Abstract
Salinomycin is widely used in animal husbandry as a food additive due to its antibacterial and anticoccidial activities. However, its biosynthesis had only been studied by feeding experiments with isotope-labeled precursors. A strategy with degenerate primers based on the polyether-specific epoxidase sequences was successfully developed to clone the salinomycin gene cluster. Using this strategy, a putative epoxidase gene, slnC, was cloned from the salinomycin producer Streptomyces albus XM211. The targeted replacement of slnC and subsequent trans-complementation proved its involvement in salinomycin biosynthesis. A 127-kb DNA region containing slnC was sequenced, including genes for polyketide assembly and release, oxidative cyclization, modification, export, and regulation. In order to gain insight into the salinomycin biosynthesis mechanism, 13 gene replacements and deletions were conducted. Including slnC, 7 genes were identified as essential for salinomycin biosynthesis and putatively responsible for polyketide chain release, oxidative cyclization, modification, and regulation. Moreover, 6 genes were found to be relevant to salinomycin biosynthesis and possibly involved in precursor supply, removal of aberrant extender units, and regulation. Sequence analysis and a series of gene replacements suggest a proposed pathway for the biosynthesis of salinomycin. The information presented here expands the understanding of polyether biosynthesis mechanisms and paves the way for targeted engineering of salinomycin activity and productivity.
INTRODUCTION
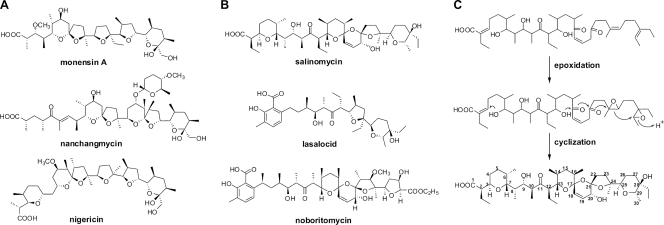
Polyether antibiotics (Fig. 1), such as monensin, nanchangmycin, lasalocid, and salinomycin, are widely used as anticoccidial drugs and growth promoters in animal husbandry (11). They have the ability to chelate metal ions selectively within a hydrophobic matrix and then transport the ions across the cell membrane, which destroys the physiological ion gradients of the cells and results in the death of Eimeria sp., the causative agent of coccidiosis (31). Depending on the arrangement of the first four biogenetic units of acetate (A) or propionate (P), polyethers are divided into two stereochemical prototypes (5). The APPA type is exemplified by monensin, nachangmycin, and nigericin (Fig. 1A), whereas the PAPA type comprises salinomycin, lasalocid, and noboritomycin (Fig. 1B).
Fig 1.
Structures of polyether antibiotics and proposed pathway for salinomycin biosynthesis. (A) APPA-type polyether antibiotics. (B) PAPA-type polyether antibiotics. (C) Hypothetical ether bond formation in salinomycin biosynthesis.
So far, five polyether biosynthesis gene clusters have been identified and characterized for the biosynthesis of monensin (27), nanchangmycin (37), lasalocid (35), nigericin (15), and tetronomycin (9). Based on in silico and experimental analysis, the polyene backbones, assembled by a type I polyketide synthase (PKS), could undergo oxidative cyclization catalyzed by an epoxidase and one or more epoxide hydrolases to generate the characteristic pattern of ether bonds. Accordingly, genes for the putative epoxidase and epoxide hydrolases have been identified in all five reported gene clusters. The epoxidase MonCI in the monensin cluster was proved to perform epoxidation by heterologous expression in Streptomyces coelicolor, using the unsaturated terpene (+/−)-linalool as a substrate (27). Moreover, the E,E,E-triene lactone isolated from the monCI mutant of Streptomyces cinnamonensis strongly implied that the E-configured double bonds are targets for MonCI to initiate oxidative cyclization (3). On the other hand, deletion of either or both of the epoxide hydrolase genes monBI and monBII abolished the production of monensin A and B (10). Recently, in vitro experiments directly demonstrated that the epoxide hydrolase Lsd19 from the lasalocid cluster could recognize the internal epoxide of the substrate and catalyze epoxide-opening and concomitant polyether ring formation cascades (24, 26, 33). Although both epoxidases and epoxide hydrolases are key enzymes in polyether biosynthesis, epoxidases are more conserved and show higher homology with each other. Moreover, epoxidases involved in polyether biosynthesis are distinct from epoxidases for nonpolyether antibiotic biosynthesis. Therefore, the existence of such an epoxidase gene might be considered a sign of polyether-producing potential (21).
Salinomycin is an important commercial polyether comprised of a unique tricyclic spiroketal ring system and an unsaturated six-membered ring (Fig. 1B). It has been used for 30 years as an anticoccidial drug and to improve nutrient absorption and feed efficiency in ruminants and swine. Recently, salinomycin has also been identified as an agent to kill epithelial cancer stem cells (12). Previous isotope-labeled feeding experiments clearly revealed that the backbone of salinomycin is assembled from six acetate, six propionate, and three butyrate units (32). In a proposed pathway for salinomycin biosynthesis (5) (Fig. 1C), unsaturated acyclic diene intermediates initially undergo oxidation to form a diepoxide, followed by an epoxide-opening cascade reaction catalyzed by epoxide hydrolases, to generate the characteristic ether ring system. However, details of the biosynthesis of salinomycin have been elusive.
In 2003, cloning of the salinomycin biosynthesis gene cluster in Streptomyces albus ATCC 21838 was first claimed through PCR amplification using degenerate ketosynthase (KS) primers. The involvement of this 4.5-kb region in salinomycin biosynthesis was further suggested through a single-crossover gene disruption (17). In 2007, another group cloned a 25.81-kb region using the same strategy from the salinomycin producer S. albus CCM 4719, which contained the previously identified 4.5-kb DNA fragment. However, gene disruption was not conducted in this strain (19).
Here, a homologous region was first cloned according to the published sequence in our salinomycin producer, S. albus XM211. However, the polyketide backbone deduced from the sequences on polyketide synthases did not fit with any portion of the salinomycin structure. Alternatively, the strategy of PCR amplification with degenerate epoxidase-specific primers was designed, and another genomic region was cloned and then confirmed as the salinomycin biosynthesis gene cluster through gene replacement and complementation.
MATERIALS AND METHODS
Bacterial strains, plasmids, culture conditions, and general techniques.
