Abstract
Background & Aims
Complement is involved in the development of alcoholic liver disease in mice; however, the mechanisms for complement activation during ethanol exposure have not been identified. C1q, the recognition subunit of the first complement component, binds to apoptotic cells, thereby activating the classical complement pathway. Since ethanol exposure increases hepatocellular apoptosis, we hypothesized that ethanol-induced apoptosis would lead to activation of complement via the classical pathway.
Methods
Wild-type and C1qa-/- mice were allowed free access to ethanol containing diets or pair-fed control diets for 4 or 25 days.
Results
Ethanol feeding for 4d increased apoptosis of Kupffer cells in both wild-type and C1qa-/- mice. Ethanol-induced deposition of C1q and C3b/iC3b/C3c was co-localized with apoptotic Kupffer cells in wild-type, but not C1qa-/-, mice. Furthermore, ethanol-induced increases in TNFα and IL-6 expression at this early time point were suppressed in C1q-deficient mice. Chronic ethanol feeding (25d) increased steatosis, hepatocyte apoptosis, and activity of serum alanine and aspartate aminotransferases in wild-type mice. These markers of hepatocyte injury were attenuated in C1qa-/- mice. In contrast, chronic ethanol (25d)-induced increases in CYP2E1 expression and oxidative stress did not differ between wild-type and C1qa-/- mice.
Conclusions
For the first time, these data indicate that ethanol activates the classical complement pathway via C1q binding to apoptotic cells in the liver and that C1q contributes to the pathogenesis of ethanol-induced liver injury.
Keywords: complement, alcoholic liver disease, macrophages, C1q, apoptosis
Introduction
Alcoholic liver disease (ALD) is characterized by the development of steatosis, inflammation, hepatocyte necrosis and apoptosis, with the eventual development of fibrosis and cirrhosis. There is a growing appreciation that chronic inflammation is an important contributor to ALD (1). In particular, different arms of the innate immune system, including cellular components (macrophages and NKT cells) and fluid-phase components (complement), make critical contributions to the progression of ALD (2). Despite improved understanding of the pathophysiology of ALD, the development of effective preventative and/or treatment strategies for ALD has remained elusive.
The complement system is an ancient part of the immune system that bridges innate and adaptive immunity (3). Complement can be activated via the classical, lectin or alternative pathways. Complement activation leads to increased expression of inflammatory cytokines and chemokines, and plays an important role in host defense, as well as wound healing and response to tissue injury (3). Ethanol feeding to mice activates complement; activation of complement occurs as early as 2-4 days after the initiation of ethanol exposure, as well as after chronic (4-6 weeks) ethanol exposure (4; 5; 6). Importantly, deficiency in C3 or C5 protects mice from chronic ethanol-induced liver injury, while mice lacking CD55/decay accelerating factor, a complement regulator, exhibit exacerbated injury (7; 5). While it is clear that complement is an important contributor to the pathophysiology of ethanol-induced liver injury, the mechanisms by which ethanol activates complement have not been investigated.
The classical pathway of complement is activated upon the binding of C1q, the recognition subunit of first component (C1) in the classical pathway of activation, to immune complexes (8). The classical pathway is also activated via the interaction of C1q with cell surface markers on apoptotic cell, including interactions with phosphatidylserine (8), surface blebs (9) or nucleic acids on the cell surface (10). The binding of C1q to apoptotic cells likely plays a dual role in mediating the clearance of apoptotic cells. First, C1q binding leads to activation of C1/classical pathway and production of C3 fragments that opsonize apoptotic cells and mark them for clearance (11). Second, C1q on the surface of apoptotic cells likely serves as a bridging molecule, interacting with receptors on the surface of macrophages involved in the clearance of apoptotic cells (12).
One of the hallmarks of ethanol-induced liver injury is hepatocellular apoptosis; ethanol sensitizes hepatocytes to apoptosis both in vivo (13; 14) and in vitro (15; 16). Because C1q can bind to apoptotic cells and thereby activate the classical pathway, we hypothesized that ethanol feeding would activate C1, stimulate complement activation via the classical pathway and contribute to the progression of ethanol-induced liver injury. Through the use of wild-type and C1qa-/- mice, we have identified a novel interaction between ethanol-induced apoptosis of Kupffer cells, the resident macrophage in the liver, and a C1q-dependent activation of complement in the early phase of the hepatic response to ethanol exposure. C1q-deficient mice were protected from chronic ethanol-induced liver injury, indicated by a reduced accumulation of hepatic triglycerides, hepatocellular apoptosis and reduced activity of serum alanine (ALT) and aspartate (AST) amino transferase activity These data demonstrate for the first time that ethanol exposure activates the classical complement pathway and that C1q contributes to the development of ethanol-induced liver injury in mice.
