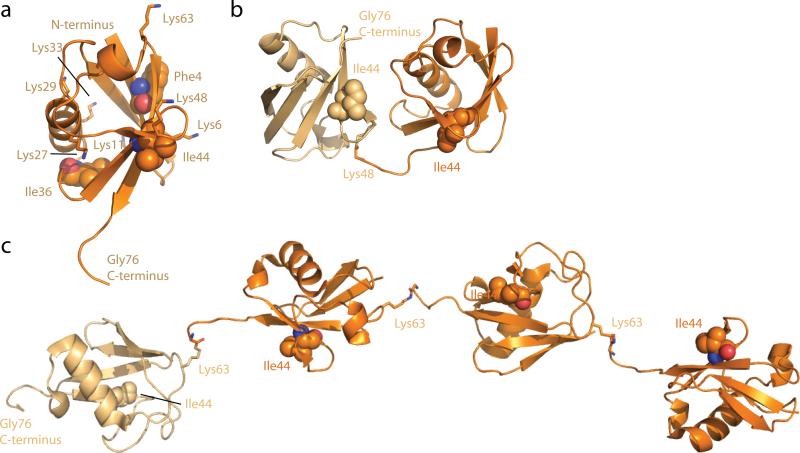
The ubiquitin fold consists of a five-stranded β-sheet and a single α-helix (121) (Figure 2a). The N- and C-termini are on opposite ends of the three-dimensional structure. The N-terminus is well-ordered, while the last six residues of the C-terminus extend beyond the structural core and are flexible. This latter feature of the structure is critically important, because the C-terminal carboxylate at Gly76 is the point of covalent attachment to cargo. As such, it must be flexible enough to undergo reactions catalyzed by ubiquitin ligases and DUBs. Ubiquitin has three major hydrophobic patches (106), one surrounding Phe4 near the N-terminus, one around Ile36, and the third surrounding Ile44, which is near the C-terminus in three dimensions. The latter patch is involved in every recognition event by the membrane trafficking machinery with only one known exception (42). The ubiquitin core is relatively rigid, but the intrinsic flexibility of the side-chains of a subset of the key recognition residues allows ubiquitin to bind a wide range of different partners (60, 86).
Figure 2.
Structure of ubiquitin and selected ubiquitin chains. (a) Structure (1UBQ) and major functional features of ubiquitin. The seven Lys residues are shown in a stick model, and the three hydrophobic residues at the center of the main hydrophobic patches are highlighted in space-filling spheres. (b) Structure of a K48-linked diubiquitin chain (1TBE). (c) Structure of a Lys63-linked tetraubiquitin chain (3HM3). In (b) and (c), the Lys involved in the linkage is shown in a stick model, and Ile44 of each ubiquitin monomer is shown in space-filling spheres. The proximal ubiquitin (the one that would be linked directly to cargo) is colored light orange, while other moieties are colored orange.
Polymeric ubiquitin chains are formed primarily by conjugation of the ubiquitin C-terminus to either the α–amino group of the polypeptide chain or the ε-amino groups of any one of the seven Lys residues of ubiquitin (89) (Figure 2a). The best studied ubiquitin chains are linked through Lys48, located next to the Ile44 hydrophobic patch and relatively close to the C-terminal flexible region, and through Lys63, located in three dimensions near the N-terminus.
Excluding branched chains, eight types of peptide or isopeptide chains occur, and four of these have been characterized structurally. The Lys11 and Lys48-linked ubiquitin chains are linked at points close together in the three-dimensional structure, and thus have little conformational freedom and prefer closed conformations (Figure 2b). The C- to N-terminally linked or “linear” ubiquitin chain and the Lys63-linked chain are connected at the two ends of the structure farthest from one another, and consequently adopt open conformations (Figure 2c).
MECHANISMS OF CARGO UBIQUITINATION
The Nedd4/Rsp5 family
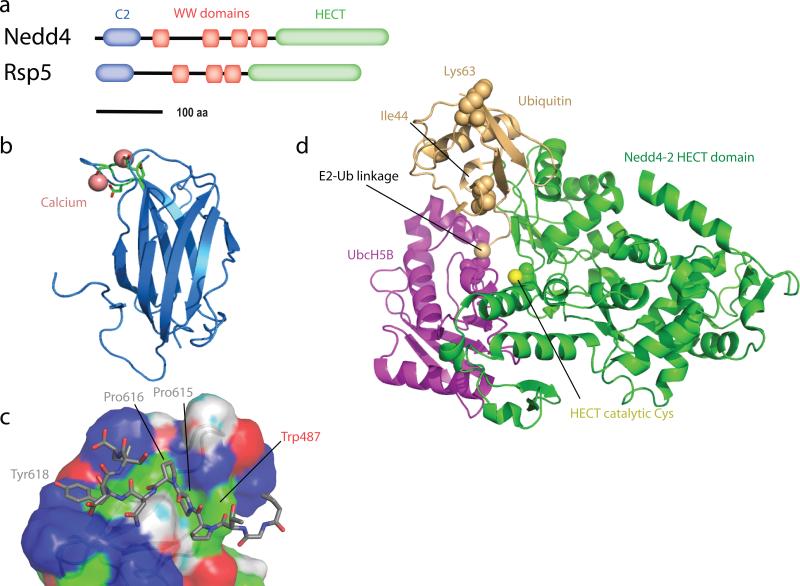
The human genome codes for nine Rsp5 orthologs, which comprise the Nedd4 ubiquitin ligase family (44). Rsp5 and the Nedd4 family members have a domain architecture consisting of an N-terminal C2 domain, two to four WW domains with connecting linkers, and a C-terminal HECT domain (Figure 3a). C2 (Protein kinase C conserved region 2) domains are β-sandwiches that come in two different structural permutations (78). The Nedd4 class of C2 domains belong to type II (Figure 3b). C2 domains also come in two flavors, those that bind Ca2+ and those that don't (78). Typically, Ca2+ binding promotes binding to membranes, and sometimes dimerization or binding to other proteins. The Nedd4 family C2 domains bind Ca2+ (Figure 3b). Ca2+ binding to the Rsp5 C2 domain promotes its binding to acidic lipids (21), following the conventional pattern.
Figure 3.
Rsp5 and the Nedd4 Family of ubiquitin ligases. (a) Domain architecture of Nedd4 and Rsp5. (b) Structure of the C2 domain of Nedd4 (2NSQ). Ca2+ ions and their ligands are shown as salmon-colored spheres and sticks, respectively. (c) Structure of the third WW domain of Nedd4 bound to the PPXY motif of EnaC (1I5H). (d) Structure of the Nedd4-2 HECT domain (3JW0; green, with catalytic Cys highlighted in space-filling spheres) in complex with an UbcH5B (magenta)~Ubiquitin (orange) adduct. The Ub-UbcH5B bond is highlighted with a space-filling sphere. Ile44 and Lys63 are highlighted to orient the viewer.
WW domains are compact 3-stranded β-sheet structures of just ~40 residues, one of the smallest functional protein domains known (69). WW domains are best known for binding to PPXY motifs and other Pro-rich recognition sequences, but in some cases also bind to phosphorylated Ser (pSer) and Thr (pThr) containing sequences (66). Nedd4 binds to some substrates, such as the EGFR-associated kinase ACK, via binding of its third WW domain (WW3) to a classical PPXY motif in ACK (65). Nedd4-2 WW3 binds to ENaC via the sequence PPPNYDSL, which comprises a C-terminally extended variant of the PPXY motif (49) (Figure 3c). Many other substrates of Nedd4 family ubiquitin ligases do not possess PPXY motifs. In some of these cases, adaptor proteins that possess PPXY motifs may serve as bridge to Nedd4 family membrane through their WW domains (103). In many others, alternative combinations of phosphorylated residues and PY elements are responsible. The Nedd4-2 WW2 binds to pT-PY peptides of Smad2/3 with high micromolar affinity (27). The structural mechanism for pT binding to the WW2 domain is not clear, because a key Arg residue involved in phosphopeptide binding in certain other WW domains (119) is not conserved in Nedd4 family WW domains.