The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material.
S. albus XM211, here called XM211, is the producer of salinomycin and was isolated from a mangrove forest in Xiamen, China. For sporulation, S. albus XM211 and its derivatives were grown at 30°C on ISP 4 agar plates (BD) (34) for 8 to 10 days. ISP 4 medium was also used for conjugation between Escherichia coli and Streptomyces. For salinomycin isolation, the seed culture was grown in a 250-ml flask containing 50 ml medium (4% glucose, 3% soy flour, 1% yeast extract, 0.2% calcium carbonate) on a rotary shaker at 200 rpm and 32°C for 26 to 32 h. Then, 5 ml of the seed culture was used to inoculate 50 ml fermentation medium (3% glucose, 1% casein hydrolysate, 0.2% sodium chloride, 0.2% potassium chloride, 0.5% ammonium sulfate, 0.02% dipotassium hydrogen phosphate, 0.01% magnesium sulfate, 0.01% calcium chloride, 0.5% calcium carbonate). The fermentation was continued at 32°C for 5 to 6 days. For isolation of the total DNA, S. albus XM211 was cultivated in Trypticase soy broth (TSB) medium (18) supplemented with 10.3% sucrose and 1% yeast extract.
E. coli DH10B (Invitrogen) was used as cloning host, and E. coli ET12567(pUZ8002) (28) was used as a conjugation host. The E. coli strains were grown at 37°C in LB broth and LB agar. Plasmid pMD18-T was used for cloning and sequencing of PCR-amplified products. pJTU824, a derivative of pSET152, was used for gene complementation in the mutants (40).
Conjugation between E. coli and S. albus XM211 was performed as described by Kieser et al. (18). DNA fragments were recovered from agarose gels using a Gel Recovery Kit (Omega).
Isolation and analysis of salinomycin.
Liquid fermentation cultures of the wild-type strain and mutants were extracted twice with 2 volumes of ethyl acetate. The organic fraction was evaporated in vacuo, and the residue was redissolved in methanol for analysis. Samples were applied to liquid chromatography-tandem mass spectrometry (LC–MS-MS) (Agilent 1100 series LC/MSD Trap system) after passing through 0.2-μm filters. The liquid chromatograph was operated at a flow rate of 0.3 ml/min with an Agilent Eclipse TC-C18 column (4.6 by 250 mm; particle size, 5 μm) with UV detection at 210 nm (4). The mobile phase was acetonitrile-2% acetic acid (92:8 [vol/vol]). The ion-trap mass spectrometer was operated with an electrospray ionization source in positive ion mode. The drying gas flow was 10 liters/min with a temperature of 325°C, and the nebulizer pressure was 30 lb/in2.
PCR amplification with degenerate epoxidase-specific primers.
The degenerate primers Epo-110-F-2/Epo-1030-R-1, designed according to conserved domains of reported epoxidases, are listed in Table S4 in the supplemental material. GC buffer (Takara Biotechnology, Dalian, China) was used for amplification with templates of Streptomyces DNA. DNA fragments of the desired sizes were cloned into the pMD18-T vector and sequenced. A pair of specific primers, sln1-F/sln1-R (see Table S4 in the supplemental material), was then designed to screen the XM211 genomic library.
Sequencing and analysis of the salinomycin biosynthesis gene cluster.
The XM211 genomic library was first screened by PCR amplification using sln1-F/sln1-R. For subsequent chromosome walking, three pairs of specific primers based on sequenced fosmids were used, i.e., sln2-F/sln2-R, sln3-F/sln3-R, and sln4-F/sln4-R (see Table S4 in the supplemental material). Fosmids 12H8, 25E8, 10B8, and 6G11 and 3.58-kb and 7.93-kb BamHI fragments of 19H1 were sequenced at Majorbio Biotech Co., Ltd. The raw sequence data were analyzed with FramePlot 4.0beta (16) and Glimmer (7) online programs. Annotation analysis was performed using the BLAST algorithm (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (1). The assembly of the PKS region was analyzed with the online NRPS-PKS program (2). Sequence alignments were performed with BioEdit Sequence Alignment Editor.
Gene replacement and trans-complementation of slnC.
Gene replacement of slnC was carried out by Redirect Technology (13). Using the gel-purified 1,384-bp EcoRI/HindIII fragments from pIJ773 as templates, 1,449-bp products carrying the apramycin-resistant gene aac(3)IV and oriT were amplified by PCR with primers 12H8tar-F and 12H8tar-R (see Table S4 in the supplemental material). This amplified fragment was used to replace the 1,416-bp region of slnC in pJTU5359 cloned with a 9.0-kb SpeI/BglII DNA fragment from fosmid 12H8, giving rise to pJTU5360. Subsequent transfer of the 9.1-kb BglII fragment from pJTU5360 to pJTU412 (36) resulted in the construction of pJTU5361 for slnC inactivation in XM211.
Then, pJTU5361 was transferred into strain XM211 by conjugation from E. coli ET12567(pUZ8002). The thiostrepton-sensitive and apramycin-resistant (Thios Aprr) recombinants were selected from the initial Aprr exconjugants after several rounds of nonselective growth. For further verification, the mutant candidates were verified by PCR amplification with primers JCY16-F and JCY16-R (see Table S4 in the supplemental material) and SacI digestion of the amplified products.
To complement the slnC mutant JCY16, a pJTU824-derived construct, pJTU5376, was used. This plasmid contained the complete slnC gene under the control of the ermE* promoter (an upregulated variant of the ermE promoter.). The slnC gene was amplified from fosmid 12H8 using the primers epoxidase-F and epoxidase-R (see Table S4 in the supplemental material). The 1.45-kb PCR product was purified, cloned into the pMD18-T vector, and sequenced. The slnC gene was transferred to pJTU824 digested with NdeI and EcoRI, and an integrative plasmid, pJTU5376, with the intact slnC gene under the control of the ermE* promoter was constructed. Plasmid pJTU5376 was introduced into JCY16 through conjugation, and apramycin-resistant and thiostrepton-resistant (Thior Aprr) exconjugants were selected.
Replacements of other genes in this study are described in the supplemental material.
Nucleotide sequence accession number.
The sequence of the salinomycin biosynthesis gene cluster was deposited in GenBank with accession number JN033543.