Materials and Methods
Materials
Female C57BL/6 mice were purchased from Jackson Labs (Bar Harbor, Maine). C1qa-/- mice were originally developed by Marina Botto (17) and back-crossed on a C57BL/6 background (18). A breeding pair, provided to us by Dr. Michael Carroll at Center for Blood Research, Harvard University, Boston, was then used to establish a breeding colony at the Cleveland Clinic. Lieber-DeCarli high-fat ethanol liquid diet was purchased from Dyets (Bethlehem, PA). Antibodies were purchased from the following sources: CYP2E1 (Research Diagnostics, Inc. Flanders, NJ); hsc 70 (Santa Cruz Biotechnology, Inc, Santa Cruz, CA); C3b-iC3b/C3c (C3b) and C1q (Hycult Biotechnology, Uden, The Netherlands); 4-HNE (Alpha Diagnostics Intl. Inc., San Antonio, TX); caspase-generated fragment of cytokeratin-18 (M30) (Roche, Mannheim, Germany); F4/80 (Serotec, Raleigh, NC); tumor necrosis factor α (TNFα) antibody for immunohistochemistry was purchased from R&D Systems (Minneapolis, MN); TNFα and IL-6 ELISA antibody pairs (BioLegend, San Diego, CA). TUNEL was visualized using the ApopTag® plus In Situ Apoptosis Detection kit (S7111 for fluorescein staining, S7165 for rhodamine staining and S7101 for peroxidase staining). Chemicon International, Temula, CA). Malondialdehyde (MDA) was measured using a TBARS assay kit (Cayman Chemical Company, Ann Arbor, MI).
Ethanol-feeding
All procedures using animals were approved by the Cleveland Clinic Institutional Animal Care and Use Committee. 8-10 week old female C57BL/6 and C1qa-/- mice were housed 2 per cage in shoe-box cages with microisolator lids. Mice were randomized into either ethanol-fed or pair-fed groups. Both groups were allowed free access to the control liquid diet for 2 days and then the ethanol-fed group was allowed free access to increasing concentrations of ethanol in a complete liquid diet. Control mice were pair-fed diets which iso-calorically substituted maltose dextrins for ethanol over the entire feeding period. Ethanol concentrations were increased as follows: 5% ethanol (percent of total calories in the diet) for 2 days, followed by 11% for 2 days, followed by 22% for 7 days, then 27% for 7 days and finally 32% for 7 days. Consumption of the ethanol diets and body weights were not affected by genotype (Supplemental Table 1).
Ethanol- and pair-fed mice were euthanized at 4 days (4d/11% ethanol model, termed short-term ethanol) and 25 days (25d/32% ethanol model, termed chronic ethanol). At the end of the feeding trial, blood was collected in tubes containing EDTA. Plasma was collected and stored at -80°C. Livers were perfused with saline and excised. Portions of each liver were then either fixed in 10% formalin or frozen in optimal cutting temperature (OCT) compound (Sakura Finetek U.S.A., Inc., Torrance, CA) for histology, preserved in RNAlater (Qiagen, Valencia, CA) or flash frozen in liquid nitrogen and stored at -20 or -80°C until further analysis.
Acute ethanol exposure by intragastric gavage
Mice were exposed to 6g/kg of ethanol via an intragastric gavage. Plasma ethanol concentrations at 90 mins after the gavage were measured using an enzymatic assay kit (Diagnostic Chemicals, LTD, Oxford CT) and were not affected by genotype (data are provided in Supplemental Table 1).
Biochemical assays
Plasma samples were assayed for ALT and AST using commercially available enzymatic assay kits (Diagnostic Chemicals, LTD). Total liver triglycerides were measured biochemically using the Triglyceride Reagent Kit from Pointe Scientific Inc. (Lincoln Park, MI). Hepatic malondialdehyde (MDA) was measured in liver lysates using a TBARS assay kit following the manufacturer's instructions.
Immunohistochemistry
Immunohistochemical analysis of 4-HNE, TUNEL, a caspase-generated fragment of cytokeratin-18, F4/80 and TNFα was performed in liver sections, as previously described (19; 20; 6). C1q and C3b immunohistochemical analysis was performed in frozen liver sections (6). For C1q, both a slide without primary antibody and sections from C1qa-/- mice were used as negative controls. Images were semi-quantified using Image Pro software (Media Cybernetics, Inc., Bethesda, MD). Apoptosis was detected by deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining. TUNEL-positive nuclei relative to total DAPI stained nuclei were counted using Image Pro software. The caspase cleavage product of cytokeratin-18 (M30), a marker of hepatocyte apoptosis, was stained with monoclonal antibody recognizing the M30 fragment, according to the manufacturer's instructions. Three 10× magnification images were taken of each section and the number of positive cells per 10× frame counted.
Analysis of hepatic proteins
Frozen liver was homogenized as previously described (19). Protein concentrations were measured using a BCA kit (Pierce, Rockford, IL). Liver lysates were used for Western blot analysis (19) or used to measure the concentrations of TNFD and IL-6 by ELISA (6).
Statistical analysis
Values reported are means ± SEM. Data were analyzed by general linear models procedure (SAS, Carey, IN). Data were log transformed if needed to obtain a normal distribution. Follow-up comparisons were made by least square means testing. Adjustments for multiple group comparisons were made using the Tukey-Kramer test.
