Abstract
Accumulation of toxic metals in the environment represents a public health and wildlife concern. Bacteria resistant to toxic metals constitute an attractive biomass for the development of systems to decontaminate soils, sediments, or waters. In particular, biosorption of metals within the bacterial cell wall or secreted extracellular polymeric substances (EPS) is an emerging process for the bioremediation of contaminated water. Here the isolation of bacteria from soil, effluents, and river sediments contaminated with toxic metals permitted the selection of seven bacterial isolates tolerant to mercury and associated with a mucoid phenotype indicative of the production of EPS. Inductively coupled plasma-optical emission spectroscopy and transmission electron microscopy in conjunction with X-ray energy dispersive spectrometry revealed that bacteria incubated in the presence of HgCl2 sequestered mercury extracellularly as spherical or amorphous deposits. Killed bacterial biomass incubated in the presence of HgCl2 also generated spherical extracellular mercury deposits, with a sequestration capacity (40 to 120 mg mercury per g [dry weight] of biomass) superior to that of live bacteria (1 to 2 mg mercury per g [dry weight] of biomass). The seven strains were shown to produce EPS, which were characterized by Fourier transform-infrared (FT-IR) spectroscopy and chemical analysis of neutral-carbohydrate, uronic acid, and protein contents. The results highlight the high potential of Hg-tolerant bacteria for applications in the bioremediation of mercury through biosorption onto the biomass surface or secreted EPS.
INTRODUCTION
Throughout the twentieth century, human activities such as mining, chemical industries, and agriculture have yielded high accumulations of toxic metals in the environment. These metals, bioavailable and persistent (33), constitute a major environmental problem, adversely affecting ecosystems and public health (24). Mercury pollution is of real concern because of the high toxicity of the metal and its translocation all along the food chain: mercury is accumulated upward through the aquatic food chain and is transformed to more-toxic organic mercury forms, mainly highly neurotoxic methylmercury (24).
Toxic metals are difficult to remove from the environment, since they cannot be chemically or biologically degraded and are ultimately indestructible. Physicochemical remediation of metal-polluted sites, from incineration of soils to chemical precipitation or/and ion-exchange technologies, has been widely used but remains costly and environmentally damaging. Biological approaches based on metal-resistant microorganisms have received a great deal of attention as alternative remediation processes (20, 26). The biological methods used currently for mercury removal consist of Hg2+ reduction to volatile metal mercury by bacterial strains harboring the mer resistance operon (1, 32, 35, 48). Live or dead bacterial biomass has also been used for biosorption applications (52), which consist of passive immobilization of metals by the biomass and can rely on different physicochemical mechanisms, such as adsorption, surface complexation, ion exchange, or surface precipitation (26). Biosorption appears to be a fast and metabolism-independent process that allows the use of dead biomass, in contrast to the intracellular metal accumulation process called bioaccumulation (49). The applicability and benefits of growing bacterial/fungal/algal cells and dead biomass for metal removal through biosorption have been reviewed previously (3, 28). Both secreted extracellular polymeric substances (EPS) and cell walls have been shown to participate in this process (2). Bacteria appear to have a greater capacity to adsorb metals from solutions than any other form of life, since they display the highest surface-to-volume ratio (10). Several bacterial species have already been studied for their mercury sorption capacities (47), but the bacterial biosorption mechanisms still have to be characterized further, in parallel with technological developments in bioremediation.
In this study, seven environmental bacterial strains tolerant to mercury were isolated from soils, sediments, and effluents collected at different metal-rich sites. They were selected for their tolerance to mercury and their mucoid phenotypes, indicative of EPS production. Metal depletion capacities were quantified by inductively coupled plasma-optical emission spectroscopy (ICP-OES) analysis and were compared for live and dead bacterial biomass, which permitted differentiation between biosorption and bioprecipitation processes. Metal accumulation was observed by transmission electron microscopy (TEM) in conjunction with X-ray energy dispersive spectrometry (XEDS) analysis. The EPS associated with the cells or secreted in the supernatants were extracted and submitted to preliminary characterization.
MATERIALS AND METHODS
Isolation of arsenic- and mercury-tolerant mucoid bacteria.
As- and Hg-tolerant bacteria were isolated from soils, effluents, and river sediments collected in the Vík í Mýrdal black sand beach (volcanic area, Southern Iceland), in the Petit Saut reservoir (French Guiana), and at the edges of the Tinto and Odiel Rivers (mining area; Iberian Pyritic Belt, Spain). The soil samples were mixed volume-to-volume with a sterile saline solution (KCl, 8.5 g/liter). All the samples were sonicated briefly in an ultrasonic bath, and enrichment cultures were prepared from a 1/5 dilution of the suspensions in poor broth (PB) nutrient medium (10 g/liter Bacto tryptone, 5 g/liter NaCl [pH 7]) and were incubated for 24 h at 28°C under shaking. One hundred microliters of the enrichment cultures was plated onto PB or Luria-Bertani (LB) (10 g/liter Bacto tryptone, 5 g/liter yeast extract, 10 g/liter NaCl [pH 7]) agar plates supplemented with either 5 mM As2O5, 5 mM Na2HAsO4, or 10 μM HgCl2, and the cultures were incubated for 1 week at 28°C. Colony types differing in shape, color, and margin were isolated and streak purified. The isolated strains were subcultivated onto LB agar alone or LB agar supplemented with 3% (wt/vol) glucose for 1 week at 30°C, and strains with a mucoid phenotype were selected.
Identification of mercury-tolerant mucoid bacteria.
Bacterial isolates were observed under a phase-contrast microscope to characterize their morphology, their motility, and the presence or absence of endospores. They were further identified by combining Gram staining and 16S rRNA gene sequencing. Fragments of the 16S rRNA gene from bacterial colonies were PCR amplified with the bacterial primer set 27F and 1385R (amplification of 1.5-kb fragments) (43) or BK1F et BK1R, specific to the genus Bacillus (amplification of 1.1-kb fragments) (54) (see Table S1 in the supplemental material), using Taq DNA polymerase with ThermoPol buffer (New England BioLabs, Ozyme, Saint-Quentin-en-Yvelines, France). PCRs, performed in a Mastercycler thermal cycler (Eppendorf, Le Pecq, France), included 28 cycles, each consisting of annealing for 1 min at 51°C, elongation for 2 min at 72°C, and denaturation for 1 min at 95°C. PCR products were sequenced by Eurofins MWG Operon (Ebersberg, Germany). The sequences were matched against nucleotide sequences from GenBank using BLASTN (5).
Determination of metal tolerance levels.
Susceptibilities to HgCl2 were determined for each selected strain in 96-well plates by using the 2-fold microdilution method (15) with slight modifications. The bacteria were precultivated in LB medium for 16 h at 30°C, and a 1/100 dilution was used to inoculate 100 μl LB or PB medium supplemented with HgCl2 (2 μM to 1 mM). Each metal concentration was tested in triplicate. Control wells contained either HgCl2 or the bacterium in the culture medium considered. After 48 h of incubation at 30°C under shaking (180 rpm), growth was monitored by measuring the optical density at 595 nm on a Multiskan FC plate reader (Thermo Fisher Scientific, Illkirch, France) and subtracting the signals measured for the metal salts alone.
Metal depletion in culture supernatants and metal accumulation in cell pellets.