RESULTS AND DISCUSSION
Attempted cloning of the salinomycin biosynthesis gene cluster based on the previously reported sequences.
Based on previously published sequences (GenBank accession number AB087998), a pair of primers, SalPrimerF1/SalPrimerR1 (see Table S4 in the supplemental material), for amplifying the acyl carrier protein (ACP)-KS region were designed for screening an S. albus XM211 genomic library. The sequence of the amplified 0.20-kb fragment matches the predicted sequence. Finally, fosmid 37F1 was chosen to be sequenced, and the previously reported 4.50-kb DNA region was shown to be included in its sequence (17).
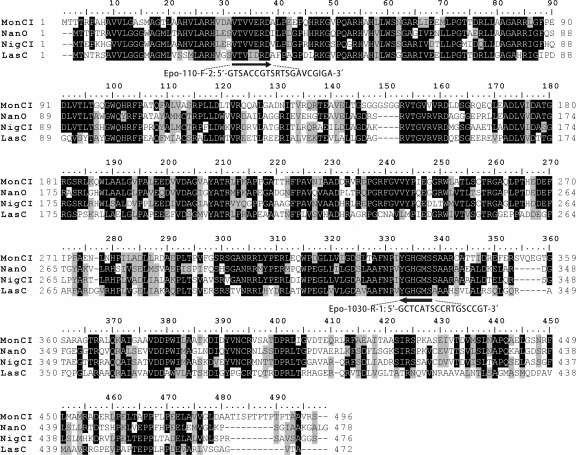
Sequence analysis identified 11 potential open reading frames (ORFs), as shown in Table S2 and Fig. S2 in the supplemental material. Three large ORFs, designated samK1 to samK3, encode typical multifunctional type I PKSs. SamK1 consists of extension module K1-1 (KS-ATp [methylmalonyl-coenzyme A {CoA}-specific acyltransferase]-KR [ketoreductase]-ACP), extension module K1-2 (KS-ATa [malonyl-CoA-specific acyltransferase]-DH [dehydratase]-KRa-ACP, where a represents an inactive domain), and one integrated thioesterase (TE) domain. SamK2 contains extension module K2-1 (KS-ATa-DHa-KRa-ACP) and module K2-2 (KS-ATa-KR-ACP), and SamK3 represents extension modules K3-1 (KS-ATa-KRa-ACP) and K3-2 (KS-ATa-KR-ACP). In fact, however, two sets of ). Several sets of degenerate primers were designed based on conserved blocks using the PaBaLiS (29) program. To validate the efficiency of these primers, the detection of epoxidase genes was first conducted with DNA isolated from polyether producer strains of monensin, nanchangmycin, nigericin, and lasalocid. PCR amplification with the primers Epo-110-F-2/Epo-1030-R-1, designed according to two conserved blocks, VTVV(E/D)RD and YGHGMS (Fig. 2), gave a 0.90-kb product with genomic DNA from each of the four strains. All the amplified products were confirmed by sequencing to be polyether-specific epoxidase genes (see Fig. S3 in the supplemental material).
Fig 2.
Multiple alignments of amino acid sequences of known epoxidases from polyether biosynthesis gene clusters. The arrows indicate the regions selected for the design of primers Epo-110-F-2 and Epo-1030-R-1. Black shading indicates identical amino acids, and gray shading indicates similar amino acids.
The successful application of the degenerate primers Epo-110-F-2/Epo-1030-R-1 encouraged us to clone the salinomycin-specific epoxidase gene using the same strategy. PCR amplification of S. albus XM211 genomic DNA gave a 0.90-kb product (see Fig. S3 in the supplemental material), which was then cloned and sequenced. This portion was named slnC, and its product showed 54% and 52% identities with epoxidase LasC, encoded in the lasalocid gene cluster, and MonCI, encoded in the monensin gene cluster, respectively. Based on this newly obtained sequence, a pair of primers, sln1-F and sln1-R, were designed and used to screen the whole XM211 genomic library. Twelve overlapping fosmids, including 12H8, were found to give the expected 0.6-kb PCR products (data not shown).
Additionally, two 0.90-kb expected PCR products were also obtained from the BL580Δ (see Fig. S1 in the supplemental material) producer Streptomyces sp. NRRL 8180 and the mutalomycin (see Fig. S1 in the supplemental material) producer Streptomyces sp. NRRL 8088 (see Fig. S3 in the supplemental material). Both products were confirmed to be polyether-epoxidase genes through sequencing. They are therefore likely to be involved in the biosynthesis of the corresponding polyethers.
Phylogenetic analysis of the available epoxidase sequences (see Fig. S7 in the supplemental material) revealed that the distribution of epoxidases can reflect the structural differences of polyether antibiotics. SlnC and LasC, whose corresponding polyether antibiotics are both categorized as PAPA type, form one clade. On the other hand, MonCI, NanO, NigCI, and deduced epoxidases in mutalomycin and BL580Δ biosynthesis form another clade for APPA-type polyethers. This indicates that the sequences of epoxidases are correlated with the structures of the parental polyketides of polyethers.
The epoxidase gene slnC is involved in the biosynthesis of salinomycin.
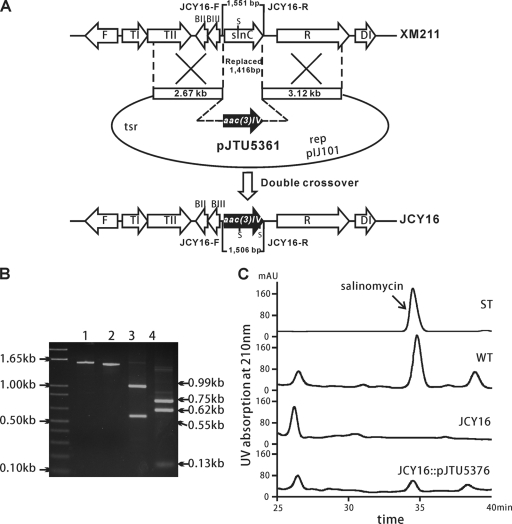
The involvement of slnC in salinomycin production was proved through gene replacement. A 1,416-bp DNA fragment of the epoxidase gene in the sequenced 12H8 fosmid was replaced by the apramycin-resistant gene aac(3)IV to generate the mutant JCY16 (Fig. 3A). Allelic replacement of slnC in JCY16 was confirmed by PCR amplification with primers JCY16-F/JCY16-R and SacI digestion. The 1.50-kb PCR product amplified from the mutant JCY16 could be digested into 0.62-kb, 0.75-kb, and 0.13-kb fragments, whereas a 1.55-kb expected product from wild-type XM211 was digested into 0.99-kb and 0.55-kb fragments (Fig. 3B). LC-MS analysis of the extracts from the mutant JCY16 displayed a complete loss of salinomycin production (Fig. 3C), suggesting that this epoxidase gene is involved in salinomycin biosynthesis.