HECT domains are ~350 residue catalytic models that form a covalent adduct with ubiquitin via a catalytic Cys residue before transferring ubiquitin to the substrate. Structures of three Nedd4 family HECT domains are known, those of WWP1 (120), Smurf2 (82), and Nedd4 (pdb 2ONI). These structures revealed flexibility between the ~250 residue N-terminal E2-binding domain and the ~100 residues C-terminal Cys-containing domain. A non-Nedd4 family HECT domain, that of E6-AP, has been crystallized in complex with an associated E2 enzyme, UbcH7 (40). This complex structure showed there was a 41 Å gap between the catalytic Cys residues of the E2 and E3 enzymes. When the HECT domain of Nedd4-2 was co-crystallized with UbcH5B~Ubiquitin(the covalent adduct between ubiquitin and the E2 enzyme, instead of the E2 enzyme alone), the HECT C-terminal domain rotated about the hinge such that the E2-Cys distance shrank to just 8 Å, nearly close enough for direct ubiquitin transfer (47) (Figure 3d).
The Cbl family
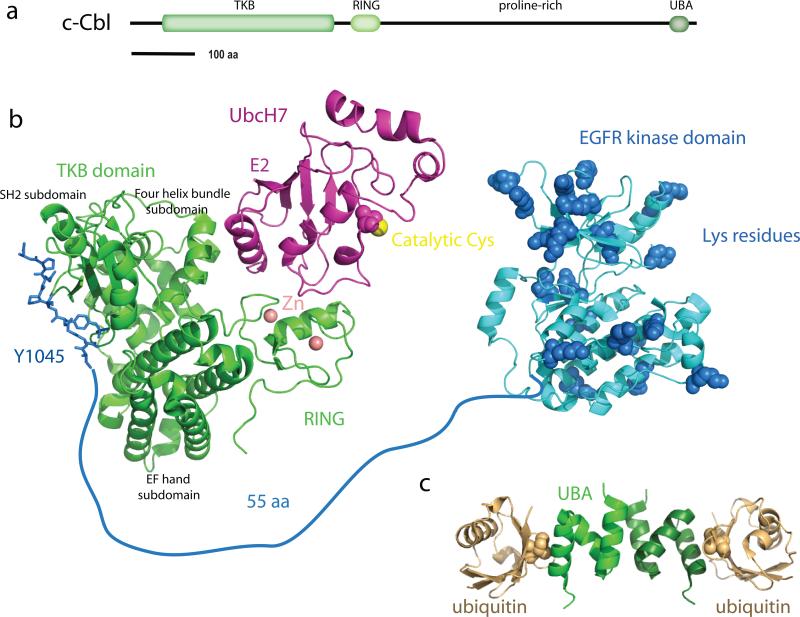
The Cbl proteins responsible for EGFR ubiquitination have similar architectures to one another: at their N-termini, a tyrosine kinase-binding (TKB) domain, followed by a RING ubiquitin ligase domain, a proline-rich domain (PRD), a Ser and Tyr-rich unstructured region, and finally a UBA domain. The architecture of Cbl-c corresponds to a truncated form of c-Cbl and Cbl-b, ending early in the PRD (Figure 4a). The TKB domain is essentially an expanded and divergent variant of the SH2 domain, which is the archetypal phosphotyrosine binding domain. The TKB domain consists of a divergent SH2 domain augmented by a four helix bundle subdomain and a Ca2+-binding EF hand subdomain (73) (Figure 4b). Although the variant SH2 domain of the TKB is missing the secondary β-sheet that contributes to peptide binding in classical SH2 domains, the pTyr-binding pocket and the overall mode of peptide binding is otherwise similar to that of classical SH2 domains (73). Ca2+-binding to the EF hand is important for the TKB to adopt the right conformation for the four-helix bundle to contribute to high-affinity pTyr peptide binding, and the integrity of all three of the subdomains is required for function (73). Following phosphorylation of Tyr1069, EGFR residues 1063-1075 fit the canonical Cbl TKB-binding motif (N/D)XpY(S/T)XXP, and the structure of this complex has been determined (79). The EGFR peptide forms an unusual internal salt bridge between the Arg immediately preceding the pTyr and the phosphate group of the pTyr (79).
Figure 4.
The Cbl family of ubiquitin ligases. (a) Domain architecture of c-Cbl. (b) Structural model for Cbl-dependent ubiquitination of the EGFR intracellular region. The structure of the TKB and RING portion of Cbl (green) in complex with UbcH7 (magenta) and a pTyr peptide (blue) from ZAP70 (1FBV) is used as a stand-in for the complex with Ube2D1-4 and the Tyr1045 region of EGFR. The catalytic domain of EGFR (3GT8) is colored blue. Lys residues of the EGFR catalytic domain are highlighted in space-filling spheres, even though not all of these Lys have been directly shown to be ubiquitinated. Structural zinc ions of the Cbl RING domain are shown as salmon-colored spheres. (c) Dimeric complex of the Cbl-b UBA domain (two shades of green) with ubiquitin (two shades of orange) (2OOB).
There are two major classes of ubiquitin ligases, the HECT domain ligases, such as the Nedd4 family discussed above, and the RING domain class, of which Cbl is a member. In contrast to HECT domain ubiquitin ligases, RING domain ubiquitin ligases do not form a covalent adduct with the ubiquitin moiety. Rather, they serve as adaptors to bring together the Ub-bound E2 enzyme together with its substrate. The function of the RING domain is to recruit the E2 enzyme, while other regions of the ligase are typically responsible for substrate binding. The crystal structure of the TKB-RING portion of c-Cbl has been determined in complex with the E2 enzymeUbcH7 and a pTyr peptide from the substrate ZAP70 (126) (Figure 4b). The RING domain itself is built around two tetrahedrally coordinated zinc ions, which have a structural rather than catalytic role. Cbl has a groove formed by the zinc-binding loops and an α-helix in which the E2 enzyme binds. Five residues from these two loops and the helix control E2 enzyme specificity. In the case of Cbl, these residues are mostly hydrophobic, notably Ile383 and Trp408. This site on the RING domains specifies an E2 that has a Phe residue on its L1 loop, Phe63 in the case of UbcH7. The Ube2D enzymes responsible for Cbl-dependent EGFR ubiquitination conform to this sequence pattern (117), thus the UbcH7-Cbl complex probably provides a reasonable model for the structures of the functional Ube2D-Cbl complexes.
c-Cbl functions in EGFR endocytosis, by virtue of its coupling to the SH3-domain containing endocytic proteins CIN85 and endophilin. Residues 902-912 of the c-Cbl PRD form a unique SH3 binding motif in which two SH3 recognition sequences are interwoven with one another (46). A similar mechanism allows the DUIM motif of Hrs to bind two ubiquitin moieties at once, as described below. Two copies of the CIN85 SH3 domain thus bind to a single eleven residue peptide motif from c-Cbl (46). This drives the dimerization of CIN85 and other proteins with similar SH3 domains. Finally, the C-terminal UBA domains of c-Cbl and Cbl-b bind Ub. The Cbl-b UBA domain binds monoubiquitin with ~60 μM affinity, and K48-Ub4 with 2 μM affinity (87). The Cbl-b UBA domain dimerizes, but only weakly in the absence of ubiquitin chains (87). Cross-linking of the UBA domains by binding to polyubiquitin promotes dimerization (Figure 4c), and in turn, activation of Cbl. How the Ub-chain induced dimerization of Cbl-b is integrated into the larger picture of Cbl physiology remains to be discovered.