The bacterial strains were grown in 5 ml LB medium supplemented with glucose (3%, wt/vol) (LB/Glc) at 30°C under shaking. After 48 h of culture, 5 ml LB/Glc was added. After an additional 24 h, 5 ml HgCl2 solution (300 μM in LB/Glc) was added, and the cultures were incubated for 24 h. The cultures were then centrifuged at 8,000 × g for 20 min. The supernatants were collected and were ultrafiltered with 100-kDa-cutoff spin columns (15 ml; Vivaspin; Sartorius). The pellets were washed once with LB medium and twice with sterile deionized water; then they were dried at 50°C and weighed. Mercury analyses were subsequently performed by ICP-OES as described below.
Metal removal by dead bacterial biomass.
For each strain, dead bacterial biomass was obtained from a 48-h 1-liter culture in LB/Glc in the absence or presence of HgCl2 at a concentration equal to the MIC value divided by 2. The dead bacterial biomass was centrifuged at 8,000 × g for 15 min. The pellets were washed twice, resuspended in sterile deionized water, and heated at 120°C at 15 lb/in2 for 45 min. The absence of live bacteria after this step was checked by plating onto LB agar plates. The mass concentration of dead bacteria was determined spectrophotometrically at 620 nm by using a calibration curve previously established from optical density measurements as a function of dry weights. Fifteen-milliliter suspensions (3.3 mg bacterial biomass/ml) were incubated at room temperature for 24 h under shaking in the presence of 1 or 2 mM HgCl2 and were then centrifuged at 8,000 × g for 15 min. The pellets were washed three times with sterile deionized water and were dried at 50°C, while the supernatants were filtered on 0.2-μm-pore-size filter cartridges.
Hg(0) volatilization assay.
The volatilization of Hg(0) was assessed using the protocol described by Nakamura and Nakahara (34). The strains were cultivated on LB agar plates supplemented with 40 μM HgCl2 for Microbacterium oxydans HG3, Ochrobactrum sp. strain HG16, Lysinibacillus sp. strain HG17, Bacillus sp. strain CM111, and Serratia marcescens HG19 and with 10 μM HgCl2 for Kocuria rosea EP1 and Bacillus cereus MM8. After incubation for 24 h at 30°C, colonies were harvested and were resuspended in 100 μl 0.07 M phosphate buffer (pH 7), containing 0.5 mM EDTA, 0.2 mM magnesium acetate, and 5 mM sodium thioglycolate, either without HgCl2 or with HgCl2 at concentrations up to 100 μM. The suspensions were diluted with buffer to give an optical density of 0.2 at 620 nm. The suspensions were transferred to a microplate, covered with a sensitive film used for TEM photography (SO-163; Kodak), and cultivated for 4 h at 30°C in the dark. The foggy areas on the film, due to reduction of the Ag+ emulsion of the film by the mercury vapor, were interpreted as Hg(0) volatilization. Escherichia coli strain DU1040, which carries the mer plasmid R831 (46), was used as a positive control. Sterile LB medium, alone or with HgCl2 at concentrations up to 100 μM, was used as a control to assess the level of mercury volatilization due to the culture medium.
ICP-OES.
Each sample of dry bacterial biomass was suspended in 1 ml 70% nitric acid and was heated at 60°C for 16 h. One milliliter of each 100-kDa filtrate was diluted 1:1 in 70% nitric acid and was heated at 60°C for 16 h. After appropriate dilution of the samples, mercury analyses were performed on a Vista-MPX optical emission spectrometer (Agilent, Massy-Palaiseau, France). The ICP-OES system was calibrated by serial dilutions of mercury standards within limits of detection ranging from 50 μg/liter to 50 mg/liter. The emission lines used for the analyses were 184.887 and 194.164 nm.
Statistical analysis.
Statistical analysis was performed using R (http://cran.r-project.org/). Means and standard deviations of values for adsorbed Hg were calculated from triplicates of biomass-metal incubations and subsequent ICP-OES measurements. To assess the significance of differences in biosorption between strains, Kruskal-Wallis and Wilcoxon-Mann-Whitney nonparametric tests were carried out. The significance level was set at a P value less than 0.05.
TEM and XEDS.
The bacterial cultures or dead bacterial biomass suspensions (in the absence or presence of metal) were suspended in sterile water and were deposited on carbon Formvar-coated electron microscope grids (Electron Microscopy Sciences, Euromedex, Souffelweyersheim, France) without staining. A JEM-2100 transmission electron microscope (JEOL, Croissy-sur-Seine, France), equipped with a LaB6 electron source and operated at 200 kV, was used to observe the bacteria. The images were acquired with a 2K (2,000 pixel-by-2,000 pixel) charge-coupled device (CCD) camera (Ultrascan 1000; Gatan). An X-ray energy dispersive spectrometer (XEDS) detector was used to acquire the X-ray spectra. X-ray mapping was performed using XEDS in conjunction with a scanning transmission electron microscopy (STEM) module. Tomography experimental data were acquired with a tomography module monitored by Digital Micrograph software (Gatan).
Isolation and chemical analysis of the EPS.
Six-day-culture broths (200 ml in LB/Glc) were centrifuged (6,000 × g, 30 min, 4°C). The EPS were isolated either (i) from bacterial pellets or (ii) from supernatants. (i) The cell pellets from the culture were suspended in 10 ml NaCl (20 g/liter) in water and were centrifuged (6,000 × g, 30 min, 4°C). The resulting pellets were suspended in 10 ml (10-g/liter) NaCl in water and were dialyzed against water for 48 h (Spectra/Por dialysis membrane; molecular size cutoff, 12 to 14 kDa). The dialyzed samples were centrifuged (6,000 × g, 30 min, 4°C), and the supernatants were lyophilized and stored at 4°C. (ii) The culture supernatants were pressure-filtered through cellulose nitrate filters (pore sizes, 5 μm, 3 μm, and 0.45 μm; Whatman). After the addition of cold ethanol (ethanol/filtrate ratio, 2:1), the solutions were kept at 4°C overnight. The EPS precipitates were recovered by centrifugation (6,000 × g, 30 min, 4°C). The pellets were suspended in water and were dialyzed against water (Spectra/Por dialysis membrane; molecular size cutoff, 12 to 14 kDa). The dialyzed samples were lyophilized and stored at 4°C. The protein contents of EPS were determined by the Bradford method (12), using bovine serum albumin as a standard (Roti-Quant; Carl Roth, Lauterbourg, France). The total neutral-carbohydrate contents were determined by the phenol-sulfuric acid method (16), using d-glucose as a standard. The uronic acid contents were determined by the meta-hydroxydiphenyl method (11), using d-glucuronic acid as a standard. Cultures, extractions, and determinations of protein, carbohydrate, and uronic acid contents were performed in triplicate.
Gel electrophoresis and mass spectrometry analysis of the EPS.