Fig 3.
Replacement and complementation of epoxidase gene slnC. (A) Schematic representation of the replacement of slnC. A 1,416-bp region of slnC was replaced by the 1,371-bp aac(3)IV gene through double crossover. S, SacL. (B) Confirmation of the mutant JCY16 through PCR amplification and SacI digestion. An approximately 1.50-kb product was amplified using primers JCY16-F/JCY16-R. After SacI digestion, the electrophoresis gel revealed 0.62-kb, 0.75-kb, and 0.13-kb bands using the mutant genomic DNA as a template (lane 4) instead of the 0.55-kb and 0.99-kb bands of the wild type (lane 3). (C) HPLC analysis of the wild type (WT), slnC mutant JCY16, and complementation strain JCY16::pJTU5376. ST, salinomycin standard; AU, absorbance units.
To confirm that the loss of salinomycin productivity was caused by the replacement of slnC, a trans-complementation experiment using pJTU5376 was performed. This plasmid, harboring the intact epoxidase gene slnC under the control of the ermE* promoter, was introduced into JCY16 by conjugation. The high-performance liquid chromatography (HPLC) profile of the extract of the complemented strain revealed the restoration of salinomycin, albeit at a reduced level (∼20%) compared to the wild-type strain (Fig. 3C; see Fig. S4 in the supplemental material). Thus, slnC was proved to be related to salinomycin biosynthesis.
Cloning and sequence analysis of the salinomycin biosynthesis gene cluster.
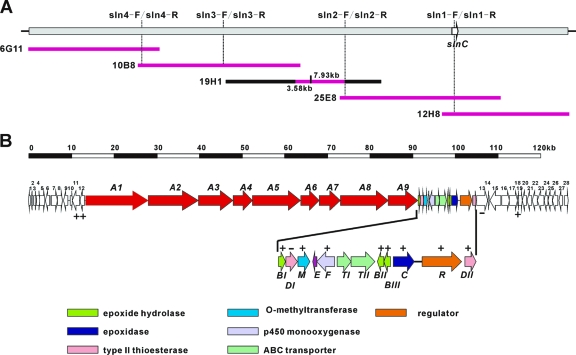
DNA sequence analysis of fosmid 12H8 shows that the epoxidase gene slnC is located at the left end, and most ORFs in the right side seem not to be related to salinomycin biosynthesis. In order to clone the entire gene cluster, sequential chromosome walking was initiated from the left end of 12H8, revealing a continuous ∼127-kb DNA region (Fig. 4A). Computer-assisted analysis of the cloned sequence led to the identification of the constituent genes (Fig. 4B). The putative functions of these genes are listed in Table 1 and in Table S3 in the supplemental material, including genes encoding a type I PKS, oxidative cyclization, hydroxylation, regulation, and export. Including the replacement of slnC, a total of 14 gene replacements were performed to investigate boundaries and gene functions of the salinomycin biosynthesis gene cluster (Fig. 4B).
Fig 4.
Gene organization of the salinomycin biosynthesis gene cluster. (A) Overlapping fosmids encompassing the sequenced region. Primers sln1-F and sln1-R were used to screen for the salinomycin gene cluster from the S. albus XM211 genomic library. The other primers were used for chromosome walking. The pink lines represent the sequenced region, including 12H8, 25E8, 10B8, 6G11, and 3.58-kb and 7.93-kb BamHI fragments from 19H1. (B) Gene organization of the salinomycin biosynthesis gene cluster. −, gene products not involved in salinomycin biosynthesis, as confirmed through gene replacement; +, gene products involved in salinomycin biosynthesis, as confirmed through gene replacement and/or trans-complementation to the corresponding mutants.
Table 1.
Deduced functions of ORFs in the salinomycin biosynthesis gene cluster
| ORF | No. of amino acids | Proposed functiona | Sequence similarity (protein, origin) | % identity/similarity | Accession no. |
|---|---|---|---|---|---|
| Orf11 | 572 | 3-Hydroxybutyryl-CoA dehydrogenase | StrviDRAFT_8389, Streptomyces violaceusniger Tü 4113 | 61/70 | ZP07610704 |
| Orf12 | 342 | 3-Oxoacyl-(acyl-carrier-protein) synthase III | SSNG_07461, S. treptomyces sp. C | 77/86 | ZP07291840 |
| SlnA1 | 4,925 | PKS | LasA1, Streptomyces lasaliensis | 56/68 | CAQ64686 |
| Loading | KSQ-ATa-ACP | ||||
| Module 1 | KS-ATb-DH-KR-ACP | ||||
| Module 2 | KS-ATa-DH-ER-KR-ACP | ||||
| SlnA2 | 3,917 | PKS | NanA5, Streptomyces nanchangensis NS3226 | 51/63 | AAP42859 |
| Module 3 | KS-ATp-DH-KR-ACP | ||||
| Module 4 | KS-ATa-DH-ER-KR-ACP | ||||
| SlnA3 | 2,692 | PKS | MeiA2, S. nanchangensis NS3226 | 52/64 | ADC45534 |
| Module 5 | KS-ATa-ACP | ||||
| Module 6 | KS-ATa-KR-ACP | ||||
| SlnA4 | 1,642 | PKS | NigAVI, S. violaceusniger DSM4137 | 51/62 | DQ354110 |
| Module 7 | KS-ATp-KRa- ACP | ||||
| SlnA5 | 3,750 | PKS | LasAII, S. lasaliensis | 56/68 | CAQ64687 |
| Module 8 | KS-ATp-DH-ER-KR-ACP | ||||
| Module 9 | KS-ATb-KR-ACP | ||||
| SlnA6 | 1,435 | PKS | LasAVI, S. lasaliensis | 58/71 | CAQ64691 |