Results
Hepatic C1q and C3b deposition in mice after short-term ethanol
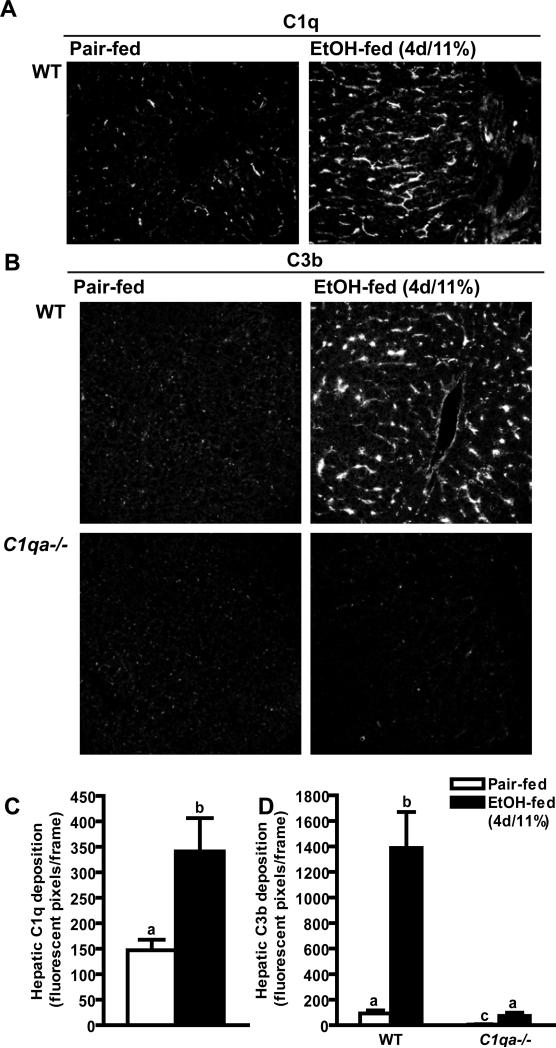
Complement activation contributes to the progression of ethanol-induced liver injury (7; 6; 5); however, the specific complement activation pathways involved in the response to ethanol are not known. (7; 6). In wild-type mice, ethanol feeding for 4 days increases the deposition of C3b/iC3b/C3c (abbreviated here to C3b for convenience) in hepatic sinusoids (6). If the classical pathway of complement activation contributes to ethanol-induced complement activation, then C1q protein would also deposit in the liver in response to ethanol. While a minimal amount of immunoreactive C1q protein was detected in liver of pair-fed wild-type mice, after short-term ethanol feeding (4d/11% ethanol), immunoreactive C1q was 2.5-fold higher when compared to pair-fed control mice (Figure 1A and C). C1q was deposited with a sinusoidal distribution (Figure 1A).
Figure 1. Hepatic C1q and C3b deposition in mice after short-term ethanol.
Wild-type and C1qa-/- mice were allowed free access to ethanol-containing diets (4d/11% ethanol) or pair-fed control diets. Immunoreactive (A) C1q and (B) C3b were visualized by immunohisotochemistry in frozen liver sections. Images are shown at 200X magnification. (C/D) The total number of fluorescent pixels per 200X field was determined using Image Pro software. Values represent means ± SEM. Pair-fed n=4, ethanol-fed n=6. Values with different superscripts are significantly different from each other, C) p<0.047, D) p<0.001.
Short-term ethanol did not increase C3b deposition in C1q-deficient mice
If activation of complement by ethanol is dependent on C1q, then ethanol-induced complement activation should be abrogated in C1q-deficient mice. Complement activation in the liver was assessed using an antibody that recognizes neo-epitopes on C3b/iC3b/C3c that are revealed after cleavage of C3 (21). C3b was not detected in the livers of pair-fed mice. Short-term ethanol feeding (4d/11% ethanol) increased immunoreactive C3b in liver of wild-type mice in a sinusoidal distribution (Figure 1B and D), but not in C1q-deficient mice (Figure 1B and D), suggesting that C1q is required for early ethanol-induced activation of complement.
Short-term ethanol increased apoptosis of Kupffer cells in both wild-type and C1q-deficient mice
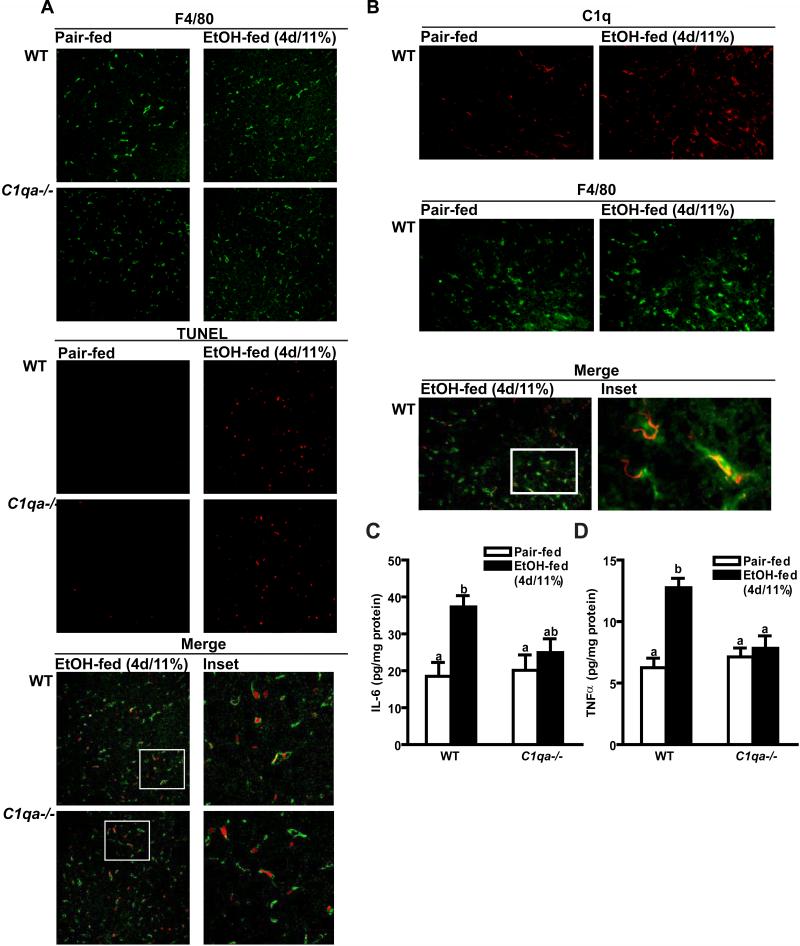
One important mechanism for activation of complement by the classical pathway is via the interaction of C1q with molecules on the surface of apoptotic cells (8). Long-term ethanol exposure, as well as exposure to high concentrations of ethanol, increases hepatocellular apoptosis; the extent of apoptosis is positively associated with severity of liver injury (13). However, it is not known whether exposure to low concentrations of ethanol, such as those used in the short-term ethanol model (4d/11% ethanol), also results in hepatocellular apoptosis. The number of TUNEL-positive nuclei increased in both wild-type and C1qa-/- mice after short-term ethanol exposure when compared to pair-fed controls. This increase in TUNEL staining after short-term ethanol feeding was not affected by genotype (Figure 2A).