HOW THE UBIQUITIN TAG IS RECOGNIZED
Virtually all of the known effects of ubiquitination on protein sorting are mediated by binding of the ubiquitin tags to specific ubiquitin binding domains (UBDs) (17, 34, 42). The past decade has seen rapid advances in the structure, function, and biology of UBDs in both the trafficking area and in other fields. UBDs as a class have been reviewed elsewhere (17, 34, 42), and the supplementary section of the online version of this article at http://www.annualreviews.org/ describes the specifics of UBD function in the trafficking of the cargoes described above.
REMOVAL OF THE UBIQUITIN TAG
DUBs cleave the isopeptide bond between the ubiquitin C-terminal Gly and the Lys residue of the ubiquitinated protein (14, 57). DUBs are therefore proteases, and use the same chemical reaction mechanims as conventional proteases. There are five structural classes of DUBs (57). Of these, two families, the JAMM (JAB1/MPN/MOV34) family and the USP (Ub specific protease) family, are the most extensively implicated in regulating the cargoes described earlier in this review.
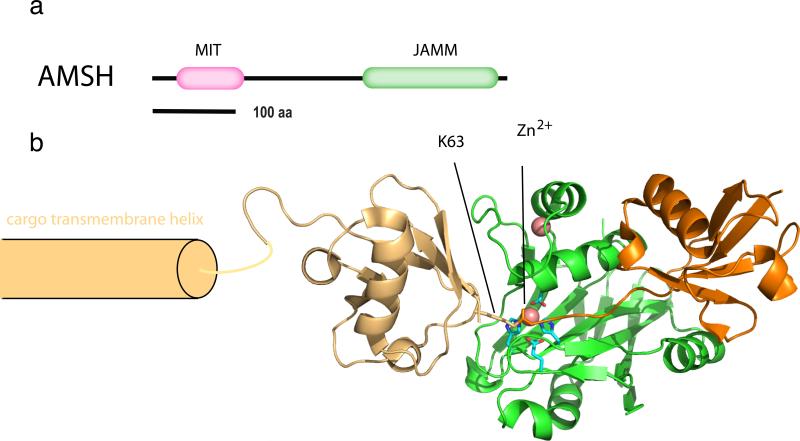
The JAMM family members AMSH and AMSH-LP
The human proteome includes eight JAMM family members. Two of these, AMSH and AMSH-LP, are implicated in regulation of membrane traffic, and both contain N-terminal microtubule interacting and transport (MIT) domains (43, 116) (Figure 5a). Many MIT domains, including the MIT domain of AMSH, bind to MIT-interacting motifs (MIMs), short sequences at the C-termini of ESCRT-III proteins that bind to grooves between the three helices of the MIT domain helical bundle (54, 81, 115). The AMSH MIT domain binds to CHMP1A, 1B, and 3 by multiple reports (68, 97, 116, 125). These are MIM1-containing proteins, and the MIM1 binding site of the structurally-characterized VPS4-MIT is mostly preserved in AMSH-MIT. The structurally defined Vps4-MIT-MIM1 interaction (81, 115) is a reasonable model for the AMSH MIT-CHMP3 interaction. It will still be important to know the experimental structure of the latter complex, since AMSH-MIT, unlike VPS4 MIT, does not bind to CHMP2 isoforms.
Figure 5.
The zinc isopeptidase AMSH. (a) Domain architecture of AMSH. (b) Structural model for the catalytic complex of AMSH-LP bound to a Lys63-Ub2-modified cargo. The structural model was derived by superimposing the structure of active, zinc-bound, Ub-free form of AMSH-LP (2ZNR) on the structure of the Lys63-Ub2 complex (2ZVN; two shades of orange for the two moieties) with an inactivated mutant lacking the catalytic zinc ion. To illustrate the positioning of the proximal and distal moieties of the Lys63-Ub, the cargo is modeled as a single pass transmembrane protein with the ubiquitin conjugated close to the transmembrane domain. Zinc ions are shown as salmon-colored space-filling spheres.
JAMM domain DUBs are zinc metalloproteases that are mechanistically related (although not homologous to) the archetypal metalloprotease thermolysin. Not all JAMM domain proteins are DUBs. As compared to non-DUB JAMM domain, the JAMM domain of AMSH-LP has two AMSH family-specific sequence insertions, Ins-1 and Ins-2 (101). The AMSH-family JAMM domains bind two zinc ions, one a structural zinc ion bound to the conserved JAMM core, and the second a catalytic zinc ion bound by residues of Ins-2. The structure of a catalytically inactivated AMSH-LP JAMM domain in a stable complex with Lys63-Ub2 has been determined (Figure 5b), yielding remarkably detailed insight into the specificity of AMSH and AMSH-LP for Lys63-linked ubiquitin chains (101). Lys63-Ub2 is bound in a highly open conformation, similar to the open structure of Lys63 chains crystallized by themselves or with Lys63-specific recognition domains (15, 58, 59, 100). The Gln62-Lys63-Glu64 tripeptide of the proximal ubiquitin moiety (corresponding to the moiety attached to a Lys63-Ub2 modified cargo protein) makes multiple hydrogen bonds directly with the Ins-2 region of the JAMM domain (101). These hydrogen bonds align Lys63-ubiquitin for catalytically productive isopeptide bond hydrolysis, while other types of linkages would fail to make these interactions. Thus Lys63 specificity appears to be hard-wired into the architecture of the AMSH subfamily of JAMM domains.
USP family members USP8 and Doa4
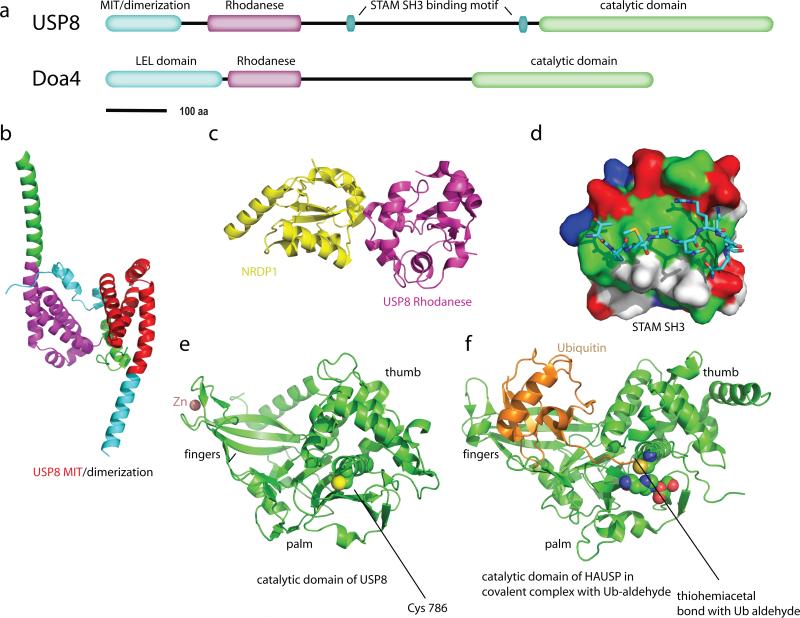
With the exception of the JAMM domain family described above, the other four families of DUBs are all Cys proteases (57). The Cys thiol is activated by participation in a Cys-His-Asp/Asn triad similar to that of other Ser and Cys proteases. The unprotonated Cys thiol group acts as a nucleophile to attack the carbon atom of the isopeptide bond. This leads to formation of an acylenzyme intermediate and ejection of the Lys residue. The electrostatic environment of the enzyme active site acts to stabilize the negative charge on the acylenzyme intermediate, which is the hydrolyzed by an attacking water molecule. UCH-L3, responsible for deubiquination of EnaC, is an ubiquitin C-terminal hydrolase (UCH) family member and has been crystallized (45). The largest family of DUBs in the human proteome is the ubiquitin-specific protease (USP) family, of which human USP8 is a member. USP8 contains an N-terminal MIT domain, a rhodanese homology domain, a ~500 residue linker, and a catalytic domain (Figure 6a). Yeast Doa4, responsible for deubiquitinating Cps1 and other yeast MVB cargoes, has a similar domain architecture to USP8, with both Rhodanese and Cys-containing catalytic domains.