The EPS extracted from Lysinibacillus sp. HG17 were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), according to the method of Laemmli (25), on a 10% polyacrylamide gel. A broad-range protein marker (New England BioLabs) was used as a molecular weight (MW) reference. The proteins were visualized by Coomassie blue staining. The major band was excised from the gel and was washed successively, twice with 25 mM NH4HCO3 (pH 8), once with 50% acetonitrile in 25 mM NH4HCO3 (pH 8), once with 25 mM NH4HCO3 (pH 8), and once with water, before being vacuum dried. The gel slices were rehydrated with 50 μl digestion buffer (25 mM NH4HCO3, 5 mM CaCl2 [pH 8.0], containing 20 ng/μl trypsin from bovine pancreas [T8642; Sigma]) and were incubated for 16 h at 37°C under shaking. Supernatants were collected. Gels were washed once with 0.1% formic acid in water and once with acetonitrile, and the extracts were combined with the supernatants that had been collected formerly. The pooled supernatants were vacuum dried, resuspended in 50 μl 0.1% formic acid in water-acetonitrile (90:10), and analyzed by liquid chromatography-mass spectrometry (LC-MS). The LC-MS analysis was conducted on an Ultimate 3000 Micro-HPLC system (Dionex, Voisins-le-Bretonneux, France) connected to an electrospray ionization (ESI)-hybrid quadrupole time-of-flight (QqTOF) QSTAR Pulsar mass spectrometer (Applied Biosystems, Les Ulis, France) equipped with an ion spray source. Separation was achieved in a Strategy C18(2) column (internal diameter, 1 mm; length, 150 mm; pore size, 100 Å; particle size, 5 μm) (Interchim, Montluçon, France) by using a gradient of water plus 0.1% formic acid (solvent A) and acetonitrile (solvent B) at 40°C and 40 μl · min−1, with a linear increase from 10% to 70% solvent B within 25 min, followed by a linear increase to 100% solvent B in 5 min. The MS instrument was operated in positive mode using information-dependent acquisition (IDA). This mode completes a series of 1-s survey scans (m/z detection range, 250 to 1,300) followed by two 2-s tandem MS (MS-MS) experiments on the two most intense peaks from the survey scan, as long as they have not been fragmented in the last 2 min (m/z detection range, 50 to 1,500). The data generated were analyzed with the MASCOT MS-MS ion search engine (41).
FT-IR spectroscopy.
Pellets for Fourier transformed-infrared (FT-IR) analysis were obtained by grinding a mixture of 2 mg EPS with 200 mg dry KBr and then pressing the mixture into a 16-mm-diameter mold. The FT-IR spectra were recorded on an 8400S spectrometer (Shimadzu, Champs-sur-Marne, France) with a resolution of 1 cm−1, in the 4,000- to 400-cm−1 region.
Nucleotide sequence accession numbers.
The sequences determined in this study were deposited in GenBank under accession numbers FJ481923, FJ228159, FJ228161, FJ228157, FJ228156, FJ228160, and FJ228158.
RESULTS
Isolation and identification of mercury-tolerant bacteria.
Mercury-tolerant bacteria from soil, sediments, and effluent samples collected in Southern Iceland and French Guiana were isolated on poor broth (PB) or Luria Bertani (LB) medium in the presence of 10 μM HgCl2, a typical concentration for the isolation of resistant strains (37, 44). Eighty bacterial isolates were obtained after a 1-week incubation at 30°C. In addition, 40 strains isolated for their resistance or tolerance to arsenic from soil and sediment samples collected at the edges of the Tinto and Odiel Rivers were screened for their levels of resistance/tolerance to mercury. The mercury MICs (i.e., the lowest concentrations that caused 100% growth inhibition) were measured in LB culture. One hundred five strains revealed tolerance to 10 μM HgCl2. Seven strains were further selected for showing both mercury tolerance levels in the 20 to 100 μM range and enhanced mucoid phenotypes when grown on LB agar medium supplemented with glucose (Table 1). It must be noted that the MICs for these strains in PB were generally lower than those measured in LB, with values in the 15 to 30 μM range. The mucoid phenotype was considered an indicator of a propensity to produce EPS. The bacteria were identified by 16S rRNA gene sequencing (Table 1) (primers are listed in Table S1 in the supplemental material, and a phylogenic tree is shown in Fig. S1 in the supplemental material), and the sequences were deposited in GenBank. Of the seven strains selected, all except HG16 and HG19 were Gram positive, and three were Bacillus or Lysinibacillus species (MM8, CM111, and HG17).
Table 1.
Names, origins, and identificationa of the mercury-tolerant strains selected in this study
| Isolate | Environmental sample | Most similar species (% identity) | GenBank accession no. | Gram reactionb | HgCl2 MIC (μM) in the following mediumc: |
|
|---|---|---|---|---|---|---|
| LB | PB | |||||
| MM8 | Tinto River mining area (water + sediments, pH 3) | Bacillus cereus (99) | FJ481923 | + | 30 | 15 |
| HG17 | Petit Saut, French Guiana (sediments) | Lysinibacillus sp. (99) | FJ228157 | + | 60 | 15 |
| CM111 | Odiel River (water, pH 3.5) | Bacillus sp. (99) | FJ228158 | + | 100 | 30 |
| EP1 | Odiel River (sludge) | Kocuria rosea (99) | FJ228160 | + | 20 | 20 |
| HG3 | Vik, Iceland (black sand) | Microbacterium oxydans (99) | FJ228159 | + | 100 | 30 |
| HG19 | Petit Saut, French Guiana (sediments) | Serratia marcescens (98) | FJ228156 | − | 60 | 30 |
| HG16 | Petit Saut, French Guiana (sediments) | Ochrobactrum sp. (99) | FJ228161 | − | 60 | 30 |
Based on 16S rRNA gene sequences and HgCl2 MICs.
+, Gram positive; −, Gram negative.
The following MICs are given for comparison: for E. coli DU1040 (carrying plasmid R831 harboring the mer operon and therefore resistant to mercury through reduction and volatilization), 60 μM in LB and 30 μM in PB medium; for Bacillus sp. MM3 (isolated in this study and considered nonresistant), 10 μM in LB and 4 μM in PB medium; for Bacillus cereus GTC 02826 (reference strain), 15 μM in LB and 8 μM in PB medium; and for Bacillus subtilis subsp. subtilis DSM 10 (reference strain), 15 μM in LB and 4 μM in PB medium.
Measurements of mercury removal by growing bacteria.
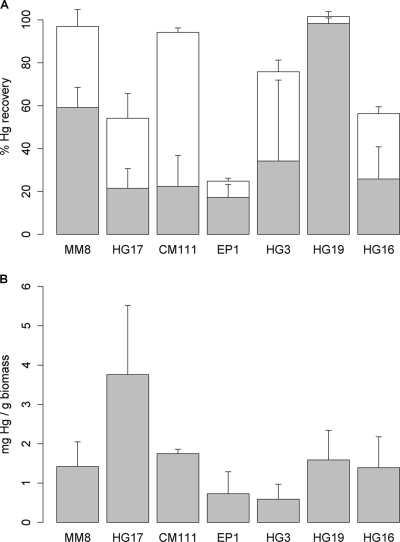
For each selected strain, the mercury removal properties were assessed by measuring metal depletion in culture supernatants and metal accumulation in cell pellets by inductively coupled plasma-optical emission spectroscopy (ICP-OES) (Fig. 1). The supernatants were filtered on 100-kDa-cutoff ultrafiltration cartridges prior to quantification in order to suppress the potential contribution of the secreted bacterial EPS to metal adsorption. Mercury removal was observed for all strains, resulting in 18 to 97% depletion of the 100 μM mercury initially present in the culture medium (Fig. 1A). The maximum ratio of sorption to cell biomass was measured for S. marcescens HG19, a strain that triggered mercury precipitation visible to the naked eye. For K. rosea EP1 and, to a minor extent, Ochrobactrum sp. HG16 and Lysinibacillus sp. HG17, the sum of the percentages of mercury in the supernatants and the bacterial pellets did not reach 100%. This trend suggests that these strains would secrete EPS that are able to bind mercury, which would be eliminated during the ultrafiltration step. The content of mercury within bacterial pellets was about 2 mg per g (dry weight) of pellet (Fig. 1B), with Lysinibacillus sp. HG17 and Bacillus sp. CM111 showing maximum values. The values measured for mercury in the pellet fraction displayed important variability, which might be due to depletion of Hg-rich material during the washing steps.
Fig 1.
