Abstract
The tight-binding I12-X86 lac repressor binds to non-operator DNA in a sequence-specific fashion. Using the DNA of the E. coli I gene we have investigated these sequence-specific interactions and compared them to the operator binding of wild-type repressor. The specific, non-operator DNA interactions are sensitive to the inducer IPTG. One strong binding site in the I gene DNA was found to be one of two expected on the basis of their homology with the lac operator. The binding of I12-X86 repressor to this site was visualized using the footprinting technique, and found to be consistent with an operator-like binding configuration. The protection pattern extends into an adjacent sequence suggesting that two repressor tetramers are bound in tandem.
Full text
PDF

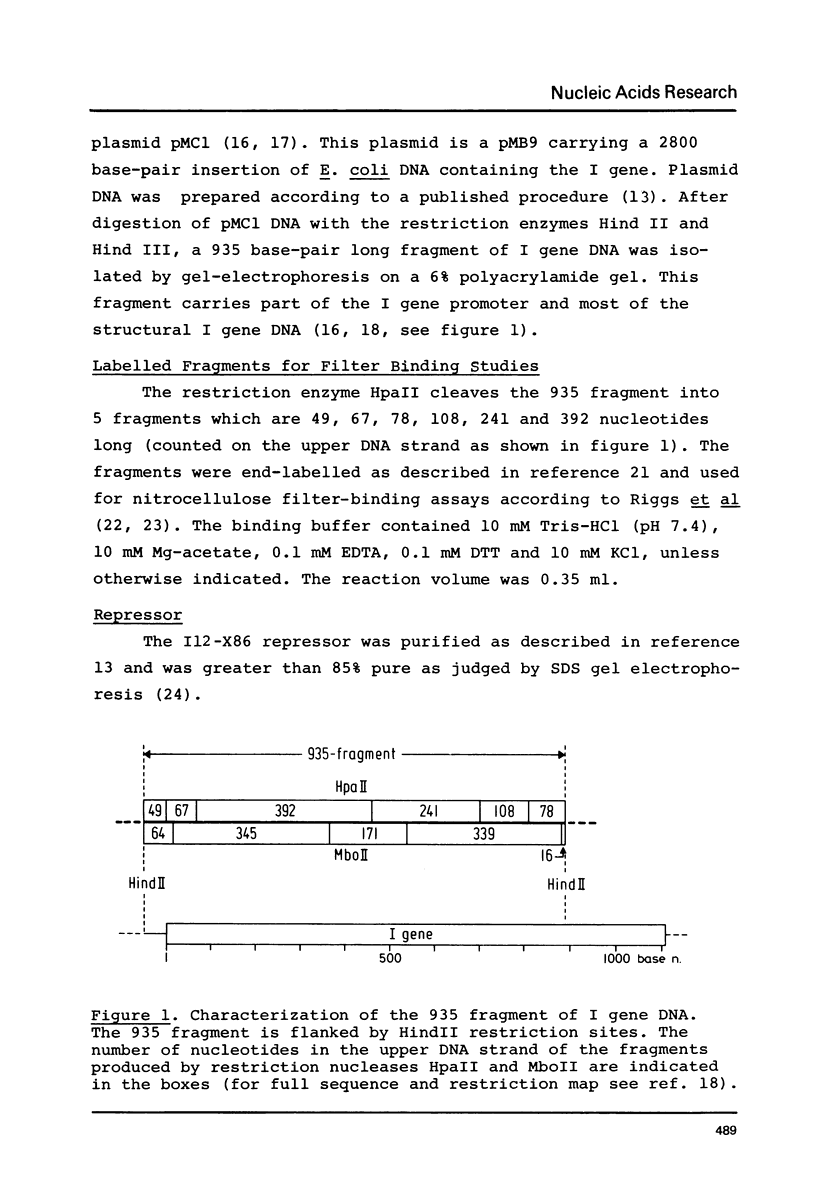

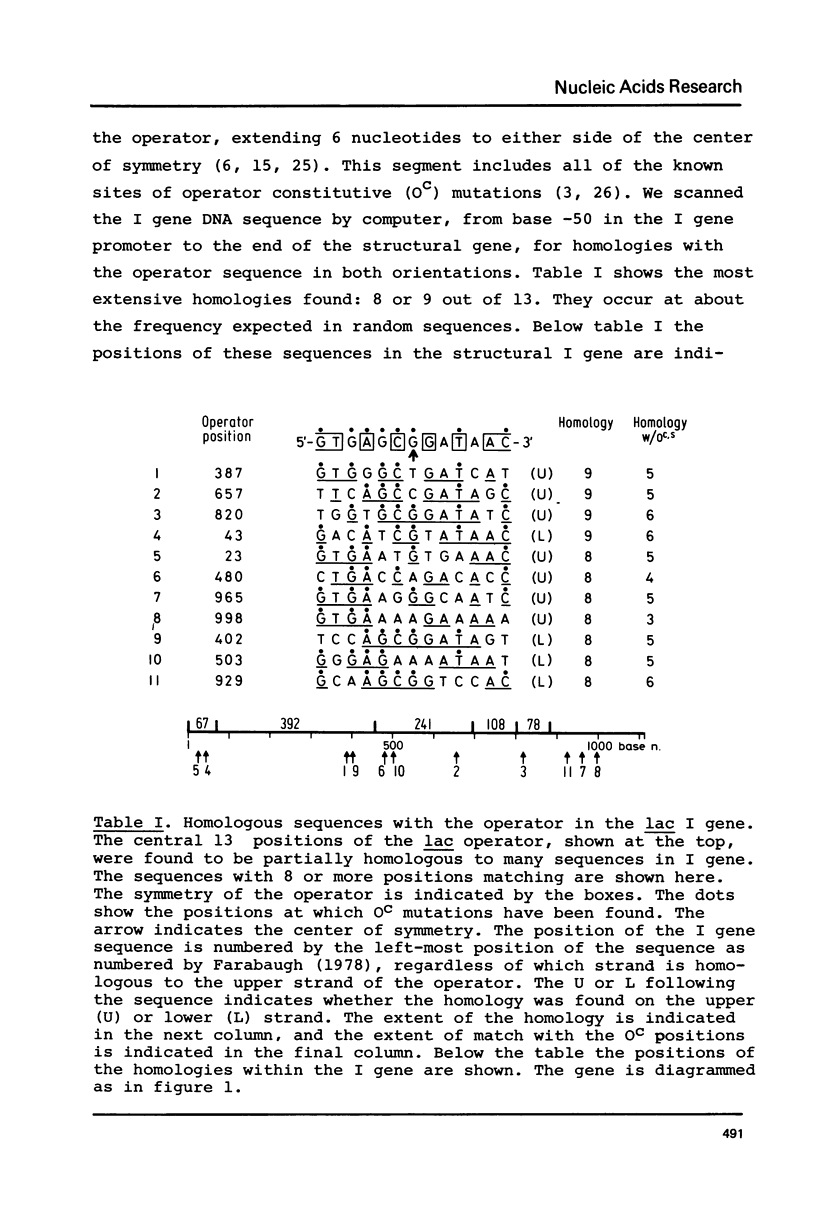


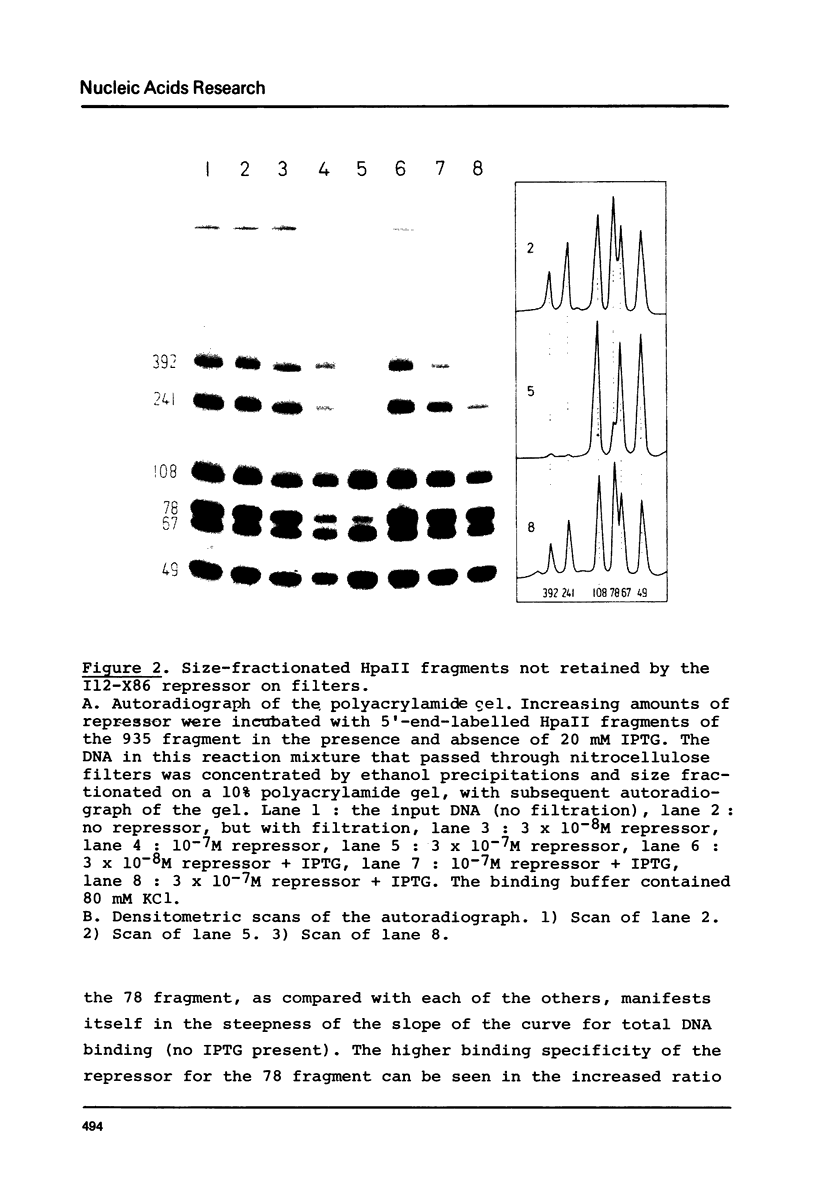
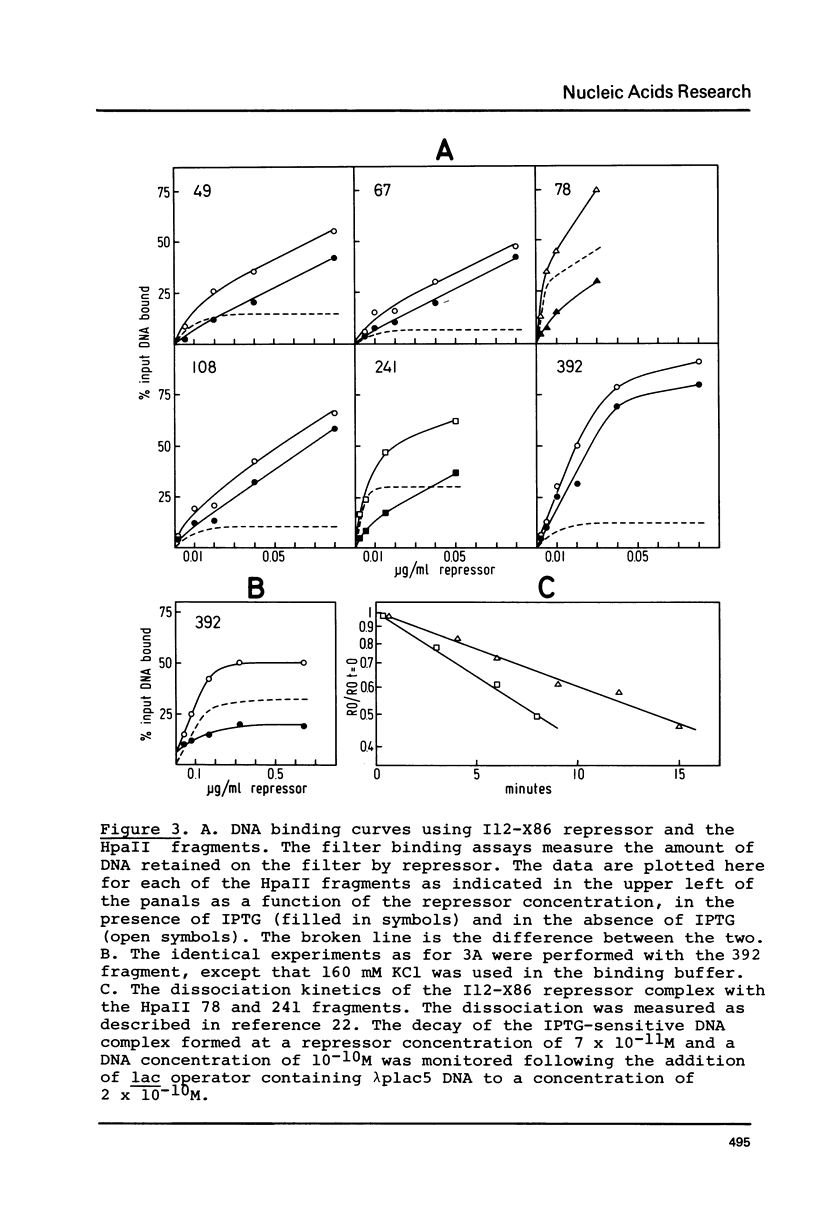





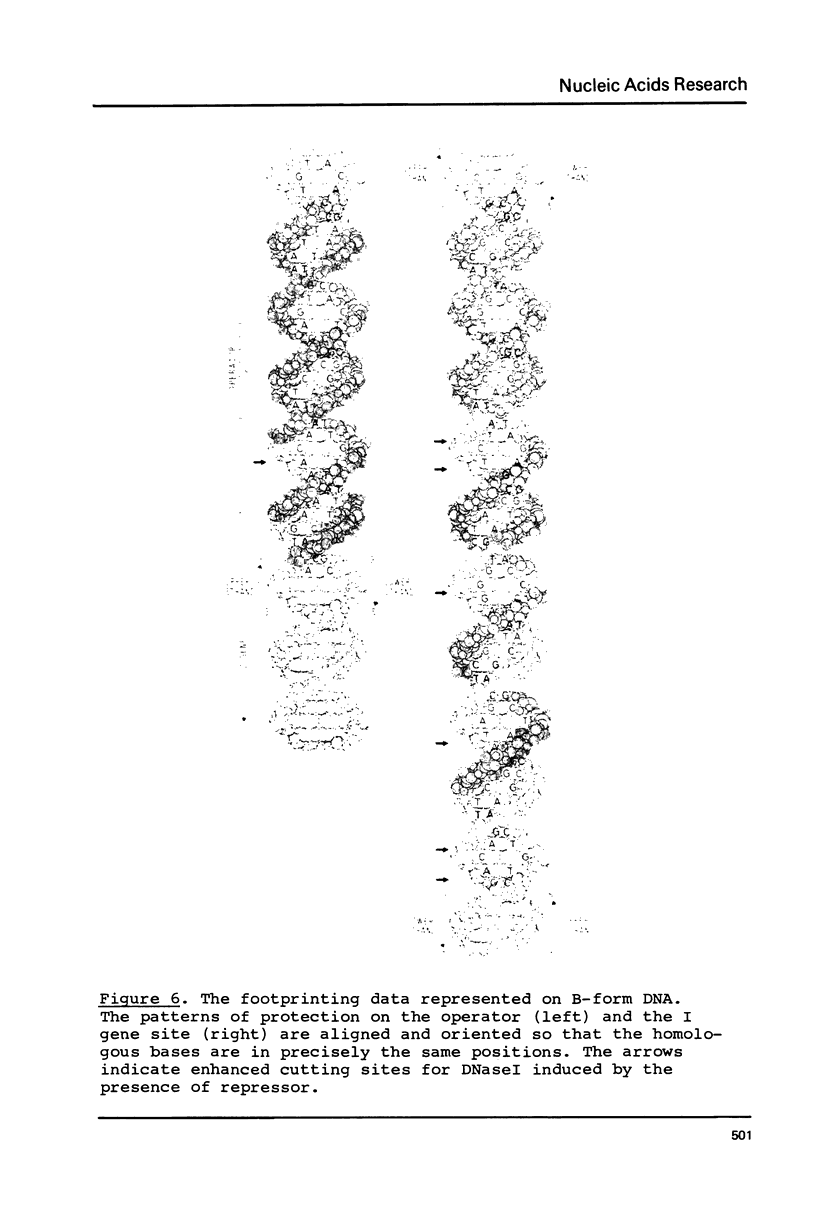





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Betz J. L., Sadler J. R. Tight-binding repressors of the lactose operon. J Mol Biol. 1976 Aug 5;105(2):293–319. doi: 10.1016/0022-2836(76)90113-3. [DOI] [PubMed] [Google Scholar]