Figure 6.
The Cys isopeptideases USP8 and Doa4. (a) Domain architecture of USP8 and Doa4. (b) Dimeric structure of the N-terminal domain of USP8 (2A9U). The portions of the two monomers the correspond to the MIT sequence motif are colored red and magenta, and the remainder of the subunits are colored green and cyan, respectively. (c) Structure of the rhodanese domain of USP8 (2GWF; magenta) in complex with the non-catalytic C-terminal domain of the ubiquitin ligase NRDP1 (yellow). (d) A surface representation of the SH3 domain of STAM2 (1UJ0), colored green (hydrophobic residues), blue(basic residues), red (acidic residues), and white (uncharged polar residues). USP8 residues 699-709 are shown in a stick model colored by atom type. (e) Catalytic domain of USP8 in an inactive conformation (2GF0). The non-catalytic (structural) zinc ion is colored salmon. (f) Catalytic domain of HAUSP in covalent complex with Ub-aldehyde (1NBF), as a model for the active form of USP8. The residues of the Cys-His-Asp catalytic triad are shown in space filling spheres and colored by atom type.
The USP8 MIT domain binds tightly to CHMP1B and more weakly to a subset of other CHMPs (97). Unlike other MIT domains, which are stand-alone three-helix bundles, the USP8 MIT is part of a larger dimeric seven-helical domain (5) (Figure 6b). The dimer interface is extensive, but it has not been established that it is required for USP8 function, nor it is known how dimerization might relate to ESCRT-III binding. Doa4 contains an N-terminal domain with 5-7 predicted helices that is also implicated in recruitment to endosomes by ESCRT-III (3). While not detectably related by sequence homology, their similar size, function, helical structure, and placement in the domain architecture suggests an underlying structural commonality between these regions of USP8 and Doa4. The USP8 and Doa4 sequences contain a rhodanese domain just C-terminal to their ESCRT-III binding domain. Classical rhodanese domains use a catalytic Cys to carry out phosphatase or sulfurtransferase reactions, while non-catalytic variants lack the catalytic Cys and have collapsed active sites. The USP8 rhodanese domain belongs to the inactive category and is involved in protein-protein interactions, including a structurally characterized complex with the non-catalytic C-terminal domain of the E3 ligase NRDP1 (5) (Figure 6c). Residues 699-709 of the long rhodanese-UCH linker region bind to the STAM SH3 with ~ 30 μM affinity, and the structure of the complex has been determined (48) (Figure 6d). These residues, TPMVNRENKP, do not conform to an SH3-binding consensus motif, but the non-ideal nature of the sequence is partially compensated by electrostatic interactions between the Arg and Lys residues of the motif with acidic pockets on the SH3 domain (48).
The USP8 catalytic domain has been crystallized in an inactive conformation in the absence of ubiquitin (5) (Figure 6e). The structure of another USP protein, HAUSP, in a covalent complex with Ub-aldehyde (37) (Figure 6f), provides a reasonable model for the mechanism of deubiquitination across the USP family. The catalytic domain has been compared to a hand, with the fingers grasping the bulk of the ubiquitin moiety, while the C-terminal tail of ubiquitin is threaded between the palm and the thumb. The catalytic Cys-His-Asp triad is located between the thumb and palm such that a covalent bond can be formed between the C-terminal carboxylate of ubiquitin and the catalytic Cys. This places the cargo protein (or a more proximal ubiquitin moiety) on the distal side of the thumb region with respect to the ubiquitin moiety being cleaved (Figure 6f).
CONCLUDING REMARKS AND PERSPECTIVES
Since the concept of ubiquitin-dependent membrane traffic was born in the mid-nineties, substantial progress has been made in understanding the molecular details of how cargo proteins are ubiquitinated by E3 ubiquitin ligases, how sorting components such as GGAs and ESCRT subunits recognize the ubiquitinated cargoes, how ubiquitin recognition controls the intracellular itineraries of membrane proteins, and how DUBs are recruited to remove ubiquitin moieties. Even though sufficient data are available to allow a broad overview of ubiquitin as a tag for protein trafficking, several outstanding questions remain to be addressed before such processes can be modelled in detail.
One of the topics that awaits further clarification is the importance of Lys63-linked polyubiquitin chains as a sorting signal. In mammalian systems there is evidence that Lys63-linked polyubiquitin via the E2s Ube2D1-4 and the E3 Cbl is crucial for endolysosomal sorting of EGFRs (117), and that Lys63-linked polyubiquitination via the E2 Ubc13 and the E3 K3 is required for Kaposi's sarcoma associated Herpes virus-induced endocytosis and endolysosomal sorting of MHC class I molecules (19, 20). Likewise, Lys63-linked polyubiquitination appears to be required for vacuolar targeting of the yeast amino acid permease Gap1 (61). On the other hand, there is convincing evidence that monoubiquitin is a sufficient signal for endocytosis of several yeast membrane proteins such as Gap1, the maltose transporter Mal1, and the mating factor receptor Ste2 (61). Moreover, Lys63-linked diubiquitination appears sufficient to target Cps1 to the vacuole (61). It will be important to establish whether the differential requirements for Lys63 polyubiquitination may be related to the types of sorting components that are involved. A few measurements have been made of the affinities and avidities of sorting machinery for ubiquitin chains (59, 94), but more data are needed, particularly in the context of cargo and membranes.
The cargoes described in this review were selected because they have been so well studied compared to most others. Examples such as the specific recognition of phosphorylated EGFR by the TKB domain of Cbl (79) beautifully illustrate the specific targeting and regulation of the ubiquitination machinery. Such clear cut examples are still more the exception than the rule. Chain specificity remains just as poorly characterized in most cases, with a few spectacular exceptions such as the mechanism of Lys63-ubiquitin specific cleavage by AMSH (100). More insights into these mechanisms will be eagerly awaited.
Ubiquitin signals and ubiquitin receptors operate at multiple sequential steps in trafficking. It remains to be understood how ubiquitinated cargoes are handed off at each step. Simple models, such as the possibility of a gradient of sorting complex UBD ubiquitin affinities that follows the directionality of sorting, do not seem sufficient to account for cargo hand-off. In this connection, it is intriguing that at certain steps in the pathway, ubiquitin ligases, DUBs, and ubiquitin-binding proteins appear to form assemblies with one another. For example, the yeast ESCRTs interact, directly or indirectly, with both the ligase Rsp5 (10, 93) and the DUB Doa4 (3, 67). This raises a host of possibilities for ubiquitin chain remodeling and compartmentalized, complete ubiquitination/deubiquitination cycles that have barely begun to be explored. The molecular gymnastics of the ubiquitin system in membrane trafficking should continue to offer insights and surprises for years to come.
Supplementary Material
SUMMARY POINTS.
S. cerevisiae Cps1 is the prototypical ubiquitin-dependent biosynthetic cargo. Its sorting from the Golgi to the MVB depends on its ubiquitination by the HECT domain E3 ligase Rsp5 and its recognition of the ubiquitin moieties by ubiquitin binding domains of the ESCRT complexes. Ubiquitin is recycled by the Cys-catalytic triad DUB Doa4.
The level of ENaC at the apical membrane of epithelial cells is controlled by Nedd4-2 dependent ubiquitination and internalization to early endosomes. Nedd4-2 is a HECT family E3 ligase that binds to ENaC through its third WW domain. At the early endosome, deubiquitination by the Cys-dependent DUBs UCH-L3 and USP-45 directs ENaC recycling to the apical membrane. In the absence of DUB action, ENaC is directed to the lysosome for its degradation via the ESCRT complexes.