ICP-OES quantification of mercury removal by living bacterial biomass. (A) Quantification of mercury contents (expressed as percentages of the initial metal content) of bacterial pellets (shaded areas of bars) and culture supernatants (open areas of bars) after cultivation of bacteria in the presence of 100 μM HgCl2. (B) Mass of mercury (expressed as milligrams per gram [dry weight] of bacterial biomass) after cultivation in the presence of 100 μM HgCl2. Error bars represent standard deviations for triplicate experiments.
Observation of metal accumulation within bacterial biomass.
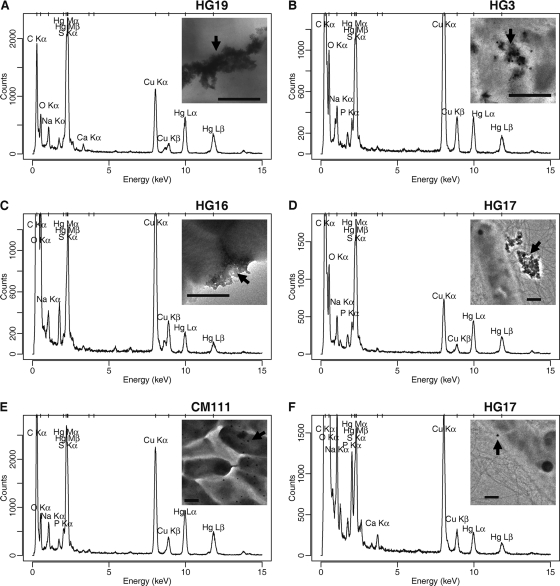
The selected bacteria were observed by TEM after incubation in the presence of HgCl2, and XEDS was used to characterize their elemental composition (Fig. 2; see also Fig. S2 and S3 in the supplemental material). XEDS clearly showed the presence of mercury, through the characteristic Lα and Lβ emission peaks (9.99 and 11.82 keV, respectively), together with the Mα and Mβ peaks (2.19 and 2.28 keV, respectively), the latter overlapping the Kα peak of the element S (2.31 keV). For S. marcescens HG19 and M. oxydans HG3, black precipitates were visible to the naked eye within the bacterial cultures. The corresponding TEM observations showed that these were amorphous extracellular precipitates (Fig. 2A and B; see also Fig. S2E2 and F2 in the supplemental material). The other mercury deposits observed consisted either of amorphous extracellular aggregates located next to the bacterial surface, for Ochrobactrum sp. HG16 and Lysinibacillus sp. HG17 (Fig. 2C and D; see also Fig. S2B2 and G1 in the supplemental material) or of spherical deposits, for every strain except S. marcescens HG19 (Fig. 2E and F; see also Fig. S2A2, B2, C2, D2, E1, and G2 in the supplemental material), with a 10- to 100-nm diameter (data not shown). The chemical composition of the mercury deposits was difficult to assign given the contribution of the spectra of the surrounding bacteria. X-ray cartography experiments permitted us to better localize the mercury within the bacterial biomass, showing a concentration inside amorphous and spherical deposits and a diffuse localization over the bacteria (see Fig. S3 in the supplemental material for K. rosea EP1). In order to assess the intra- or extracellular localization of the spherical mercury deposits, tomography experiments were carried out. They consist in tilting the grid and recording successive TEM photographs, thereby making possible a 3-dimensional (3D) visualization of the bacteria. Such experiments suggested an extracellular localization of the spherical deposits (see Movie S5, recorded for K. rosea EP1, in the supplemental material). However, this trend is clear only for the deposits located on the sides of the pictures generated. Therefore, intracellular localization of the metal cannot be completely ruled out.
Fig 2.
XEDS analyses of mercury deposits observed for bacteria cultivated in the presence of HgCl2. (A and B) Extracellular precipitates; (C and D) extracellular deposits of mercury within EPS; (E and F) regions rich in spherical precipitates. XEDS spectra obtained for S. marcescens HG19 (A), M. oxydans HG3 (B), Ochrobactrum sp. HG16 (C), Lysinibacillus sp. HG17 (D), Bacillus sp. CM111 (E), and Lysinibacillus sp. HG17 (F) are shown. (Insets) TEM pictures showing the deposits analyzed (indicated by arrows). Bars, 0.5 μm.
Absence of resistance based on Hg(0) volatilization.
The usual resistance system of bacteria toward mercury is conferred by the mer operon and relies on mercury reduction and volatilization (9, 35). It involves the conversion of Hg(II) to Hg(0) catalyzed by the mercuric ion reductase MerA, followed by the passive diffusion of Hg(0) out of the cell. Our objective was to select bacteria with an alternative resistance mechanism, based on biosorption. Thus, we checked the absence of mercury resistance based on reduction and volatilization by using a functional test carried out to detect the release of Hg(0) by bacteria growing in the presence of HgCl2 (see Fig. S4 in the supplemental material). Fogging of the X-ray film appeared for the positive control Escherichia coli DU1040 but either was not detected or was negligible for the seven strains tested. These results indicated that the tolerance of the tested bacteria to mercury could rely on biosorption or precipitation of the metal rather than on Hg(0) volatilization.
Mercury biosorption by dead bacterial biomass.
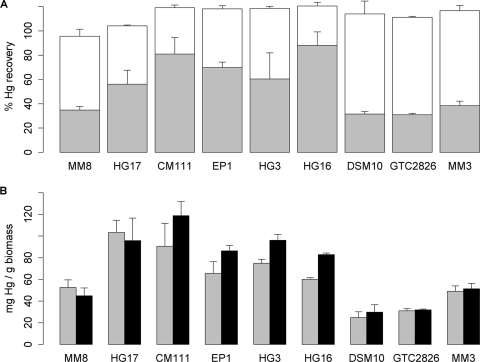
The selected bacteria, cultivated in the absence or in the presence of 20 μM HgCl2, were killed and incubated in the presence of HgCl2 in order to assess the mercury removal capacity of the dead bacterial biomass. The presence of mercury during cultivation did not modify the mercury removal capacity of dead bacterial biomass (data not shown). Therefore, only the results obtained for bacteria cultivated in the absence of mercury are presented here. With dead bacterial biomass, the HgCl2 concentration was not limited by the tolerance of bacteria for the toxic metal. Therefore, the removal capacities of dead bacterial biomass were measured after incubation in the presence of 1 and 2 mM HgCl2. Measurements were carried out for all the strains except S. marcescens HG19, given the absence of mercury deposits observed by TEM-XEDS for this strain (see below). For the other six strains, dead biomass induced 35 to 85% depletion of the initial 2 mM HgCl2 (Fig. 3A). The mercury content was 40 to 120 mg per g (dry weight) of dead biomass (Fig. 3B). The best adsorption capacities were found for Lysinibacillus sp. HG17 and Bacillus sp. CM111, as with the growing bacteria. In order to help ascertain the specificity of the properties described for the selected strains, the biosorption capacity of dead bacterial biomass was measured for three additional strains: Bacillus sp. strain MM3, isolated in this study (GenBank accession number FJ228146.1), and two Bacillus reference strains, B. cereus GTC 02826 and Bacillus subtilis subsp. subtilis DSM 10 (see Fig. S1 in the supplemental material). These strains were considered nontolerant to mercury as determined by MIC measurements (Table 1). The mercury biosorption capacities of these strains appeared close to that of Bacillus cereus MM8 and lower than those of the other strains, although statistical analysis did not permit us to show significant differences between strains.
Fig 3.