| Module 10 | KS-ATp-DH*- ACP | ||||
| SlnA7 | 1,644 | PKS | LasAIV, S. lasaliensis | 56/68 | CAQ64689 |
| Module 11 | KS-ATp-KR-ACP | ||||
| SlnA8 | 3,724 | PKS | MeiA1, S. nanchangensis NS3226 | 52/64 | ADC45586 |
| Module 12 | KS-ATp-KR-ACP | ||||
| Module 13 | KS-ATa-DH-ER-KR-ACP | ||||
| SlnA9 | 2,312 | PKS | AmphI, Streptomyces nodosus | 52/66 | AAK73501 |
| Module 14 | KS-ATb-KR-ACP | ||||
| KS | KS | ||||
| SlnBI | 128 | Epoxide hydrolase | NigBI, S. violaceusniger DSM4137 | 32/50 | ABC84467 |
| SlnDI | 265 | Thioesterase | MonAX, S. cinnamonensis | 50/65 | AA065810 |
| SlnM | 271 | O-Methyltransferase | TcmP, Streptomyces glaucescens | 39/59 | AAA67510 |
| SlnE | 93 | Ferredoxin | Tcs_55098_011, Streptomyces sp. ATCC 55098 | 53/70 | ADU56364 |
| SlnF | 407 | Cytochrome P450 | NigD, S. violaceusniger DSM4137 | 35/52 | ABC84463 |
| SlnTI | 319 | ABC transporter ATP-binding protein | TnrB2, S. longisporoflavus | 64/77 | CAA52012 |
| SlnTII | 546 | ABC transporter efflux pump | TnrB3, S. longisporoflavus | 39/53 | CAA52013 |
| SlnBII | 149 | Epoxide hydrolase | NigBI, S. violaceusniger DSM4137 | 47/64 | ABC84467 |
| SlnBIII | 153 | Epoxide hydrolase | NigBI, S. violaceusniger DSM4137 | 52/72 | ABC84467 |
| SlnC | 484 | Epoxidase | NigCI, S. violaceusniger DSM4137 | 51/67 | ABC84466 |
| SlnR | 906 | LuxR family transcriptional regulator | Las4, S. lasaliensis | 42/52 | CAQ64683 |
| SlnDII | 253 | Thioesterase | MonAX, S. cinnamonensis | 40/54 | AA065810 |
| Orf13 | 862 | Peptide synthetase | AcmC, Streptomyces anulatus | 48/61 | ADG27359 |
| Orf14 | 82 | Hypothetical protein | SSNG_00262, Streptomyces sp. C | 74/80 | ZP07284641 |
| Orf15 | 855 | SARP family transcriptional regulator | Sros_3791, Streptomyces roseum DSM 43021 | 33/45 | YP003339454 |
| Orf16 | 597 | Acyl-CoA synthetase | PRK09192, Streptomyces viridochromogenes | 43/54 | AEF16021 |
| Orf17 | 572 | Acyl-CoA dehydrogenase | SSCG_02872, Streptomyces clavuligerus ATCC 27064 | 40/52 | ZP05005545 |
| Orf18 | 96 | Peptidyl carrier protein | SSCG_02873, S. clavuligerus ATCC 27064 | 58/75 | ZP05005546 |
| Orf19 | 279 | 4′-Phosphopantetheinyl transferase | SCLAV_0457, S. clavuligerus ATCC 27064 | 50/61 | ZP06769934 |
*, redundant domain; a, inactive domain; ER, enoyl reductase.
Determining the boundaries of the salinomycin biosynthesis gene cluster.
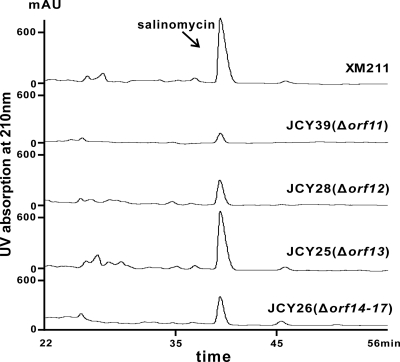
To the left of the PKS gene slnA1, two genes, orf11 and orf12, are divergently transcribed (Fig. 4B) and annotated as 3-hydroxybutyryl-CoA dehydrogenase and 3-oxoacyl-(acyl carrier protein) synthase III genes, respectively. Genes orf11 and orf12 are thought to be involved in the dedicated supply of the butyrate extender unit for salinomycin biosynthesis (25, 35). Genes orf11 and orf12 were individually replaced with aac(3)IV-oriT, and the salinomycin yields of the orf11 mutant JCY39 and the orf12 mutant JCY28 were found to be 10% and 36% of the wild type, respectively (Fig. 5; see Fig. S5 in the supplemental material), suggesting the involvement of orf11 and orf12 in salinomycin biosynthesis, possibly through the supply of a butyrate extender units.
Fig 5.
HPLC analysis of mutants of S. albus XM211 for boundary determination.
To the right of gene slnDII, gene orf13, putatively encoding a peptide synthetase, was first replaced by aac(3)IV. The salinomycin productivity was not changed, as expected, in the mutant JCY25 (Fig. 5). Further deletion of orf14 to orf17, however, resulted in a moderate drop in salinomycin production in the mutant JCY26 to 45% of the wild type (Fig. 5; see Fig. S5 in the supplemental material). Among these four genes, the deletion of the SARP family transcriptional regulator gene orf15 probably reduced the transcription of salinomycin biosynthesis genes.
PKS genes.
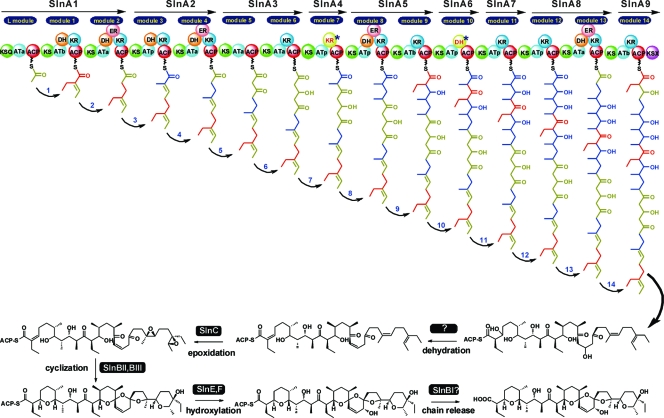
Nine large adjacent genes (slnA1 to slnA9) are colinearly arranged in the cluster (Fig. 4B). Four of these (slnA4, slnA6, slnA7, and slnA9) each encode a single extension module, while the other five (slnA1, slnA2, slnA3, slnA5, and slnA8) harbor two extension modules, in agreement with the 14 condensation steps required for the biosynthesis of the salinomycin polyketide skeleton. The linear polyketide is synthesized by the loading of an acetate starter unit and incorporation of 5 acetate, 6 propionate, and 3 butyrate units (see Fig. 7).