- Bickle T. A., Pirrotta V., Imber R. A simple, general procedure for purifying restriction endonucleases. Nucleic Acids Res. 1977 Aug;4(8):2561–2572. doi: 10.1093/nar/4.8.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A. P., Revzin A., von Hippel P. H. Molecular parameters characterizing the interaction of Escherichia coli lac repressor with non-operator DNA and inducer. Biochemistry. 1977 Nov 1;16(22):4757–4768. doi: 10.1021/bi00641a001. [DOI] [PubMed] [Google Scholar]
- Calos M. P. DNA sequence for a low-level promoter of the lac repressor gene and an 'up' promoter mutation. Nature. 1978 Aug 24;274(5673):762–765. doi: 10.1038/274762a0. [DOI] [PubMed] [Google Scholar]
- Calos M. P., Johnsrud L., Miller J. H. DNA sequence at the integration sites of the insertion element IS1. Cell. 1978 Mar;13(3):411–418. doi: 10.1016/0092-8674(78)90315-x. [DOI] [PubMed] [Google Scholar]
- Chamness G. C., Willson C. D. An unusual lac repressor mutant. J Mol Biol. 1970 Nov 14;53(3):561–565. doi: 10.1016/0022-2836(70)90084-7. [DOI] [PubMed] [Google Scholar]