ICP-OES quantification of mercury removal by dead bacterial biomass. (A) Quantification of the mercury contents (expressed as percentages of the initial metal content) in bacterial pellets (gray areas of bars) and supernatants (open areas of bars) after incubation of dead bacterial biomass in the presence of 2 mM HgCl2. (B) Mass of mercury (expressed as milligrams per gram [dry weight] of bacterial biomass) after incubation of dead bacterial biomass in the presence of 1 mM (shaded bars) or 2 mM (filled bars) HgCl2. Error bars represent standard deviations for triplicate experiments. Although the Kruskal-Wallis test performed on the whole set of strains revealed a nonhomogeneous distribution of the mass (mg/g) and content (percentage) of Hg within the pellets for all strains (P = 0.003), no significant difference between strains was found by using the pairwise Wilcoxon-Mann-Whitney test.
TEM observations revealed a high content of mercury within dense spherical deposits for the killed bacterial biomass of every strain incubated in the presence of 100 μM HgCl2, except S. marcescens HG19 (see Fig. S2A3 to G3 in the supplemental material). The number of deposits per bacterium was particularly high for Bacillus sp. CM111 (see Fig. S2C3). The precipitates observed for the live bacteria S. marcescens HG19 and M. oxydans HG3 were absent for the dead biomass, revealing that these precipitates depend on a specific metabolic process.
Characterization of the production of EPS.
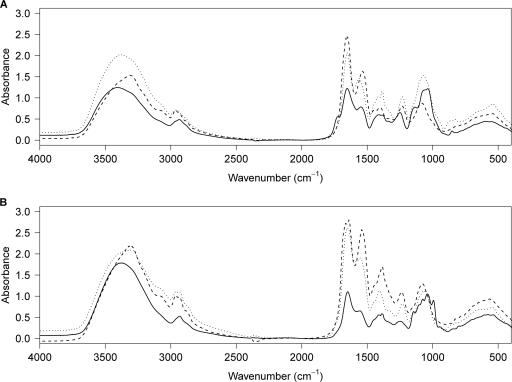
The mucoid phenotype and the mercury biosorption capacity of the selected strains suggested that they produced EPS. Therefore, we searched for EPS in both the culture pellets and the supernatants of the seven selected strains, and colorimetric quantification of the proteins, neutral carbohydrates, and uronic acids was performed on the isolated materials (Table 2). Production of EPS was observed for every strain, although it was very weak for B. cereus MM8 and M. oxydans HG3. The most abundant secreted EPS were observed for Ochrobactrum sp. HG16, K. rosea EP1, and Lysinibacillus sp. HG17, while S. marcescens HG19 and Bacillus sp. CM111 showed the most abundant cell-associated EPS (Table 2). A capsule was observed for Bacillus sp. CM111 by India ink staining (data not shown) and scanning electron microscopy (see Fig. S6 in the supplemental material). The EPS from Bacillus sp. CM111 and S. marcescens HG19, which appeared mainly as bacterium-associated EPS (Table 2), showed reduced contents of protein and neutral carbohydrates, suggesting the presence of other entities, such as lipids and/or modified carbohydrates. The secreted EPS of Ochrobactrum sp. HG16, K. rosea EP1, and Lysinibacillus sp. HG17 were composed mainly of neutral carbohydrates, with various protein contents (highest for HG17, intermediate for EP1, and lowest for HG16). The highest protein content was measured in the EPS isolated from both the supernatants and the pellets of Lysinibacillus sp. HG17. The EPS extracted were further characterized by Fourier transform-infrared (FT-IR) spectroscopy (Fig. 4), which showed the typical absorption bands for carbohydrates (a broad O—H elongation band in the 3,200- to 3,700-cm−1 range; bands in the 1,000- to 1,200-cm−1 range assigned to C—O elongation), as well as bands characteristic of proteins (elongation of amide C O between 1,680 and 1,630 cm−1; bending N—H amide band between 1,550 and 1650 cm−1). The relative intensities of the protein and carbohydrate bands were in agreement with the colorimetric quantification measurements. Finally, the stretching P
O between 1,680 and 1,630 cm−1; bending N—H amide band between 1,550 and 1650 cm−1). The relative intensities of the protein and carbohydrate bands were in agreement with the colorimetric quantification measurements. Finally, the stretching P O band at 1,210 to 1,140 cm−1 permitted us to determine the presence of phosphate in the EPS extracted from both the pellets and the supernatants of Lysinibacillus sp. HG17 and Bacillus sp. CM111. The EPS extracted from Lysinibacillus sp. HG17, which displayed the highest protein content, were further characterized by SDS-PAGE, followed by in-gel digestion and LC-MS analysis (see Fig. S7 in the supplemental material). A major band corresponding to a protein with a molecular size of 120 kDa was detected on the SDS gel for the EPS extracted from both the supernatant and the pellet (see Fig. S7A). In-gel digestion of this protein and LC-MS analysis of the digest in the IDA mode, which permits multiple data-dependent MS-MS experiments, were conducted. The data generated permitted the identification of 9 peptides corresponding to SbpA, an S-layer protein from Lysinibacillus sphaericus CCM2177 (see Fig. S7B) (GenBank accession number AAF22978.1). S-layers are exoproteins that form bidimensional crystalline arrays, which cover the bacterial cell surface and can participate in cell protection and cell adhesion (45). The peptides that were matched to the SbpA sequence are located in the N-terminal region of the protein (see Fig. S7C), which contains three S-layer homology (SLH) domains. These domains are conserved regions that permit noncovalent anchoring of S-layer proteins and other exoproteins to the cell surface through interaction with cell wall carbohydrates (31). Therefore, these results permit us to propose that the EPS produced by Lysinibacillus sp. HG17 consists of an S-layer protein.
O band at 1,210 to 1,140 cm−1 permitted us to determine the presence of phosphate in the EPS extracted from both the pellets and the supernatants of Lysinibacillus sp. HG17 and Bacillus sp. CM111. The EPS extracted from Lysinibacillus sp. HG17, which displayed the highest protein content, were further characterized by SDS-PAGE, followed by in-gel digestion and LC-MS analysis (see Fig. S7 in the supplemental material). A major band corresponding to a protein with a molecular size of 120 kDa was detected on the SDS gel for the EPS extracted from both the supernatant and the pellet (see Fig. S7A). In-gel digestion of this protein and LC-MS analysis of the digest in the IDA mode, which permits multiple data-dependent MS-MS experiments, were conducted. The data generated permitted the identification of 9 peptides corresponding to SbpA, an S-layer protein from Lysinibacillus sphaericus CCM2177 (see Fig. S7B) (GenBank accession number AAF22978.1). S-layers are exoproteins that form bidimensional crystalline arrays, which cover the bacterial cell surface and can participate in cell protection and cell adhesion (45). The peptides that were matched to the SbpA sequence are located in the N-terminal region of the protein (see Fig. S7C), which contains three S-layer homology (SLH) domains. These domains are conserved regions that permit noncovalent anchoring of S-layer proteins and other exoproteins to the cell surface through interaction with cell wall carbohydrates (31). Therefore, these results permit us to propose that the EPS produced by Lysinibacillus sp. HG17 consists of an S-layer protein.
Table 2.