Fig 7.
Proposed biosynthesis pathway for salinomycin.
SlnA1 contains a loading module (KSQ-ATa-ACP [where KSQ is the KS domain in the loading module with active-site cysteine replaced by glutamine]) for the initiation of salinomycin polyketide synthesis and modules 1 and 2 for polyketide chain extension from C-25 to C-30. The AT domains of the loading module and module 2 have the characteristic motif HAFH for recognizing acetate (malonyl-CoA) extender units (ATa) (8, 38). The AT domain of module 1 is predicted to incorporate a butyrate (ethylmalonyl-CoA) unit due to the presence of a VASH motif (25, 35). SlnA2 putatively encodes modules 3 and 4 and is responsible for the extension of C-21 to C-24 through incorporation of one propionate (methylmalonyl-CoA) unit (8, 38) and one acetate unit.
SlnA3 putatively encodes modules 5 and 6 and is responsible for the incorporation of C-17 to C-20. According to the hypothetical salinomycin biosynthesis pathway (5), an unsaturated double bond at C-18 and C-19 could be formed during polyketide elongation. Surprisingly, module 6 has a KS-ATa-KR-ACP domain structure but lacks a DH domain for the required dehydration. SlnA4 contains only module 7, presumably for the incorporation of C-15 and C-16. KR7 has four changes in the conserved NADP(H) binding motif GXGXXGXXXA and in the essential residues K-S-Y-N (30) and thus appears to be inactive. This is in full agreement with the presence of a keto group at C-17 in the polyketide chain. SlnA5, -A6, -A7, and -A8 putatively correspond to modules 8 to 13 for the incorporation of C-3 to C-14.
The last ORF, SlnA9, contains module 14 for the incorporation of C-1 and C-2; an apparently intact C-terminal KS domain, SlnKSX; and an inactive AT domain (137 amino acids [aa] shorter than AT14). Similar to module 6, module 14 has a KS-ATb [ethylmalonyl-CoA-specific acyltransferase]-KR-ACP domain assembly, which does not match the predicted double bond at C-2–C-3 required for the formation of the tetrahydrofuran. This discrepancy suggests the possibility that the required dehydration reactions may take place after polyketide chain assembly catalyzed by a discrete dehydratase. There is no putative dehydratase, however, in the cloned region, and these genes might be located in another region of the chromosome.
Genes possibly involved in polyketide chain elongation and release.
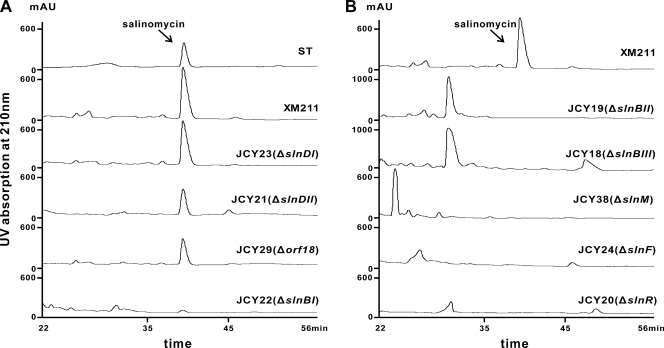
Due to the absence of a conventional TE domain within the PKS or a NanE-like thioesterase (22, 23), there are three candidates for the requisite thioesterase activity in the salinomycin biosynthesis gene cluster. Genes slnDI and slnDII, encoding discrete type II TEs, display high similarity to monAIX and monAX in the monensin gene cluster (14). The slnDI disruption mutant JCY23 produced salinomycin at a level similar to that of wild-type XM211 (Fig. 6A), whereas the slnDII disruption mutant JCY21 had a 40 to 50% drop in salinomycin production (Fig. 6A). SlnDI and SlnDII are therefore predicted to serve a conventional editing function with the ability to remove nonelongateable residues from unprocessed PKS proteins, thus ensuring the efficiency of polyketide biosynthesis.
Fig 6.
Comparison of the S. albus XM211 wild type and its various mutants by HPLC. ST, salinomycin standard purchased from Sigma.
In the pathway to monensin, the discrete MonKSX is proposed to be capable of catalyzing the transfer of the linear polyketide chain to the MonACPX, on which oxidative cyclization occurs (15). The extra SlnKSX domain fused downstream of ACP14 in the last module of the salinomycin PKS may catalyze the transfer (Fig. 7). However, the required discrete ACP gene is not present in an adjacent region. In the whole sequenced region, only orf18 is predicted to encode a peptidyl carrier protein to potentially perform such a function. Targeted inactivation of orf18 was therefore carried out to create a mutant JCY29. Interestingly, analysis of the mutant revealed a 50 to 60% drop in salinomycin production compared to wild-type XM211 (Fig. 6A). Thus, this result implies that Orf18 may be relevant to salinomycin biosynthesis but not essential to the polyketide chain release.
Immediately downstream of the last PKS gene, slnA9, lies a putative epoxide hydrolase gene, slnBI. However, sequence alignments (see Fig. S6 in the supplemental material) reveal that SlnBI has an isoleucine in place of the conserved glutamic acid residue in an acidic amino acid pair (D38-E65), which was proven to play a critical role in stepwise epoxide opening and subsequent cyclization (26). Although SlnBI is much smaller than NanE-like enzymes and shows no significant similarity to them, its relationship with the chain release was investigated through gene replacement. In the slnBI mutant JCY22, the yield of salinomycin was dramatically decreased to less than 5% of that of the wild type (Fig. 6A). Since no other convincing candidate enzyme was found for this role in the cloned region, SlnBI merits consideration as a potential enzyme with thioesterase activity.
Genes involved in epoxide-opening cascades.