- Files J. G. Direct identification of the amino acid changes in two mutant lac repressors. J Mol Biol. 1978 Aug 15;123(3):454–456. [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeddel D. V., Yansura D. G., Caruthers M. H. How lac repressor recognizes lac operator. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3578–3582. doi: 10.1073/pnas.75.8.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobe A., Bourgeois S. The lac repressor-operator interaction. VII. A repressor with unique binding properties: the X86 repressor. J Mol Biol. 1972 Dec 14;72(1):139–152. doi: 10.1016/0022-2836(72)90075-7. [DOI] [PubMed] [Google Scholar]
- Klug A., Jack A., Viswamitra M. A., Kennard O., Shakked Z., Steitz T. A. A hypothesis on a specific sequence-dependent conformation of DNA and its relation to the binding of the lac-repressor protein. J Mol Biol. 1979 Jul 15;131(4):669–680. doi: 10.1016/0022-2836(79)90196-7. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H., Coulondre C., Farabaugh P. J. Correlation of nonsense sites in the lacI gene with specific codons in the nucleotide sequence. Nature. 1978 Aug 24;274(5673):770–775. doi: 10.1038/274770a0. [DOI] [PubMed] [Google Scholar]
- Ogata R. T., Gilbert W. An amino-terminal fragment of lac repressor binds specifically to lac operator. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5851–5854. doi: 10.1073/pnas.75.12.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata R., Gilbert W. Contacts between the lac repressor and the thymines in the lac operator. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4973–4976. doi: 10.1073/pnas.74.11.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfahl M. lac Repressor-operator interaction. Analysis of the X86 repressor mutant. J Mol Biol. 1976 Sep 25;106(3):857–869. doi: 10.1016/0022-2836(76)90269-2. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, deHaseth P. L., Lohman T. M. Interpretation of monovalent and divalent cation effects on the lac repressor-operator interaction. Biochemistry. 1977 Nov 1;16(22):4791–4796. doi: 10.1021/bi00641a005. [DOI] [PubMed] [Google Scholar]
- Reznikoff W. S., Winter R. B., Hurley C. K. The location of the repressor binding sites in the lac operon. Proc Natl Acad Sci U S A. 1974 Jun;71(6):2314–2318. doi: 10.1073/pnas.71.6.