Figure 2. TUNEL, F4/80 and C1q staining and inflammatory cytokine protein concentration in mouse livers after short-term ethanol feeding.
Wild-type and C1qa-/- mice were allowed free access to ethanol-containing diets (4d/11% ethanol) or pair-fed control diets. (A) TUNEL-positive nuclei and F4/80-positive cells were visualized by immunohistochemistry in formalin-fixed liver sections. (B) F4/80 and C1q-positive cells were visualized by immunohistochemistry in frozen liver sections. Images are representative of at least 4 mice per group and are shown at 200X magnification. (C/D) Livers lysates were prepared and quantity of IL-6 (C) and TNFα (D) protein was measured by ELISA. Values represent means ± SEM. Pair-fed n=4, ethanol-fed n=6. Values with different superscripts are significantly different from each other, C) p<0.001, D) p<0.003.
Hepatocytes are important targets for ethanol-induced apoptosis after long-term/high dose ethanol (13; 14). However, the quantity of M30, a caspase-generated fragment of cytokeratin-18 used as a marker of early hepatocyte apoptosis (20), was not increased after short-term ethanol feeding in either wild-type or C1q-deficient mice (data not shown), suggesting that short-term ethanol exposure targeted non-parenchymal cells for apoptosis. Because of the sinusoidal distribution of C1q and C3b, we next investigated whether Kupffer cells were undergoing apoptosis at this early period of ethanol exposure. Liver sections were co-labeled with TUNEL (Figure 2A red) and F4/80 (Figure 2A green), a marker for mouse macrophages. In both wild-type and C1q-deficient mice, TUNEL staining was almost completely restricted to F4/80-positive cells (Figure 2A merge). Further, after short term ethanol feeding, TUNEL-positive cells, visualized using DAB staining, exhibited characteristic macrophage-like morphology (Supplemental Figure 1). In contrast, when liver sections were co-labeled with TUNEL and stabilin-2, a marker for sinusoidal endothelial cells, minimal (<10%) of the TUNEL-positive cells co-localized with stabilin-2 positive cells (data not shown). C1q opsonizes apoptotic cells, marking them for clearance by phagocytosis (22). Therefore, we hypothesized that the ethanol-induced deposition of C1q in the liver would be localized primarily to macrophages. When liver sections were co-labeled with antibodies specific to C1q (Figure 2B red) and F4/80 (Figure 2B green), the localization of C1q was almost completely restricted to F4/80-positive cells (Figure 2B merge), suggesting that C1q marked apoptotic macrophages for clearance.
C1qa-/- mice were protected from short-term ethanol-induced increases in inflammatory cytokine expression
Short-term ethanol exposure increases expression of TNFα in liver of wild-type mice; this early increase is dependent on complement activation (6). Since C1q was required for complement activation in response to short-term ethanol exposure, we hypothesized that C1qa-/- mice would be protected from ethanol-induced increases in inflammatory cytokine expression. In wild-type mice, short-term ethanol exposure increased expression of hepatic IL-6 and TNFα protein by 2-3 fold over pair-fed mice (Figure 2 C/D). This early ethanol-induced increase in inflammatory cytokine expression was prevented in C1qa-/- mice (Figure 2 C/D).