Masses and compositions of the EPS extracted from the supernatants and the pellets of the bacteria cultivated in LB/GIca
| Strain | Supernatant |
Pellet |
||||||
|---|---|---|---|---|---|---|---|---|
| Mass (mg/200 ml of culture) | Content (μg/g) of: |
Mass (mg/200 ml of culture) | Content (μg/g) of: |
|||||
| Protein | Carbohydrate | Uronic acid | Protein | Carbohydrate | Uronic acid | |||
| B. cereus MM8 | 34 | 15.9 (1.6) | 520.8 (7.9) | 11.7 (2.8) | 1 | 156.1 (51.7) | 46.3 (9.0) | 7.0 (1.4) |
| Lysinibacillus sp. HG17 | 59 | 212.0 (6.7) | 361.5 (54.6) | 9.7 (2.1) | 6 | 365.3 (28.7) | 337.9 (33.4) | 11.5 (2.1) |
| Bacillus sp. CM111 | 42 | 22.9 (2.2) | 383.0 (6.8) | 10.3 (2.2) | 48 | 128.3 (21.2) | 70.7 (5.9) | 9.7 (2.0) |
| K. rosea EP1 | 67 | 96.6 (13.2) | 377.9 (72.1) | 35.7 (3.7) | 6 | 351.8 (74.0) | 227.6 (15.0) | 23.1 (4.1) |
| M. oxydans HG3 | 25 | 28.3 (7.4) | 389.4 (31.0) | 12.6 (1.2) | 2 | 59.4 (6.5) | 163.3 (12.3) | 34.5 (3.8) |
| S. marcescens HG19 | 38 | 27.4 (9.0) | 467.2 (61.0) | 25.6 (4.9) | 214 | 47.2 (6.6) | 152.6 (16.8) | 23.7 (5.9) |
| Ochrobactrum sp. HG16 | 115 | 15.5 (3.1) | 519.6 (29.6) | 10.6 (2.1) | 3 | 166.4 (34.6) | 508.9 (69.1) | 95.1 (12.7) |
Values in parentheses are standard deviations for triplicate extractions.
Fig 4.
FT-IR spectra of the EPS extracted from supernatants (A) and pellets (B) of mercury-tolerant bacteria. The spectra of the EPS extracted from K. rosea EP1, Lysinibacillus sp. HG17, and Bacillus sp. CM111 are shown as solid, dashed, and dotted lines, respectively.
DISCUSSION
This study aimed at selecting environmental bacteria tolerant to mercury through extracellular sequestration by using two selection criteria: (i) tolerance to mercury and (ii) a mucoid phenotype, indicative of the production of EPS. The production of EPS is intimately linked to adherence processes, such as the formation of biofilms, and is one of the mechanisms associated with antibiotic and metal resistance (7). Biofilm EPS can play a substantial role in the biosorption and biomineralization of metal ions (40). In particular, secreted exopolysaccharides have been recognized for several decades as important contributors to metal biosorption by microorganisms (13, 36).
The strains selected here displayed MICs ranging from 20 to 100 μM in LB medium and from 15 to 30 μM in PB medium, ranges typical for mercury-tolerant or -resistant bacteria (8, 51). The metal sequestration capacities of live bacterial biomass were about 1 to 2 mg per g (dry weight) of bacteria. Previous studies reported similar ranges of mercury biosorption capacities for wild-type strains of Bacillus (22) and Pseudomonas (38) and for an engineered strain of Escherichia coli producing the mer-regulatory protein MerR on the cell surface (6). As determined by TEM-XEDS analyses, all the selected strains grown in the presence of 100 μM HgCl2 displayed extracellular mercury deposits within precipitates, amorphous extracellular aggregates, and/or spherical deposits (Fig. 2; see also Fig. S2 in the supplemental material), together with diffuse deposits at the bacterial surface (see Fig. S3 in the supplemental material for Kocuria rosea EP1).
Mercury deposits associated with dead bacterial biomass (see Fig. S2A3 to G3 in the supplemental material) are indicative of a passive process, i.e., biosorption at the bacterial surface and/or through cell wall-associated EPS, as proposed for the metal-binding capacities of a collection of Bacillus sphaericus strains (50). The observation of extracellular spherical deposits for all dead bacteria except HG19 suggested that for these strains, the bacterial cell wall or cell wall-associated EPS involved in the biosorption process were preserved after the killing step (the secreted EPS were eliminated during the washing of the dead bacterial biomass). The similar aspects of the deposits observed for all the selected bacteria suggested a nonspecific biosorption process. Bacterial cell walls have high affinities for dissolved metals, and generally, metal binding occurs after initial metal complexation and neutralization of the chemically active sites (10). Cell walls of microbial biomass offer particularly abundant metal-binding functional groups, such as carboxylate, hydroxyl, sulfate, phosphate, and amino groups. In addition, the cell wall peptidoglycan and/or cell wall-associated EPS, such as components of capsules and slimes, or proteinaceous S-layers, can constitute effective biosorption matrices (52). Three of the seven strains selected here belong to the genus Bacillus or Lysinibacillus, whose metal biosorption properties have already been reported (22, 27, 42, 50) and which display a high propensity to produce a capsule (29) and/or secreted exopolysaccharides (55) and/or an S-layer (30, 42) with potential biosorption capacities. The high quantity of EPS extracted from the pellet of Bacillus sp. CM111 (Table 2) and the important deposition of mercury at the bacterial surface (see Fig. S2C3 in the supplemental material) suggest that for this strain, biosorption of mercury would rely on surface EPS. The observation of a capsule for this strain (see Fig. S6 in the supplemental material) corroborates this hypothesis. A surprising result is the large amount of EPS extracted from the pellet of S. marcescens HG19 (Table 2), although this strain showed no mercury deposition in TEM observations (see Fig. S2F3 in the supplemental material). This result could be explained by the fact that the Serratia marcescens membrane is very hydrophobic and is composed mainly of lipopolysaccharide (LPS) (23), which might not adsorb mercury under the conditions tested or might be altered during the killing step.
Based on our results, we proposed for each strain a main tolerance mechanism and a hypothesis on the nature of EPS (Table 3). For S. marcescens HG19 and M. oxydans HG3, a tolerance mechanism relying on extracellular precipitation was identified. In contrast to the other strains, S. marcescens HG19 showed no accumulation other than precipitates, and its dead biomass showed no metal sequestration. Thus, this strain can sequester mercury only through precipitation, not through biosorption. Precipitation of mercury with sulfide or organosulfur compounds is a detoxification mechanism described as a mercury tolerance mechanism rather than an effective resistance process (4, 21). Precipitation of HgS by bacteria releasing H2S has been proposed as a method for mercury removal (4, 17–18). Mercury precipitation can also result from the production of volatile organosulfur compounds, yielding precipitates containing mercury together with sulfur and carbon (17). Since the release of dimethyl disulfide has been reported previously for a Serratia marcescens strain (40), a precipitate containing Hg, S, and C can be proposed here for S. marcescens HG19. When Ochrobactrum sp. strain HG16 and K. rosea EP1 were incubated in the presence of mercury, they did not display precipitates but rather mercury accumulations within spherical deposits or amorphous aggregates, attributed to biosorption processes (see Fig. S2D2, G1, and G2 in the supplemental material). Furthermore, the low retention of mercury within the bacterial biomass (Fig. 1A) and the large amounts of EPS extracted from the supernatants of these strains (Table 2) suggested a mechanism based on biosorption by secreted EPS. Bacteria of the genus Kocuria have been detected previously in waste industry samples contaminated with heavy metals (19), but the metal tolerance/resistance mechanisms of these bacteria remain unknown. Bacteria of the genus Ochrobactrum have been reported to display metal biosorption properties through the production of exopolysaccharides (39), which supports our hypothesis. The biosorption of mercury within secreted EPS for K. rosea EP1, Ochrobactrum sp. HG16, and Lysinibacillus sp. HG17 could explain the high variability of the quantification of mercury within live bacterial biomass (Fig. 1B), which would result from variations in the depletion of extracellular substances loaded with mercury during the ultrafiltration and washing steps. Lysinibacillus sp. strain HG17 cultivated in the presence of 100 μM HgCl2 displayed mercury deposits within amorphous aggregates or spherical deposits (see Fig. S2B2 in the supplemental material), while only spherical deposits were observed after incubation of the dead bacterial biomass with mercury (see Fig. S2B3), suggesting a dual biosorption process involving both secreted EPS and the cell wall or cell wall-associated EPS. The high protein content of the EPS suggests that they consist mainly of an exoprotein. The characterization of peptides corresponding to the N-terminal region of SbpA, an S-layer protein from Lysinibacillus sphaericus CCM 2177, permitted us to propose that Lysinibacillus sp. HG17 produces an S-layer. S-layer proteins contain a well-conserved N-terminal region, are highly variable in their central and C-terminal regions, and can harbor posttranslational modifications, such as glycosylation and phosphorylation (45). This can explain why only peptides corresponding to the N-terminal region of the Lysinibacillus sp. HG17 S-layer were identified. Both live and dead biomass of Lysinibacillus sphaericus strains has been reported to show toxic-metal biosorption properties (50), which have been attributed to the binding of metal to S-layers (30, 42).