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs A. D., Bourgeois S., Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J Mol Biol. 1970 Nov 14;53(3):401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Suzuki H., Bourgeois S. Lac repressor-operator interaction. I. Equilibrium studies. J Mol Biol. 1970 Feb 28;48(1):67–83. doi: 10.1016/0022-2836(70)90219-6. [DOI] [PubMed] [Google Scholar]
- Ross W., Landy A., Kikuchi Y., Nash H. Interaction of int protein with specific sites on lambda att DNA. Cell. 1979 Oct;18(2):297–307. doi: 10.1016/0092-8674(79)90049-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz A., Galas D. J. The interaction of RNA polymerase and lac repressor with the lac control region. Nucleic Acids Res. 1979 Jan;6(1):111–137. doi: 10.1093/nar/6.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. F., Sadler J. R. The nature of lactose operator constitive mutations. J Mol Biol. 1971 Jul 28;59(2):273–305. doi: 10.1016/0022-2836(71)90051-9. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Weisblum B. Construction of a colicin E1-R factor composite plasmid in vitro: means for amplification of deoxyribonucleic acid. J Bacteriol. 1975 Jan;121(1):354–362. doi: 10.1128/jb.121.1.354-362.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T., O'Neill M., de Crombrugghe B. Interaction site of Escherichia coli cyclic AMP receptor protein on DNA of galactose operon promoters. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5090–5094. doi: 10.1073/pnas.76.10.5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C., Barkley M. D., Bourgeois S. Measurements of unwinding of lac operator by repressor. Nature. 1974 Sep 20;251(5472):247–249. doi: 10.1038/251247a0. [DOI] [PubMed] [Google Scholar]
- Yansura D. G., Goeddel D. V., Kundu A., Caruthers M. H. Studied on gene control regions IX. The effect of hypoxanthine-substituted lac operators on the lac operator--lac repressor interaction. J Mol Biol. 1979 Sep 5;133(1):117–135. doi: 10.1016/0022-2836(79)90253-5. [DOI] [PubMed] [Google Scholar]
- Zingsheim H. P., Geisler N., Weber K., Mayer F. Complexes of Escherichia coli lac-repressor with non-operator DNA revealed by electron microscopy: two repressor molecules can share the same segment of DNA. J Mol Biol. 1977 Sep 25;115(3):565–570. doi: 10.1016/0022-2836(77)90171-1. [DOI] [PubMed] [Google Scholar]