Chronic ethanol-induced liver injury was attenuated in C1qa-/- mice
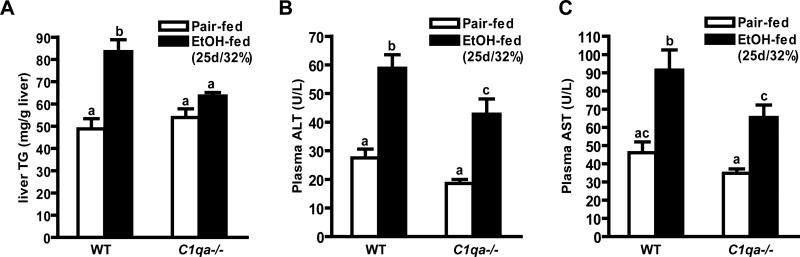
Early ethanol-induced increases in complement activation and inflammatory cytokine expression are required for the subsequent development of ethanol-induced liver injury (6). Since short-term ethanol did not activate complement or increase inflammatory cytokine expression in C1q-deficient mice, these mice should be protected from the progression of chronic ethanol-induced liver injury. To test this hypothesis, wild-type and C1qa-/- mice were allowed free access to increasing concentrations of ethanol in the diet for 25 days (25d/32% ethanol) or pair-fed control diets. Wild-type and C1qa-/- mice consumed an equivalent quantity of ethanol containing diets and body weights were not affected by genotype (Supplemental Table 1). Chronic ethanol feeding (25d/32% ethanol) increased hepatic triglycerides in wild-type mice, but not in C1qa-/- mice (Figure 3A). Chronic ethanol feeding also increased the activity of plasma ALT and AST by 2-fold in wild-type mice compared to pair-fed controls (Figure 3B and C). These increases were attenuated by 40-50% in C1qa-/- mice (Figure 3B and C).
Figure 3. Hepatic triglycerides and plasma ALT and AST activity in mice after chronic ethanol feeding.
Wild-type and C1qa-/- mice were allowed free access to ethanol-containing diets (25d/32% ethanol) or pair-fed control diets. (A) Hepatic triglyceride concentrations, and (B) ALT and (C) AST in plasma were measured. Values represent means ± SEM. Pair-fed n=8, ethanol-fed n=12. Values with different superscripts are significantly different from each other, A) p<0.005, B) p<0.03, C) p<0.04.
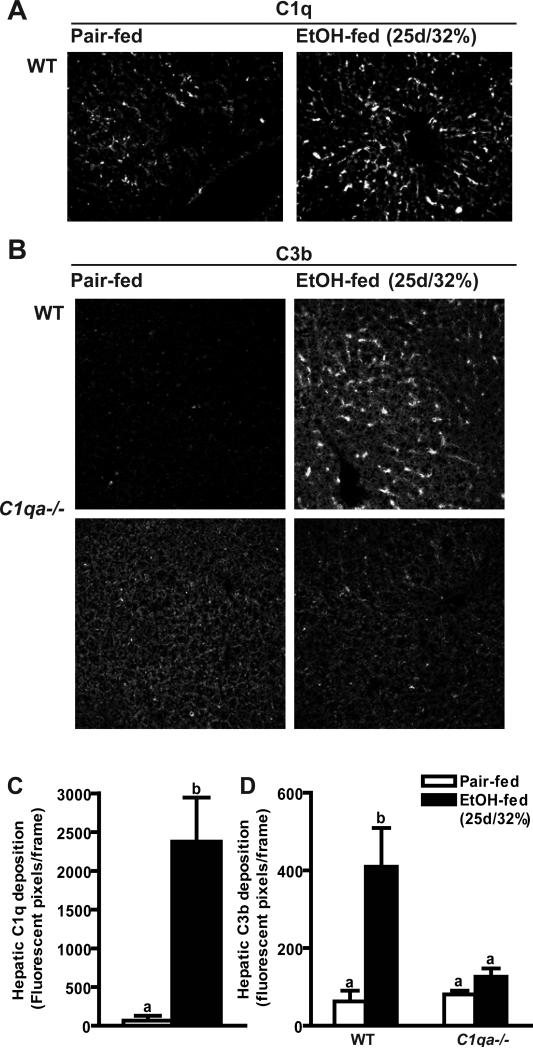
To better understand the specific mechanisms by which C1q contributes to the pathophysiology of liver injury, it was next asked whether C1q was required for complement activation after chronic ethanol exposure. Similar to the effects of short-term ethanol exposure, both C1q (Figure 4A) and C3b (Figure 4B) protein deposited in livers from wild-type mice after chronic ethanol feeding. In contrast to the sinusoidal distribution of C1q/C3b in response to short-term ethanol exposure (Figure 1), after chronic ethanol exposure, C1q/C3b deposition was shifted to the parenchyma in wild-type mice (Figure 4A and B). Deposition of C3b in response to chronic ethanol was ameliorated in C1qa-/- mice, indicating a continued role for C1q in ethanol-mediated complement activation after chronic ethanol exposure (Figure 4B and D).
Figure 4. Hepatic C1q and C3b deposition in mice after chronic ethanol feeding.
Wild-type and C1qa-/- mice were allowed free access to ethanol-containing diets (25d/32% ethanol) or pair-fed control diets. Immunoreactive (A) C1q and (B) C3b were visualized by immunohisotochemistry in frozen liver sections. (C/D) Total number of fluorescent pixels per 200X field was determined using Image Pro software. Values represent means ± SEM. Pair-fed n=4, ethanol-fed n=6. Values with different superscripts are significantly different from each other, C) p<0.001, D) p<0.05.