Table 3.
Main mercury tolerance mechanisms and hypotheses on the nature of the EPS produced by the selected strains
| Isolate | Main tolerance mechanism | Nature of EPS |
|---|---|---|
| Bacillus cereus MM8 | Biosorption | Secreted exopolysaccharide |
| Lysinibacillus sp. HG17 | Biosorption | EPS with high protein content (S-layer) |
| Bacillus sp. CM111 | Biosorption | Surface EPS (capsule) |
| Kocuria rosea EP1 | Biosorption | Secreted exopolysaccharide |
| Microbacterium oxydans HG3 | Mercury precipitation | Secreted exopolysaccharide |
| Serratia marcescens HG19 | Mercury precipitation | Surface EPS (LPS?) |
| Ochrobactrum sp. HG16 | Biosorption | Secreted exopolysaccharide |
High metal concentrations or extreme pHs are limiting factors for the use of live bacterial cells as a tool for bioremediation. In addition, the use of dead bacterial biomass avoids mercury volatilization by bacterial strains harboring the mer operon. For all seven selected strains except S. marcescens HG19, the biosorption capacities of the dead bacterial biomass (40 to 120 mg per g [dry weight] of biomass) were in the range reported for resistant or tolerant bacterial and yeast strains: as much as 400 mg per g for Pseudomonas aeruginosa and cyanobacteria (14) and 65 mg/g for Saccharomyces cerevisiae (53). All the bacteria selected here, except S. marcescens HG19, are potential mercury biosorbents that could be used as dead bacterial biomass or as EPS secretors. These bacteria thus appear to be promising tools for bioremediation applications in a sustainable-development context, since their dead biomass can mostly be considered an inert support that is easy to use without the need for conservation conditions and without the dissemination of live bacteria in the environment. In addition, four strains display biotechnological potential owing to the molecules involved in mercury biosorption: secreted exopolysaccharides (K. rosea EP1, Ochrobactrum sp. HG16), S-layer protein (Lysinibacillus sp. HG17), and capsule components (Bacillus sp. CM111).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the Region Ile-de-France in the framework of C'Nano IdF for support for the JEOL 2100 cryoelectron microscope installed at IMPMC UMR 7590. C'Nano-IdF is the nanoscience competence center of the Paris Region, supported by CNRS, CEA, MESR, and Région Ile-de-France. We thank Jean Guézennec (Ifremer, Brest, France) for fruitful discussions, Bastien Nay (UMR 7245 CNRS-MNHN, Paris, France) for assistance with the FT-IR measurements, the electron microscopy platform of the MNHN for access to the scanning electron microscope, and the mass spectrometry platform of the MNHN for access to the ESI-QqTOF instrument.
Footnotes
Published ahead of print 9 December 2011
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Abou-Shanab RA, van Berkum P, Angle JS. 2007. Heavy metal resistance and genotypic analysis of metal resistance genes in gram-positive and gram-negative bacteria present in Ni-rich serpentine soil and in the rhizosphere of Alyssum murale. Chemosphere 68:360–367 [DOI] [PubMed] [Google Scholar]
- 2. Aguilera A, Souza-Egipsy V, Martin-Uriz PS, Amils R. 2008. Extracellular matrix assembly in extreme acidic eukaryotic biofilms and their possible implications in heavy metal adsorption. Aquat. Toxicol. 88:257–266 [DOI] [PubMed] [Google Scholar]
- 3. Ahluwalia SS, Goyal D. 2007. Microbial and plant derived biomass for removal of heavy metals from wastewater. Bioresour. Technol. 98:2243–2257 [DOI] [PubMed] [Google Scholar]
- 4. Aiking H, Govers H, van 't Riet J. 1985. Detoxification of mercury, cadmium, and lead in Klebsiella aerogenes NCTC 418 growing in continuous culture. Appl. Environ. Microbiol. 50:1262–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Altschul SF, Lipman DJ. 1990. Protein database searches for multiple alignments. Proc. Natl. Acad. Sci. U. S. A. 87:5509–5513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bae W, Wu CH, Kostal J, Mulchandani A, Chen W. 2003. Enhanced mercury biosorption by bacterial cells with surface-displayed MerR. Appl. Environ. Microbiol. 69:3176–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV. 2006. Co-selection of antibiotic and metal resistance. Trends Microbiol. 14:176–182 [DOI] [PubMed] [Google Scholar]
- 8. Ball MM, Carrero P, Castro D, Yarzabal LA. 2007. Mercury resistance in bacterial strains isolated from tailing ponds in a gold mining area near El Callao (Bolivar State, Venezuela). Curr. Microbiol. 54:149–154 [DOI] [PubMed] [Google Scholar]
- 9. Barkay T, Miller SM, Summers AO. 2003. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 27:355–384 [DOI] [PubMed] [Google Scholar]
- 10. Beveridge TJ. 1989. Role of cellular design in bacterial metal accumulation and mineralization. Annu. Rev. Microbiol. 43:147–171 [DOI] [PubMed] [Google Scholar]
- 11. Blumenkrantz N, Asboe-Hansen G. 1973. New method for quantitative determination of uronic acids. Anal. Biochem. 54:484–489 [DOI] [PubMed] [Google Scholar]
- 12. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 13. Brown MJ, Lester JN. 1979. Metal removal in activated sludge: the role of bacterial extracellular polymers. Water Res. 13:817–837 [Google Scholar]
- 14. Cain A, Vannela R, Woo LK. 2008. Cyanobacteria as a biosorbent for mercuric ion. Bioresour. Technol. 99:6578–6586 [DOI] [PubMed] [Google Scholar]
- 15. CLSI 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7–A7, 7th ed. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 16. Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 28:350–356 [Google Scholar]
- 17. Essa AM, Creamer NJ, Brown NL, Macaskie LE. 2006. A new approach to the remediation of heavy metal liquid wastes via off-gases produced by Klebsiella pneumoniae M426. Biotechnol. Bioeng. 95:574–583 [DOI] [PubMed] [Google Scholar]
- 18. Essa AM, Macaskie LE, Brown NL. 2005. A new method for mercury removal. Biotechnol. Lett. 27:1649–1655 [DOI] [PubMed] [Google Scholar]
- 19. Freitas DB, et al. 2008. Molecular characterization of early colonizer bacteria from wastes in a steel plant. Lett. Appl. Microbiol. 47:241–249 [DOI] [PubMed] [Google Scholar]
- 20. Gadd GM. 2010. Metals, minerals and microbes: geomicrobiology and bioremediation. Microbiology 156:609–643 [DOI] [PubMed] [Google Scholar]
- 21. Glendinning KJ, Macaskie LE, Brown NL. 2005. Mercury tolerance of thermophilic Bacillus sp. and Ureibacillus sp. Biotechnol. Lett. 27:1657–1662 [DOI] [PubMed] [Google Scholar]