Following EGF binding and transphosphorylation of its C-terminus, EGFR is ubiquitinated by the RING domain E3 ligase Cbl. At the early endosome, ubiquitin signals are read out by Rabex-5, leading to RAB5 activation, a key event in endosome maturation. EGRF can be recycled following deubiquitination by the zinc-dependent DUB AMSH, or it can bind to GGA3 and the ESCRT complexes and enter the MVB pathway.
The key structural features of ubiquitin for membrane traffic are its Ile44 patch, which binds to nearly all of the components of ubiquitin-dependent sorting; its flexible C-terminus, which enables acrobatic ubiquitination and deubiquitination reactions and conjugation to diverse protein substrates; and Lys63, which is involved in forming open, flexible polyubiquitin chains.
Most ubiquitination events in trafficking are carried out by Nedd4 family E3 ligases, which binds to proline rich and/or phosphorylated sequences in their cargo substrates.
Tyrosine phosphorylated substrates are recognized by Cbl through its TKB domain.
DUBs are targeted to the trafficking machinery mainly through interactions between their MIT domains and the C-termini of ESCRT-III subunits, and between SH3 domains and Pro-rich sequences.
Certain components of the sorting machinery, including AMSH and the NZF2 domain of S. cerevisiae ESCRT-II, are highly specific for Lys63-linked polyubiquitin. Lys63-linked ubiquitin is abundant on substrates of membrane trafficking such as EGFR. Yet there is little evidence for Lys63-specific E2 enzymes in trafficking pathways, and definitive evidence that Lys63 linkages are required for trafficking has been elusive.
ACKNOWLEDGMENTS
We thank Y. Ye and R. Stanley for comments on the manuscript. This research was supported by the intramural program of the NIH, NIDDK to JHH, and by the Research Council of Norway, the Norwegian Cancer Society, and the European Research Council to HS.
MINI-GLOSSARY
- Ubiquitination
The covalent conjugation of the C-terminal carboxylate to a protein substrate, usually, but not always, via an isopeptide bond with the e-amino nitrogen of Lys residues
- Deubiquitination
The hydrolytic cleavage of the above mentioned isopeptide bond. Polyubiquitin. Covalently conjugated chains of ubiquitin moieties formed by ubiquitination reactions
ACRONYMS
- ESCRT
Endosomal sorting complex required for transport
- DUB
deubiquitinating enzyme
- UBD
ubiquitin binding domain
- EGFR
epidermal growth factor receptor
- USP
ubiquitin specific peptidase
- TKB
tyrosine kinase binding
- MIT
microtubule interaction and transport
- VHS
Vps27, Hrs, and STAM
- GAT
GGAs and TOM
- UIM
ubiquitin interaction motif
LITERATURE CITED
- 1.Almaca J, Kongsuphol P, Hieke B, Ousingsawat J, Viollet B, et al. AMPK controls epithelial Na(+) channels through Nedd4-2 and causes an epithelial phenotype when mutated. Pflugers Arch. 2009;458:713. doi: 10.1007/s00424-009-0660-4. [DOI] [PubMed] [Google Scholar]
- 2.Alwan HA, van Leeuwen JE. UBPY-mediated epidermal growth factor receptor (EGFR) de-ubiquitination promotes EGFR degradation. J. Biol. Chem. 2007;282:1658. doi: 10.1074/jbc.M604711200. [DOI] [PubMed] [Google Scholar]
- 3.Amerik A, Sindhi N, Hochstrasser M. A conserved late endosome-targeting signal required for Doa4 deubiquitylating enzyme function. J. Cell Biol. 2006;175:825. doi: 10.1083/jcb.200605134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amerik AY, Nowak J, Swaminathan S, Hochstrasser M. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol. Biol. Cell. 2000;11:3365. doi: 10.1091/mbc.11.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avvakumov GV, Walker JR, Xue S, Finerty PJ, Mackenzie F, et al. Amino-terminal dimerization, NRDP1-rhodanese interaction, and inhibited catalytic domain conformation of the ubiquitin-specific protease 8 (USP8). J. Biol. Chem. 2006;281:38061. doi: 10.1074/jbc.M606704200. [DOI] [PubMed] [Google Scholar]
- 6.Babst M, Odorizzi G, Estepa EJ, Emr SD. Mammalian tumor suceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic. 2000;1:248. doi: 10.1034/j.1600-0854.2000.010307.x. [DOI] [PubMed] [Google Scholar]
- 7.Bache KG, Raiborg C, Mehlum A, Stenmark H. STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J. Biol. Chem. 2003;278:12513. doi: 10.1074/jbc.M210843200. [DOI] [PubMed] [Google Scholar]
- 8.Berlin I, Schwartz H, Nash PD. Regulation of the epidermal growth factor receptor ubiquitination and trafficking by the USP8/STAM complex. J. Biol. Chem. 2010 doi: 10.1074/jbc.M109.016287. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bishop N, Horman A, Woodman P. Mammalian class E vps proteins recognize ubiquitin and act in the removal of endosomal protein-ubiquitin conjugates. J. Cell Biol. 2002;157:91. doi: 10.1083/jcb.200112080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowers K, Lottridge J, Helliwell SB, Goldthwaite LM, Luzio JP, Stevens TH. Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic. 2004;5:194. doi: 10.1111/j.1600-0854.2004.00169.x. [DOI] [PubMed] [Google Scholar]
- 11.Butterworth MB. Regulation of the epithelial sodium channel (ENaC) by membrane trafficking. Biochim.Biophys.Acta. 2010 doi: 10.1016/j.bbadis.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butterworth MB, Edinger RS, Ovaa H, Burg D, Johnson JP, Frizzell RA. The deubiquitinating enzyme UCH-L3 regulates the apical membrane recycling of the epithelial sodium channel. J. Biol. Chem. 2007;282:37885. doi: 10.1074/jbc.M707989200. [DOI] [PubMed] [Google Scholar]
- 13.Carpenter G, Cohen S. 125I-labeled human epidermal growth factor. Binding, internalization, and degradation in human fibroblasts. J. Cell Biol. 1976;71:159. doi: 10.1083/jcb.71.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clague MJ, Urbe S. Endocytosis: the DUB version. Trends Cell Biol. 2006;16:551. doi: 10.1016/j.tcb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Datta AB, Hura GL, Wolberger C. The structure and conformation of Lys63-linked tetraubiquitin. J. Mol. Biol. 2009;392:1117. doi: 10.1016/j.jmb.2009.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Debonneville C, Flores SY, Kamynina E, Plant PJ, Tauxe C, et al. Phosphorylation of Nedd4-2 by Sgk1 regulates epithelial Na(+) channel cell surface expression. EMBO J. 2001;20:7052. doi: 10.1093/emboj/20.24.7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nature Reviews Molecular Cell Biology. 2009;10:659. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dinudom A, Fotia AB, Lefkowitz RJ, Young JA, Kumar S, Cook DI. The kinase Grk2 regulates Nedd4/Nedd4-2-dependent control of epithelial Na+ channels. Proc.Natl.Acad.Sci.U.S.A. 2004;101:11886. doi: 10.1073/pnas.0402178101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duncan LM, Nathan JA, Lehner PJ. Stabilization of an E3 ligase-E2-ubiquitin complex increases cell surfaces MHC class I expression. J. Immunol. 2010;184:6978. doi: 10.4049/jimmunol.0904154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duncan LM, Piper S, Dodd RB, Saville MK, Sanderson CM, et al. Lysine-63-linked ubiquitination is required for endolysosomal degradation of class I molecules. EMBO J. 2006;25:1635. doi: 10.1038/sj.emboj.7601056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunn R, Klos DA, Adler AS, Hicke L. The C2 domain of the Rsp5 ubiquitin ligase binds membrane phosphoinositides and directs ubiquitination of endosomal cargo. J. Cell Biol. 2004;165:135. doi: 10.1083/jcb.200309026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fakitsas P, Adam G, Daidie D, van Bemmelen MX, Fouladkou F, et al. Early aldosterone-induced gene product regulates the epithelial sodium channel by deubiquitylation. J.Am.Soc.Nephrol. 2007;18:1084. doi: 10.1681/ASN.2006080902. [DOI] [PubMed] [Google Scholar]