Genes slnBII and slnBIII, encoding polypeptides of 149 aa and 153 aa, respectively, are putative epoxide hydrolase genes. slnBII and slnBIII overlap for 8 bp and are significantly similar to nigBI in the nigericin gene cluster, with 47% and 42% identity, respectively. In order to study the roles of these two genes in the biosynthesis of salinomycin, corresponding mutants were created. When analyzed by LC-MS, no trace of salinomycin was detected in either the slnBII mutant JCY19 or the slnBIII mutant JCY18, confirming their involvement in salinomycin biosynthesis (Fig. 6B). Moreover, LC-MS revealed a new peak with a shorter retention time (in both JCY18 and JCY19; m/z 734.5 [M+H]+) and another new peak with a longer retention time (in JCY18 only; m/z 735.5 [M+H]+) than salinomycin (data not shown). The chemical structures of these two novel salinomycin derivatives need to be further confirmed.
Other ancillary genes.
The P450 monooxygenase SlnF, homologous to NigD in the nigericin biosynthesis gene cluster with 35% identity and 52% similarity, and the cognate ferredoxin SlnE are likely to accomplish the hydroxylation at C-20. However, gene replacement of slnF resulted in a mutant JCY24, which completely lost salinomycin productivity (Fig. 6B), suggesting that the hydroxylation by SlnF may take place prior to the release of the mature polyketide chain from ACP.
SlnM is predicted to be an O-methyltransferase homologous to TcmP in tetracenomycin biosynthesis (37% identity, 59% similarity), which catalyzes methylation of a carboxyl group (6). Although methylated salinomycin was not detectable in XM211 fermentation extracts through LC-MS analysis, the function of slnM was preliminarily investigated through gene replacement. Surprisingly, the peak of salinomycin disappeared and a new peak emerged (m/z 791.5 [M+Na]+) in the slnM mutant JCY38 (Fig. 6B). The chemical structure of this novel derivative needs to be further confirmed, which may shed new light on its involvement in salinomycin biosynthesis.
The encoded product of slnR is similar to Las4 (42% identity, 52% similarity) in the lasalocid gene cluster and belongs to the LuxR family of transcriptional regulators (25, 35), most of which are positive regulators of antibiotic biosynthesis. Inactivation of slnR, through gene replacement with the aac(3)IV gene, abolished the production of salinomycin in the resulting mutant JCY20, which clearly indicated that SlnR is a positive pathway-specific regulator of salinomycin biosynthesis (Fig. 6B).
The gene products of slnTI and slnTII are significantly similar to TnrB2 and TnrB3 of the tetronasin biosynthesis gene cluster, with 64% and 39% identities, respectively. The ATP-binding protein TnrB2 and the transmemebrane protein TnrB3 are presumed to form an ATP-dependent efflux system for polyether tetronasin self-resistance in Streptomyces longisporoflavus (20). SlnTI and SlnTII probably confer salinomycin resistance on the producer, S. albus XM211.
Proposed salinomycin biosynthesis pathway.
Taking all this information into account, we have proposed a mechanism for salinomycin biosynthesis (Fig. 7). Loading of the malonyl-CoA onto SlnA1 is followed by condensation steps with 5 malonyl-CoA, 6 methylmalonyl-CoA, and 3 ethylmalonyl-CoA extender units. Dehydration at C-2–C-3 and C-18–C-19 is speculated to occur on the mature polyketide chain. While still bound to the ACP, the diene intermediate is thought to be converted to the diepoxide through the action of SlnC, which serves as the substrate for SlnBII and SlnBIII for the subsequent epoxide-opening and ring closure reactions. Afterward, SlnF probably carries out the hydroxylation at C-20 when the polyketide chain is still enzyme bound. The final release of the fully modified polyether chain from the PKS is probably accomplished by SlnBI or other, uncharacterized enzymes (Fig. 7).
Supplementary Material
ACKNOWLEDGMENTS
This work received financial support from the National Natural Science Foundation of China, the Ministry of Science and Technology (973 and 863 Programs), the Ministry of Education, and the Shanghai Municipal Council of Science and Technology.
Footnotes
Published ahead of print 9 December 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Ansari MZ, Yadav G, Gokhale RS, Mohanty D. 2004. NRPS-PKS: a knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res. 32:W405–W413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bhatt A, et al. 2005. Accumulation of an E,E,E-triene by the monensin-producing polyketide synthase when oxidative cyclization is blocked. Angew. Chem. Int. Ed. Engl. 44:7075–7078 [DOI] [PubMed] [Google Scholar]
- 4. Blazsek M, Surovcova A. 2003. LC determination of salinomycin in fermentation broths and premixes. J. Pharm. Biomed. Anal. 31:291–298 [DOI] [PubMed] [Google Scholar]
- 5. Cane DE, Celmer WD, Westley JW. 1983. Unified stereochemical model of polyether antibiotic structure and biogenesis. J. Am. Chem. Soc. 105:3594–3600 [Google Scholar]
- 6. Decker H, Motamedi H, Hutchinson CR. 1993. Nucleotide sequences and heterologous expression of tcmG and tcmP, biosynthetic genes for tetracenomycin C in Streptomyces glaucescens. J. Bacteriol. 175:3876–3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Del Vecchio F, et al. 2003. Active-site residue, domain and module swaps in modular polyketide synthases. J. Ind. Microbiol. Biotechnol. 30:489–494 [DOI] [PubMed] [Google Scholar]
- 9. Demydchuk Y, et al. 2008. Analysis of the tetronomycin gene cluster: insights into the biosynthesis of a polyether tetronate antibiotic. Chembiochem 9:1136–1145 [DOI] [PubMed] [Google Scholar]
- 10. Gallimore AR, et al. 2006. Evidence for the role of the monB genes in polyether ring formation during monensin biosynthesis. Chem. Biol. 13:453–460 [DOI] [PubMed] [Google Scholar]