- 22. Green-Ruiz C. 2006. Mercury(II) removal from aqueous solutions by nonviable Bacillus sp. from a tropical estuary. Bioresour. Technol. 97:1907–1911 [DOI] [PubMed] [Google Scholar]
- 23. Hejazi A, Falkiner FR. 1997. Serratia marcescens. J. Med. Microbiol. 46:903–912 [DOI] [PubMed] [Google Scholar]
- 24. Jarup L. 2003. Hazards of heavy metal contamination. Br. Med. Bull. 68:167–182 [DOI] [PubMed] [Google Scholar]
- 25. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 26. Le Cloirec P, Andrès Y. 2005. Bioremediation of heavy metals using microorganisms, p 97–140 In Fingerman M, Nagabhushanam R. (ed), Bioremediation of aquatic and terrestrial ecosystems. Science Publishers, Inc., Enfield, NH [Google Scholar]
- 27. Lo W, et al. 2003. Biosorption and desorption of copper (II) ions by Bacillus sp. Appl. Biochem. Biotechnol. 105-108:581–591 [DOI] [PubMed] [Google Scholar]
- 28. Malik A. 2004. Metal bioremediation through growing cells. Environ. Int. 30:261–278 [DOI] [PubMed] [Google Scholar]
- 29. McLean RJ, Beauchemin D, Clapham L, Beveridge TJ. 1990. Metal-binding characteristics of the gamma-glutamyl capsular polymer of Bacillus licheniformis ATCC 9945. Appl. Environ. Microbiol. 56:3671–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Merroun ML, et al. 2005. Complexation of uranium by cells and S-layer sheets of Bacillus sphaericus JG-A12. Appl. Environ. Microbiol. 71:5532–5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mesnage S, et al. 2000. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 19:4473–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Misra TK, et al. 1984. Mercuric ion-resistance operons of plasmid R100 and transposon Tn501: the beginning of the operon including the regulatory region and the first two structural genes. Proc. Natl. Acad. Sci. U. S. A. 81:5975–5979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moore MR. 2004. A commentary on the impacts of metals and metalloids in the environment upon the metabolism of drugs and chemicals. Toxicol. Lett. 148:153–158 [DOI] [PubMed] [Google Scholar]
- 34. Nakamura K, Nakahara H. 1988. Simplified X-ray film method for detection of bacterial volatilization of mercury chloride by Escherichia coli. Appl. Environ. Microbiol. 54:2871–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nascimento AM, Chartone-Souza E. 2003. Operon mer: bacterial resistance to mercury and potential for bioremediation of contaminated environments. Genet. Mol. Res. 2:92–101 [PubMed] [Google Scholar]
- 36. Nichols CA, Guezennec J, Bowman JP. 2005. Bacterial exopolysaccharides from extreme marine environments with special consideration of the southern ocean, sea ice, and deep-sea hydrothermal vents: a review. Mar. Biotechnol. 7:253–271 [DOI] [PubMed] [Google Scholar]
- 37. Nieto JJ, et al. 1989. Survey of metal tolerance in moderately halophilic eubacteria. Appl. Environ. Microbiol. 55:2385–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Noghabi KA, et al. 2007. Mercury absorption by Pseudomonas fluorescens BM07 grown at two different temperatures. Pol. J. Microbiol. 56:111–117 [PubMed] [Google Scholar]
- 39. Ozdemir G, Ozturk T, Ceyhan N, Isler R, Cosar T. 2003. Heavy metal biosorption by biomass of Ochrobactrum anthropi producing exopolysaccharide in activated sludge. Bioresour. Technol. 90:71–74 [DOI] [PubMed] [Google Scholar]
- 40. Pal A, Paul AK. 2008. Microbial extracellular polymeric substances: central elements in heavy metal bioremediation. Indian J. Microbiol. 48:49–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551–3567 [DOI] [PubMed] [Google Scholar]
- 42. Pollmann K, Raff J, Merroun M, Fahmy K, Selenska-Pobell S. 2006. Metal binding by bacteria from uranium mining waste piles and its technological applications. Biotechnol. Adv. 24:58–68 [DOI] [PubMed] [Google Scholar]
- 43. Rheims H, Sproer C, Rainey FA, Stackebrandt E. 1996. Molecular biological evidence for the occurrence of uncultured members of the actinomycete line of descent in different environments and geographical locations. Microbiology 142:2863–2870 [DOI] [PubMed] [Google Scholar]
- 44. Sadhukhan PC, Ghosh S, Chaudhuri J, Ghosh DK, Mandal A. 1997. Mercury and organomercurial resistance in bacteria isolated from freshwater fish of wetland fisheries around Calcutta. Environ. Pollut. 97:71–78 [DOI] [PubMed] [Google Scholar]
- 45. Sára M, Sleytr UB. 2000. S-layer proteins. J. Bacteriol. 182:859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schottel J, Mandal A, Clark D, Silver S, Hedges RW. 1974. Volatilisation of mercury and organomercurials determined by inducible R-factor systems in enteric bacteria. Nature 251:335–337 [DOI] [PubMed] [Google Scholar]
- 47. Srivastava NK, Majumder CB. 2008. Novel biofiltration methods for the treatment of heavy metals from industrial wastewater. J. Hazard. Mater. 151:1–8 [DOI] [PubMed] [Google Scholar]
- 48. Valls M, de Lorenzo V. 2002. Exploiting the genetic and biochemical capacities of bacteria for the remediation of heavy metal pollution. FEMS Microbiol. Rev. 26:327–338 [DOI] [PubMed] [Google Scholar]
- 49. Valls M, Gonzalez-Duarte R, Atrian S, De Lorenzo V. 1998. Bioaccumulation of heavy metals with protein fusions of metallothionein to bacterial OMPs. Biochimie 80:855–861 [DOI] [PubMed] [Google Scholar]
- 50. Velásquez L, Dussan J. 2009. Biosorption and bioaccumulation of heavy metals on dead and living biomass of Bacillus sphaericus. J. Hazard. Mater. 167:713–716 [DOI] [PubMed] [Google Scholar]
- 51. Vetriani C, et al. 2005. Mercury adaptation among bacteria from a deep-sea hydrothermal vent. Appl. Environ. Microbiol. 71:220–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vijayaraghavan K, Yun YS. 2008. Bacterial biosorbents and biosorption. Biotechnol. Adv. 26:266–291 [DOI] [PubMed] [Google Scholar]
- 53. Wang J, Chen C. 2006. Biosorption of heavy metals by Saccharomyces cerevisiae: a review. Biotechnol. Adv. 24:427–451 [DOI] [PubMed] [Google Scholar]
- 54. Wu XY, Walker MJ, Hornitzky M, Chin J. 2006. Development of a group-specific PCR combined with ARDRA for the identification of Bacillus species of environmental significance. J. Microbiol. Methods 64:107–119 [DOI] [PubMed] [Google Scholar]
- 55. Zheng Y, Fang X, Ye Z, Li Y, Cai W. 2008. Biosorption of Cu(II) on extracellular polymers from Bacillus sp. F19. J. Environ. Sci. 20:1288–1293 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.