- 23.Ferguson KM, Berger MB, Mendrola JM, Cho HS, Leahy DJ, Lemmon MA. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol. Cell. 2003;11:507. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 24.Firsov D, Gautschi I, Merillat AM, Rossier BC, Schild L. The heterotetrameric architecture of the epithelial sodium channel (ENaC). EMBO J. 1998;17:344. doi: 10.1093/emboj/17.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Futter CE, Pearse A, Hewlett LJ, Hopkins CR. Multivesicular endosomes containing internalized EGF-EGF receptor complexes mature and then fuse directly with lysosomes. J. Cell Biol. 1996;132:1011. doi: 10.1083/jcb.132.6.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galan JM, Moreau V, Andre B, Volland C, Haguenauer-Tsapis R. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J. Biol. Chem. 1996;271:10946. doi: 10.1074/jbc.271.18.10946. [DOI] [PubMed] [Google Scholar]
- 27.Gao S, Alarcon C, Sapkota G, Rahman S, Chen PY, et al. Ubiquitin Ligase Nedd4L Targets Activated Smad2/3 to Limit TGF-beta Signaling. Mol. Cell. 2009;36:457. doi: 10.1016/j.molcel.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goh LK, Huang F, Kim W, Gygi S, Sorkin A. Multiple mechanisms collectively regulate clathrin-mediated endocytosis of the epidermal growth factor receptor. J. Cell Biol. 2010;189:871. doi: 10.1083/jcb.201001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 2003;5:461. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- 30.Hershko A, Ciechanover A, Varshavsky A. The ubiquitin system. Nat. Med. 2000;6:1073. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- 31.Hettema EH, Valdez-Taubas J, Pelham HR. Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. EMBO J. 2004;23:1279. doi: 10.1038/sj.emboj.7600137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hicke L. Protein regulation by monoubiquitin. Nature Reviews Molecular Cell Biology. 2001;2:195. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 33.Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277. doi: 10.1016/s0092-8674(00)80982-4. [This seminal paper established the concept of ubiquitination as determinant of membrane trafficking.] [DOI] [PubMed] [Google Scholar]
- 34.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nature Reviews Molecular Cell Biology. 2005;6:610. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 35.Hiwasa T, Sakiyama S, Yokoyama S, Ha JM, Fujita J, et al. Inhibition of cathepsin L-induced degradation of epidermal growth factor receptors by c-Ha-ras gene products. Biochem. Biophys. Res. Commun. 1988;151:78. doi: 10.1016/0006-291x(88)90561-x. [DOI] [PubMed] [Google Scholar]
- 36.Horiuchi H, LippÇ R, McBride HM, Rubino M, Woodman P, et al. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 1997;90:1149. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- 37.Hu M, Li PW, Li MY, Li WY, Yao TT, et al. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 38.Huang F, Goh LK, Sorkin A. EGF receptor ubiquitination is not necessary for its internalization. Proc.Natl.Acad.Sci.U.S.A. 2007;104:16904. doi: 10.1073/pnas.0707416104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol. Cell. 2006;21:737. doi: 10.1016/j.molcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 40.Huang L, Kinnucan E, Wang GL, Beaudenon S, Howley PM, et al. Structure of an E6AP-UbcH7 complex: Insights into ubiquitination by the E2-E3 enzyme cascade. Science. 1999;286:1321. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- 41.Hummler E, Vallon V. Lessons from mouse mutants of epithelial sodium channel and its regulatory proteins. J.Am.Soc.Nephrol. 2005;16:3160. doi: 10.1681/ASN.2005040450. [DOI] [PubMed] [Google Scholar]
- 42.Hurley JH, Lee S, Prag G. Ubiquitin binding domains. Biochem. J. 2006;399:361. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hurley JH, Yang D. MIT domainia. Dev. Cell. 2008;14:6. doi: 10.1016/j.devcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 44.Ingham RJ, Gish G, Pawson T. The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene. 2004;23:1972. doi: 10.1038/sj.onc.1207436. [DOI] [PubMed] [Google Scholar]
- 45.Johnston SC, Larsen CN, Cook WJ, Wilkinson KD, Hill CP. Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 angstrom resolution. EMBO J. 1997;16:3787. doi: 10.1093/emboj/16.13.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jozic D, Cardenes N, Deribe YL, Moncalian G, Hoeller D, et al. Cbl promotes clustering of endocytic adaptor proteins. Nat. Struct. Mol. Biol. 2005;12:972. doi: 10.1038/nsmb1000. [DOI] [PubMed] [Google Scholar]
- 47.Kamadurai HB, Souphron J, Scott DC, Duda DM, Miller DJ, et al. Insights into Ubiquitin Transfer Cascades from a Structure of a UbcH5B similar to Ubiauitin-HECTNEDD4L Complex. Mol. Cell. 2009;36:1095. doi: 10.1016/j.molcel.2009.11.010. [The most complete account to date of the molecular gymnastics of NEDD4-family dependent ubiquitination.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaneko T, Kumasaka T, Ganbe T, Sato T, Miyazawa K, et al. Structural insight into modest binding of a non-PXXP ligand to the signal transducing adaptor molecule-2 Src homology 3 domain. J. Biol. Chem. 2003;278:48162. doi: 10.1074/jbc.M306677200. [DOI] [PubMed] [Google Scholar]
- 49.Kanelis V, Rotin D, Forman-Kay JD. Solution structure of a Nedd4 WW domain-ENaC peptide complex. Nat. Struct. Biol. 2001;8:407. doi: 10.1038/87562. [DOI] [PubMed] [Google Scholar]
- 50.Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145. doi: 10.1016/s0092-8674(01)00434-2. [This study established ubiquitin as the key signal for cargo sorting into multivesicular bodies, demonstrated ubiquitin-dependent sorting of Cps1, and named and characterized the first ESCRT complex.] [DOI] [PubMed] [Google Scholar]
- 51.Katzmann DJ, Sarkar S, Chu T, Audhya A, Emr SD. Multivesicular body sorting: Ubiquitin ligase Rsp5 is required for the modification and sorting of carboxypeptidase S. Mol. Biol. Cell. 2004;15:468. doi: 10.1091/mbc.E03-07-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kazazic M, Bertelsen V, Pedersen KW, Vuong TT, Grandal MV, et al. Epsin 1 is involved in recruitment of ubiquitinated EGF receptors into clathrin-coated pits. Traffic. 2009;10:235. doi: 10.1111/j.1600-0854.2008.00858.x. [DOI] [PubMed] [Google Scholar]
- 53.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell. Dev. Biol. 2006;22:159. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 54.Kieffer C, Skalicky JJ, Morita E, De Domenico I, Ward DM, et al. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev. Cell. 2008;15:62. doi: 10.1016/j.devcel.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 55.Kim HC, Huibregtse JM. Polyubiquitination by HECT E3s and the Determinants of Chain Type Specificity. Mol. Cell. Biol. 2009;29:3307. doi: 10.1128/MCB.00240-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kleyman TR, Carattino MD, Hughey RP. ENaC at the cutting edge: regulation of epithelial sodium channels by proteases. J. Biol. Chem. 2009;284:20447. doi: 10.1074/jbc.R800083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nature Reviews Molecular Cell Biology. 2009;10:550. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 58.Komander D, Reyes-Turcu F, Licchesi JDF, Odenwaelder P, Wilkinson KD, Barford D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. Embo Reports. 2009;10:466. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kulathu Y, Akutsu M, Bremm A, Hofmann K, Komander D. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat. Struct. Mol. Biol. 2009;16:1328. doi: 10.1038/nsmb.1731. [DOI] [PubMed] [Google Scholar]