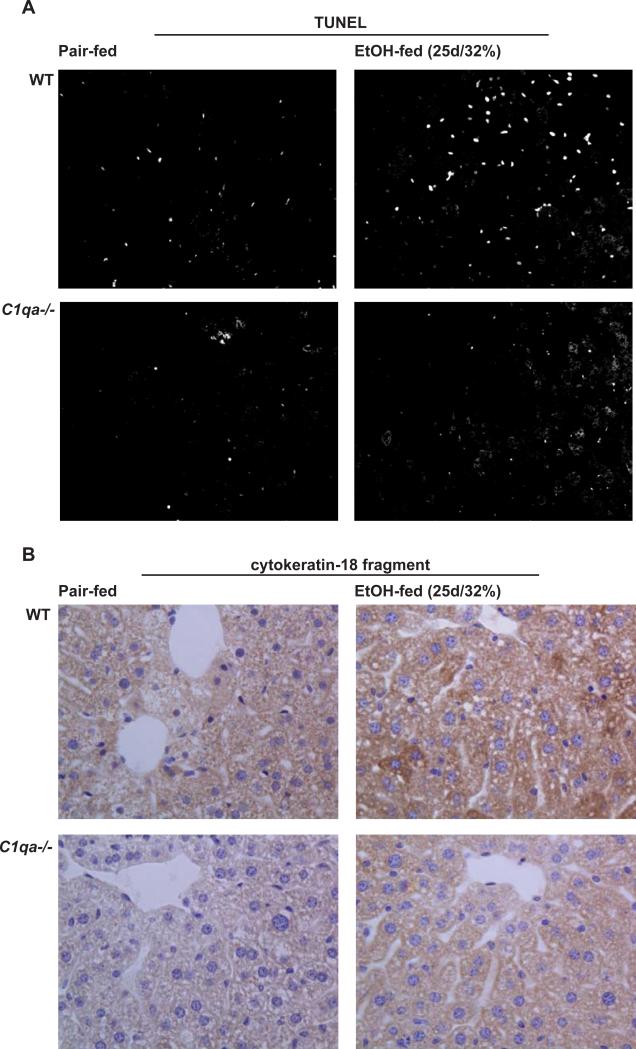
In wild-type mice, chronic ethanol feeding increased the number of TUNEL-positive nuclei compared to pair-fed controls (Figure 5A). Unlike the response to short-term ethanol feeding, the morphology of the apoptotic nuclei after chronic ethanol exposure suggested primarily hepatocyte apoptosis. Further, chronic ethanol feeding increased the quantity of M30, the caspase-generated fragment of CK-18, in wild-type mice, indicating a shift to hepatocyte apoptosis after chronic ethanol exposure (Figure 5B). Importantly, chronic ethanol-induced hepatocyte apoptosis was prevented in C1q-deficient mice (Figure 5A/B). Thus, protection from chronic ethanol-induced liver injury in the C1qa-/- mice was associated with reduced hepatocyte apoptosis.
Figure 5. Markers of apoptosis in livers of mice after chronic ethanol feeding.
Wild-type and C1qa-/- mice were allowed free access to ethanol-containing diets (25d/32% ethanol) or pair-fed control diets. (A) TUNEL-positive nuclei were visualized as in Figure 2. (B) The presence of caspase-generated cleavage fragments of cytokeratin-18 was visualized in formalin-fixed liver sections. Images were shown at 200X magnification. Red arrows point to hepatocytes staining positive for the cytokeratin-18 cleavage product. Images are representative of Pair-fed n=4, ethanol-fed n=6.
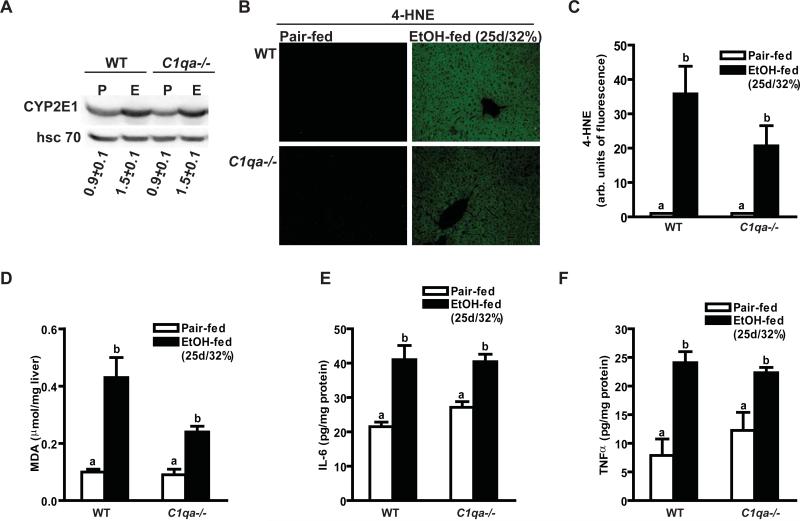
Chronic ethanol exposure increased CYP2E1 and oxidative stress in both wild-type and C1q-deficient mice
The enzyme cytochrome P450 2E1 (CYP2E1) is induced after prolonged ethanol exposure and/or exposure to high concentrations of ethanol. While short-term ethanol feeding did not increase the expression of hepatic CYP2E1 in wild-type or C1qa-/- mice (data not shown), chronic ethanol feeding induced CYP2E1 expression to an equal extent in both wild-type and C1qa-/- mice (Figure 6A). Chronic ethanol feeding also increased accumulation of oxidized lipids in the liver (Figure 6B-D). Quantity of hepatic 4-HNE, assessed by immunohistochemistry (Figure 6B/C), as well as malondialdehyde, measured by ELISA (Figure 6D), were increased after chronic ethanol feeding; this response was independent of genotype (Figure 6B-D). Another hallmark of chronic ethanol-induced liver injury is increased expression of inflammatory cytokines. Chronic ethanol exposure increased expression of hepatic IL-6 and TNFα by 2-3-fold in both wild-type and C1q-deficient mice (Figure 6D-E). Collectively, these data suggested that induction of CYP2E1, increased ROS accumulation and inflammatory cytokine expression were independent of C1q after chronic ethanol exposure.