- 11. Gumila C, Ancelin ML, Delort AM, Jeminet G, Vial HJ. 1997. Characterization of the potent in vitro and in vivo antimalarial activities of ionophore compounds. Antimicrob. Agents Chemother. 41:523–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gupta PB, et al. 2009. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 138:645–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gust B, Challis GL, Fowler K, Kieser T, Chater KF. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harvey BM, et al. 2006. Evidence that a novel thioesterase is responsible for polyketide chain release during biosynthesis of the polyether ionophore monensin. Chembiochem 7:1435–1442 [DOI] [PubMed] [Google Scholar]
- 15. Harvey BM, et al. 2007. Insights into polyether biosynthesis from analysis of the nigericin biosynthetic gene cluster in Streptomyces sp. DSM4137. Chem. Biol. 14:703–714 [DOI] [PubMed] [Google Scholar]
- 16. Ishikawa J, Hotta K. 1999. FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol. Lett. 174:251–253 [DOI] [PubMed] [Google Scholar]
- 17. Izumikawa M, Murata M, Tachibana K, Ebizuka Y, Fujii I. 2003. Cloning of modular type I polyketide synthase genes from salinomycin producing strain of Streptomyces albus. Bioorg. Med. Chem. 11:3401–3405 [DOI] [PubMed] [Google Scholar]
- 18. Kieser T, Bibb MJ, Buttner MJ, Charter KF, Hopwood DA. (ed). 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 19. Knirschova R, et al. 2007. Multiple regulatory genes in the salinomycin biosynthetic gene cluster of Streptomyces albus CCM 4719. Folia Microbiol. (Praha) 52:359–365 [DOI] [PubMed] [Google Scholar]
- 20. Linton KJ, Cooper HN, Hunter IS, Leadlay PF. 1994. An ABC-transporter from Streptomyces longisporoflavus confers resistance to the polyether-ionophore antibiotic tetronasin. Mol. Microbiol. 11:777–785 [DOI] [PubMed] [Google Scholar]
- 21. Liu T, Cane DE, Deng Z. 2009. The enzymology of polyether biosynthesis. Methods Enzymol. 459:187–214 [DOI] [PubMed] [Google Scholar]
- 22. Liu T, Lin X, Zhou X, Deng Z, Cane DE. 2008. Mechanism of thioesterase-catalyzed chain release in the biosynthesis of the polyether antibiotic nanchangmycin. Chem. Biol. 15:449–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu T, et al. 2006. Identification of NanE as the thioesterase for polyether chain release in nanchangmycin biosynthesis. Chem. Biol. 13:945–955 [DOI] [PubMed] [Google Scholar]
- 24. Matsuura Y, et al. 2010. Intriguing substrate tolerance of epoxide hydrolase Lsd19 involved in biosynthesis of the ionophore antibiotic lasalocid A. Org. Lett. 12:2226–2229 [DOI] [PubMed] [Google Scholar]
- 25. Migita A, et al. 2009. Identification of a gene cluster of polyether antibiotic lasalocid from Streptomyces lasaliensis. Biosci. Biotechnol. Biochem. 73:169–176 [DOI] [PubMed] [Google Scholar]
- 26. Minami A, et al. 2011. Enzymatic epoxide-opening cascades catalyzed by a pair of epoxide hydrolases in the ionophore polyether biosynthesis. Org. Lett. 13:1638–1641 [DOI] [PubMed] [Google Scholar]
- 27. Oliynyk M, et al. 2003. Analysis of the biosynthetic gene cluster for the polyether antibiotic monensin in Streptomyces cinnamonensis and evidence for the role of monB and monC genes in oxidative cyclization. Mol. Microbiol. 49:1179–1190 [DOI] [PubMed] [Google Scholar]
- 28. Paget MSB, Chamberlin L, Atrih A, Foster SJ, Buttner MJ. 1999. Evidence that the extracytoplasmic function sigma factor sigma(E) is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pan Z, Barry R, Lipkin A, Soloviev M. 2007. Selection strategy and the design of hybrid oligonucleotide primers for RACE-PCR: cloning a family of toxin-like sequences from Agelena orientalis. BMC Mol. Biol. 8:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reid R, et al. 2003. A model of structure and catalysis for ketoreductase domains in modular polyketide synthases. Biochemistry 42:72–79 [DOI] [PubMed] [Google Scholar]
- 31. Riddell FG. 2002. Structure, conformation, and mechanism in the membrane transport of alkali metal ions by ionophoric antibiotics. Chirality 14:121–125 [DOI] [PubMed] [Google Scholar]
- 32. Seto H, Miyazaki Y, Fujita K, Otake N. 1977. Studies on the ionophorous antibiotics X. The assignment of 13C-NMR spectrum of salinomycin. Tetrahedron Lett. 18:2417–2420 [Google Scholar]
- 33. Shichijo Y, et al. 2008. Epoxide hydrolase Lsd19 for polyether formation in the biosynthesis of lasalocid A: direct experimental evidence on polyene-polyepoxide hypothesis in polyether biosynthesis. J. Am. Chem. Soc. 130:12230–12231 [DOI] [PubMed] [Google Scholar]
- 34. Shirling EB, Gottlieb D. 1966. Methods for characterization of Streptomyces species. Int. J. Syst. Evol. Microbiol. 16:313–340 [Google Scholar]
- 35. Smith L, Hong H, Spencer JB, Leadlay PF. 2008. Analysis of specific mutants in the lasalocid gene cluster: evidence for enzymatic catalysis of a disfavoured polyether ring closure. Chembiochem 9:2967–2975 [DOI] [PubMed] [Google Scholar]
- 36. Sun Y, He X, Liang J, Zhou X, Deng Z. 2009. Analysis of functions in plasmid pHZ1358 influencing its genetic and structural stability in Streptomyces lividans 1326. Appl. Microbiol. Biotechnol. 82:303–310 [DOI] [PubMed] [Google Scholar]
- 37. Sun Y, et al. 2003. A complete gene cluster from Streptomyces nanchangensis NS3226 encoding biosynthesis of the polyether ionophore nanchangmycin. Chem. Biol. 10:431–441 [DOI] [PubMed] [Google Scholar]
- 38. Tang L, Yoon YJ, Choi CY, Hutchinson CR. 1998. Characterization of the enzymatic domains in the modular polyketide synthase involved in rifamycin B biosynthesis by Amycolatopsis mediterranei Gene 216:255–265 [DOI] [PubMed] [Google Scholar]
- 39. Wang H, et al. 2011. Genetic screening strategy for rapid access to polyether ionophore producers and products in actinomycetes. Appl. Environ. Microbiol. 77:3433–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu Y, Kang Q, Shen Y, Su W, Bai L. 2011. Cloning and functional analysis of the naphthomycin biosynthetic gene cluster in Streptomyces sp. CS. Mol. Biosyst. 7:2459–2469 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.