- 60.Lange OF, Lakomek NA, Fares C, Schroder GF, Walter KFA, et al. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science. 2008;320:1471. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- 61.Lauwers E, Jacob C, Andre B. K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J. Cell Biol. 2009;185:493. doi: 10.1083/jcb.200810114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee JR, Oestreich AJ, Payne JA, Gunawan MS, Norgan AP, Katzmann DJ. The HECT domain of the ubiquitin ligase Rsp5 contributes to substrate recognition. J. Biol. Chem. 2009;284:32126. doi: 10.1074/jbc.M109.048629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell. 1999;4:1029. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 64.Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, et al. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663. doi: 10.1101/gad.12.23.3663. [This paper showed that Cbl-mediated ubiquitination targets EGFR for lysosomal degradation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin Q, Wang J, Childress C, Sudol M, Carey DJ, Yang WNA. HECT E3 Ubiquitin Ligase Nedd4-1 Ubiquitinates ACK and Regulates Epidermal Growth Factor (EGF)-Induced Degradation of EGF Receptor and ACK. Mol. Cell. Biol. 2010;30:1541. doi: 10.1128/MCB.00013-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu PJ, Zhou XZ, Shen MH, Lu KP. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science. 1999;283:1325. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- 67.Luhtala N, Odorizzi G. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J Cell Biol. 2004 doi: 10.1083/jcb.200403139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ma YM, Boucrot E, Villen J, Affar el B, Gygi SP, et al. Targeting of AMSH to endosomes is required for epidermal growth factor degradation. J. Biol. Chem. 2007;282:9805. doi: 10.1074/jbc.M611635200. [DOI] [PubMed] [Google Scholar]
- 69.Macias MJ, Hyvonen M, Baraldi E, Schultz J, Sudol M, et al. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature. 1996;382:646. doi: 10.1038/382646a0. [DOI] [PubMed] [Google Scholar]
- 70.Madshus IH, Stang E. Internalization and intracellular sorting of the EGF receptor: a model for understanding the mechanisms of receptor trafficking. J. Cell Sci. 2009;122:3433. doi: 10.1242/jcs.050260. [DOI] [PubMed] [Google Scholar]
- 71.Malerod L, Stuffers S, Brech A, Stenmark H. Vps22/EAP30 in ESCRT-II mediates endosomal sorting of growth factor and chemokine receptors destined for lysosomal degradation. Traffic. 2007;8:1617. doi: 10.1111/j.1600-0854.2007.00630.x. [DOI] [PubMed] [Google Scholar]
- 72.McCullough J, Clague MJ, Urbe S. AMSH is an endosome-associated ubiquitin isopeptidase. J. Cell Biol. 2004;166:487. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meng WY, Sawasdikosol S, Burakoff SJ, Eck MJ. Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase. Nature. 1999;398:84. doi: 10.1038/18050. [DOI] [PubMed] [Google Scholar]
- 74.Miller K, Beardmore J, Kanety H, Schlessinger J, Hopkins CR. Localization of the epidermal growth factor (EGF) receptor within the endosome of EGF-stimulated epidermoid carcinoma (A431) cells. J. Cell Biol. 1986;102:500. doi: 10.1083/jcb.102.2.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mizuno E, Iura T, Mukai A, Yoshimori T, Kitamura N, Komada M. Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol. Biol. Cell. 2005;16:5163. doi: 10.1091/mbc.E05-06-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morvan J, Froissard M, Haguenauer-Tsapis R, Urban-Grimal D. The ubiquitin ligase Rsp5p is required for modification and sorting of membrane proteins into multivesicular bodies. Traffic. 2004;5:383. doi: 10.1111/j.1398-9219.2004.00183.x. [DOI] [PubMed] [Google Scholar]
- 77.Mueller GM, Maarouf AB, Kinlough CL, Sheng N, Kashlan OB, et al. Cys-palmitoylation of the beta subunit modulates gating of the epithelial sodium channel. J. Biol. Chem. 2010 doi: 10.1074/jbc.M110.151845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nalefski EA, Falke JJ. The C2 domain calcium-binding motif: Structural and functional diversity. Protein Sci. 1996;5:2375. doi: 10.1002/pro.5560051201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ng C, Jackson RA, Buschdorf JP, Sun QX, Guy GR, Sivaraman J. Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates. EMBO J. 2008;27:804. doi: 10.1038/emboj.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nikko E, Andre B. Evidence for a direct role of the Doa4 deubiquitinating enzyme in protein sorting into the MVB pathway. Traffic. 2007;8:566. doi: 10.1111/j.1600-0854.2007.00553.x. [DOI] [PubMed] [Google Scholar]
- 81.Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, et al. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- 82.Ogunjimi AA, Briant DJ, Pece-Barbara N, Le Roy C, Di Guglielmo GM, et al. Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol. Cell. 2005;19:297. doi: 10.1016/j.molcel.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 83.Ohya T, Miaczynska M, Coskun U, Lommer B, Runge A, et al. Reconstitution of Rab- and SNARE-dependent membrane fusion by synthetic endosomes. Nature. 2009;459:1091. doi: 10.1038/nature08107. [DOI] [PubMed] [Google Scholar]
- 84.Penengo L, Mapelli M, Murachelli AG, Confalonieri S, Magri L, et al. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell. 2006;124:1183. doi: 10.1016/j.cell.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 85.Pennock S, Wang Z. A tale of two Cbls: interplay of c-Cbl and Cbl-b in epidermal growth factor receptor downregulation. Mol.Cell Biol. 2008;28:3020. doi: 10.1128/MCB.01809-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Perica T, Chothia C. Ubiquitin - molecular mechanisms for recognition of different structures. Curr. Opin. Struct. Biol. 2010;20:367. doi: 10.1016/j.sbi.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 87.Peschard P, Kozlov G, Lin T, Mirza A, Berghuis AM, et al. Structural basis for ubiquitin-mediated dimerization and activation of the ubiquitin protein ligase Cbl-b. Mol. Cell. 2007;27:474. doi: 10.1016/j.molcel.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 88.Pickart CM. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 89.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 2004;8:610. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 90.Puertollano R, Bonifacino JS. Interactions of GGA3 with the ubiquitin sorting machinery. Nat Cell Biol. 2004;6:244. doi: 10.1038/ncb1106. [DOI] [PubMed] [Google Scholar]
- 91.Raiborg C, Malerod L, Pedersen NM, Stenmark H. Differential functions of Hrs and ESCRT proteins in endocytic membrane trafficking. Exp. Cell Res. 2008;314:801. doi: 10.1016/j.yexcr.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 92.Reggiori F, Pelham HR. Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J. 2001;20:5176. doi: 10.1093/emboj/20.18.5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ren J, Kee Y, Huibergtse JM, Piper RC. Hse1, a Component of the Yeast Hrs-STAM Ubiquitin Sorting Complex, Associates with Ubiquitin Peptidases and a Ligase to Control Sorting Efficiency into Multivesicular Bodies. Mol Biol Cell. 2007;18:324. doi: 10.1091/mbc.E06-06-0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ren XF, Hurley JH. VHS domains of ESCRT-0 cooperate in high-avidity binding to polyubiquitinated cargo. EMBO J. 2010;29:1045. doi: 10.1038/emboj.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Richter C, West M, Odorizzi G. Dual mechanisms specify Doa4-mediated deubiquitination at multivesicular bodies. EMBO J. 2007;26:2454. doi: 10.1038/sj.emboj.7601692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rotin D. Role of the UPS in Liddle syndrome. BMC.Biochem. 2008;9(Suppl 1):S5. doi: 10.1186/1471-2091-9-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Row PE, Lui H, Hayes S, Welchman R, Charalabous P, et al. The MIT domain of UBPY constitutes a CHMP binding and endosomal localization signal required for efficient EGF receptor degradation. J. Biol. Chem. 2007;282:30929. doi: 10.1074/jbc.M704009200. [DOI] [PubMed] [Google Scholar]