Figure 6. Markers of oxidative stress and inflammatory cytokine expression after chronic ethanol feeding.
Wild-type and C1qa-/- mice were allowed free access to ethanol-containing diets (25d/32% ethanol) or pair-fed control diets. (A) Immunoreactive CYP2E1 protein in liver lysates was assessed by Western blot analysis. Values under the images represent arbitrary units of density for CYP2E1 relative to hsc 70, used as a loading control. (B/C) Immunoreactive 4-HNE adducts were visualized by immunohistochemistry in formalin-fixed liver sections and fluorescence intensity per 200X field determined using Image Pro. (D) MDA was measured biochemically using a TBARS assay kit. (E/F) IL-6 (E) and TNFα (F) protein in liver lysates was measured by ELISA. Values represent means ± SEM. Pair-fed n=4, ethanol-fed n=6, except in panel D where pair-fed n=8, ethanol-fed n=12. Values with different superscripts are significantly different from each other, C) p<0.02, D) p<0.006, E/F) p<0.02. .
Discussion
Activation of complement is required for the development of ethanol-induced liver injury in mice (5; 7); however, the specific pathways by which ethanol exposure activates complement have not been identified. Here we have identified for the first time an essential role of C1q in ethanol-induced complement activation after short-term/low dose ethanol (4d/11%) and chronic/high dose ethanol (25d/32%). Importantly, in the absence of C1q, mice were protected from hepatic pathology associated with both short- and long-term ethanol exposure. In the short term, C1q-deficiency decreased inflammatory cytokine expression in the liver, while after chronic ethanol exposure, the absence of C1q prevented ethanol-induced steatosis and hepatocyte apoptosis, as well as attenuated chronic ethanol-induced increases in ALT and AST.
Activation of the classical pathway occurs in response to the binding of C1q to immune complex (23). Over the past 15 years, accumulating evidence indicates that the classical pathway is also activated by apoptotic cells (8; 9; 10). C1q is an opsonin that binds to “eat me” signals, such as phosphatidylserine, on the surface of apoptotic cells, marking them for clearance by phagocytes (8). In humans with C1q deficiencies, apoptotic bodies accumulate and cause an autoimmune response (24). Ethanol feeding induces both complement activation (5; 7) and apoptosis (13); therefore, we hypothesized that ethanol would activate the classical complement pathway and C1q would contribute to complement activation in response to ethanol exposure. Here, we monitored the activation of complement in the liver by measuring the deposition of immunoreactive C1q and C3b. In wild-type mice, ethanol feeding increased C1q and C3b deposition in the hepatic sinusoids and C3b deposition did not occur in the absence of C1q. These are the first data demonstrating C1q deposition in response to ethanol feeding and suggest an important role for the classical pathway of complement activation following ethanol exposure.
C1q deposition during ethanol exposure was associated with ethanol-induced apoptosis in the liver. However, the relationship between C1q deposition and ethanol-induced apoptosis was complex and differed after short-term and chronic ethanol exposure. We first detected C1q deposition in the liver after ethanol feeding for 4 days at 11% ethanol, a very early period in the progression of liver injury. This short-term exposure is characterized by a transient increase in C3b deposition and inflammatory cytokine expression; these events precede the induction of CYP2E1, steatosis or increases in the release of liver enzymes (6; 25). Here we provide evidence that ethanol also increased the apoptosis of Kupffer cells at this early phase. The targeting of ethanol-induced apoptosis to Kupffer cells at this early phase of ethanol exposure contrasts with apoptosis of sinusoidal endothelial cells and hepatocytes observed after longer periods of ethanol exposure (4-7 weeks) (26; 27).
The complete mechanisms for early ethanol-induced apoptosis of Kupffer cells are not understood at this time; however, our data indicate that Kupffer cell apoptosis was not dependent on the expression of C1q. Instead, after short-term ethanol exposure, C1q protein was associated with Kupffer cells in the sinusoid, apparently marking the apoptotic Kupffer cells for clearance. Activation of C1q then increased the cleavage of C3 and expression of inflammatory cytokines in wild-type mice. We have previously demonstrated that early increases in hepatic TNF-α during ethanol feeding are dependent on the presence of Kupffer cells in the liver, as well as the receptors for the anaphylatoxins C3a and C5a, but independent of TLR4 (6). C1q may also directly increase inflammatory cytokine expression in Kupffer cells, since C1q can induce TNFα protein and receptor expression and up-regulate C3 in macrophages (28). However, in other studies, C1q actually mitigates TLR4-dependent cytokine expression in macrophages and monocytes (29; 30)
In contrast to the short-term response to ethanol, chronic ethanol exposure increased apoptosis of hepatocytes and C1q was required for chronic ethanol-induced hepatocyte apoptosis (Figure 5). C1q deposition in the liver exhibited a more parenchymal distribution after chronic ethanol feeding, consistent with the increase in hepatocyte apoptosis. Chronic ethanol exposure sensitizes hepatocytes to TNFα-dependent, as well as TNFα-independent, apoptosis (31;32). Thus, protection of C1q-deficient mice from chronic ethanol-induced hepatocyte apoptosis is likely due to the decrease in inflammatory cytokine production in C1qa-/- mice observed during earlier periods of ethanol exposure. Similarly, decreased expression of inflammatory cytokines in the C1qa-/- mice at early times of ethanol exposure also likely contributes to the protection of C1q-deficient mice from chronic ethanol-induced steatosis, as it is clear from previous work that inflammatory cytokines are critical to the accumulation of triglycerides in hepatocytes in response to long-term ethanol exposure (33).