- 98.Row PE, Prior IA, McCullough J, Clague MJ, Urbe S. The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor down-regulation. J. Biol. Chem. 2006;281:12618. doi: 10.1074/jbc.M512615200. [DOI] [PubMed] [Google Scholar]
- 99.Roxrud I, Raiborg C, Pedersen NM, Stang E, Stenmark H. An endosomally localized isoform of Eps15 interacts with Hrs to mediate degradation of epidermal growth factor receptor. J. Cell Biol. 2008;180:1205. doi: 10.1083/jcb.200708115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sato Y, Yoshikawa A, Mimura H, Yamashita M, Yamagata A, Fukai S. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J. 2009;28:2461. doi: 10.1038/emboj.2009.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, et al. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455:358. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- 102.Sato Y, Yoshikawa A, Yamashita M, Yamagata A, Fukai S. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by NZF domains of TAB2 and TAB3. EMBO J. 2009;28:3903. doi: 10.1038/emboj.2009.345. [A definitive structural explanation for the Lys63-linkage specificity of AMSH.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shearwin-Whyatt L, Dalton HE, Foot N, Kumar S. Regulation of functional diversity within the Nedd4 family by accessory and adaptor proteins. Bioessays. 2006;28:617. doi: 10.1002/bies.20422. [DOI] [PubMed] [Google Scholar]
- 104.Sigismund S, Argenzio E, Tosoni D, Cavallaro E, Polo S, Di Fiore PP. Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev.Cell. 2008;15:209. doi: 10.1016/j.devcel.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 105.Sigismund S, Woelk T, Puri C, Maspero E, Tacchetti C, et al. Clathrin-independent endocytosis of ubiquitinated cargos. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2760. doi: 10.1073/pnas.0409817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sloper-Mould KE, Jemc JC, Pickart CM, Hicke L. Distinct functional surface regions on ubiquitin. J. Biol. Chem. 2001;276:30483. doi: 10.1074/jbc.M103248200. [DOI] [PubMed] [Google Scholar]
- 107.Snyder PM, McDonald FJ, Stokes JB, Welsh MJ. Membrane topology of the amiloride-sensitive epithelial sodium channel. J. Biol. Chem. 1994;269:24379. [PubMed] [Google Scholar]
- 108.Sorkina T, Huang F, Beguinot L, Sorkin A. Effect of tyrosine kinase inhibitors on clathrin-coated pit recruitment and internalization of epidermal growth factor receptor. J. Biol. Chem. 2002;277:27433. doi: 10.1074/jbc.M201595200. [DOI] [PubMed] [Google Scholar]
- 109.Soubeyran P, Kowanetz K, Szymkiewicz I, Langdon WY, Dikic I. Cbl-CIN85-endophilin complex mediates ligand-induced downregulation of EGF receptors. Nature. 2002;416:183. doi: 10.1038/416183a. [DOI] [PubMed] [Google Scholar]
- 110.Soundararajan R, Pearce D, Hughey RP, Kleyman TR. Role of epithelial sodium channels and their regulators in hypertension. J. Biol. Chem. 2010 doi: 10.1074/jbc.R110.155341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Spormann DO, Heim J, Wolf DH. Biogenesis of the yeast vacuole (lysosome). The precursor forms of the soluble hydrolase carboxypeptidase yscS are associated with the vacuolar membrane. J. Biol. Chem. 1992;267:8021. [PubMed] [Google Scholar]
- 112.Stang E, Blystad FD, Kazazic M, Bertelsen V, Brodahl T, et al. Cbl-dependent ubiquitination is required for progression of EGF receptors into clathrin-coated pits. Mol. Biol. Cell. 2004;15:3591. doi: 10.1091/mbc.E04-01-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Staub O, Gautschi I, Ishikawa T, Breitschopf K, Ciechanover A, et al. Regulation of stability and function of the epithelial Na+ channel (ENaC) by ubiquitination. EMBO J. 1997;16:6325. doi: 10.1093/emboj/16.21.6325. [Demonstration of ubiquitin-mediated sorting of EnaC.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Strous GJ, Van Kerkhof P, Govers R, Ciechanover A, Schwartz AL. The ubiquitin conjugation system is required for ligand-induced endocytosis and degradation of the growth hormone receptor. EMBO J. 1996;15:3806. [PMC free article] [PubMed] [Google Scholar]
- 115.Stuchell-Brereton M, Skalicky J, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- 116.Tsang HTH, Connell JW, Brown SE, Thompson A, Reid E, Sanderson CM. A systematic analysis of human CHMP protein interactions: Additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex. Genomics. 2006;88:333. doi: 10.1016/j.ygeno.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 117.Umebayashi K, Stenmark H, Yoshimori T. Ubc4/5 and c-Cbl continue to ubiquitinate EGF receptor after internalization to facilitate polyubiquitination and degradation. Mol. Biol. Cell. 2008;19:3454. doi: 10.1091/mbc.E07-10-0988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Urbe S, McCullough J, Row P, Prior IA, Welchman R, Clague MJ. Control of growth factor receptor dynamics by reversible ubiquitination. Biochem. Soc. Trans. 2006;34:754. doi: 10.1042/BST0340754. [DOI] [PubMed] [Google Scholar]
- 119.Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IVWW domains. Nat. Struct. Biol. 2000;7:639. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- 120.Verdecia MA, Joazeiro CAP, Wells NJ, Ferrer JL, Bowman ME, et al. Conformational flexibility underlies ubiquitin ligation mediated by the WWP1HECT domain E3 ligase. Mol. Cell. 2003;11:249. doi: 10.1016/s1097-2765(02)00774-8. [DOI] [PubMed] [Google Scholar]
- 121.Vijaykumar S, Bugg CE, Cook WJ. STRUCTURE OF UBIQUITIN REFINED AT 1.8 A RESOLUTION. J. Mol. Biol. 1987;194:531. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 122.Weissman AM. Themes and variations on ubiquitylation. Nature Reviews Molecular Cell Biology. 2001;2:169. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 123.Witsch E, Sela M, Yarden Y. Roles for Growth Factors in Cancer Progression. Physiology. 25:85. doi: 10.1152/physiol.00045.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yarden Y. The EGFR family and its ligands in human cancer. signalling mechanisms and therapeutic opportunities. Eur.J.Cancer. 2001:S3. doi: 10.1016/s0959-8049(01)00230-1. [DOI] [PubMed] [Google Scholar]
- 125.Zamborlini A, Usami Y, Radoshitzky SR, Popova E, Palu G, Gottlinger H. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19140. doi: 10.1073/pnas.0603788103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533. doi: 10.1016/s0092-8674(00)00057-x. [This ten-year old study is still the definitive structural account of Cbl-dependent ubiquitination.] [DOI] [PubMed] [Google Scholar]
- 127.Zhou R, Kabra R, Olson DR, Piper RC, Snyder PM. Hrs controls sorting of the epithelial Na+ channel between endosomal degradation and recycling pathways. J. Biol. Chem. 2010 doi: 10.1074/jbc.M110.150755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhu H, Liang Z, Li G. Rabex-5 is a Rab22 effector and mediates a Rab22-Rab5 signaling cascade in endocytosis. Mol. Biol. Cell. 2009;20:4720. doi: 10.1091/mbc.E09-06-0453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.