While C1q-deficient mice were completely protected from chronic ethanol-induced steatosis and apoptosis, the increase in plasma ALT and AST activity, measures of hepatocyte necrosis and injury, after chronic ethanol exposure was only attenuated by 40-50%. Further, increased inflammatory cytokine expression and markers of oxidative stress, important biomarkers of liver injury, were not attenuated in C1q-deficient mice after chronic ethanol exposure. These data suggest that while C1q makes an important contribution to chronic ethanol-induced liver injury, additional pathways are also involved in mediating chronic liver disease. First, since expression of CYP2E1 was induced in both wild-type and C1qa-/- mice, it is likely that CYP2E1-dependent ROS production contributed, at least in part, to chronic ethanol-induced hepatocyte injury even in C1q-deficient mice. Interestingly, CYP2E1-dependent mechanisms did not appear to contribute to steatosis or apoptosis, as C1q-deficient mice were completely protected from chronic ethanol-induced steatosis and apoptosis. Second, after chronic ethanol exposure, increased inflammatory cytokine expression is likely due to activation of TLR4 by increases in circulating lipopolysaccharide, as well as increased expression of CYP2E1 and generation of ROS (1). Finally, it is also possible that additional pathways of complement activation may contribute to the C1q-independent component of chronic ethanol-induced liver injury. Studies are currently underway to analyze the contribution of the lectin and alternative pathways of complement activation to chronic ethanol-induced liver injury.
The modest protection of C1qa-/- mice from chronic ethanol-induced increases in hepatocyte necrosis and injury, as evidenced by the 40-50% reduction in ethanol-induced increases in ALT and AST activity in the plasma, may also be a consequence of the decreased apoptosis. There is a growing appreciation for a complex relationship between apoptosis and programmed necrosis (34), with a likely role for autophagy in the regulation of these pathways of cell death (35; 36). Because of these inter-relationships, when apoptosis is inhibited in a variety of cell types, there is an increase in necrotic cell death (35; 36). We and others have reported that inhibition of apoptosis, by treatment with recombinant thioredoxin-1, an anti-apoptotic protein (14) or pancaspase inhibitors (unpublished observations), during chronic ethanol feeding completely prevents apoptois, but only modestly decreases ALT/AST activities. Similar results have been observed in mouse models of non-alcholic steatohepatitis upon treatment with pan-caspase inhibitors (37). Therefore, future studies will be necessary to test the hypothesis that inhibition of hepatocyte apoptosis during ethanol exposure leads to increases in hepatocyte necrosis.
In conclusion, here we report for the first time that ethanol activates the classical complement pathway via C1q binding to apoptotic cells in the liver, leading to an early increase in inflammatory cytokine expression. We show that during chronic ethanol exposure, C1q contributes to the pathogenesis of ethanol-induced liver injury. These data add to our understanding of the complex dynamics of ethanol-induced liver injury. Further, since apoptosis is a common element to other forms of liver disease, including non-alcoholic fatty liver/steatosis and hepatitis C (38; 39), apoptosis-mediated activation of C1q/complement may also contribute to the progression of other forms of liver injury. Consistent with this hypothesis, Renson, et al. observed complement activation in the livers of patients with non-alcoholic fatty liver disease; the extent of complement activation was associated with the severity of liver injury (40). Taken together, these data suggest that pharmacological strategies that modulate the classical pathway of complement activation may be useful targets for therapeutic intervention in ALD, as well as other forms of liver injury.
Supplementary Material
Supplemental Table 1: Characteristics of ethanol- and pair-fed wild-type and C1qa-/- mice. Pair-fed n=4, ethanol-fed n=6. No statistical differences were found between genotypes, p>0.05.
Supplemental Figure 1: TUNEL-positive cells in mouse livers after short-term ethanol feeding. Wild-type mice were allowed free access to ethanol-containing diets (4d/11% ethanol) or pair-fed control diets. TUNEL-positive nuclei were visualized by immunohistochemistry in formalin-fixed liver sections with peroxidase labelling. Slides were counterstained with hematoxylin. Images are representative of at least 3 mice per group. Areas in the red boxes are enlarged. Red arrows point to cells with macrophage-like morphology that are TUNEL-positive and white arrows point to cells with macrophage-like morphology that are TUNEL-negative.
Acknowledgements
We would like to thank Drs Marino Botto and Michael Carroll for sharing the C1qa-/- mice. We would like to thank Emmanuelle Ogier and Brian T. Pratt for the outstanding technical assistance and Dr. Michele T. Pritchard for critical discussions of the data and manuscript.
Grant support: This work was supported in part by RO1AA016399 to LEN and 1 F31 AA016434 to JIC.
Abbreviations
- 4-HNE
4-hydroxynonenal
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- C3b
C3b/iC3b/C3c
- CYP2E1
cytochrome P450 2E1
- MDA
malondialdehyde
- ROS
reactive oxygen species
- TUNEL
Terminal Deoxynucleotidyl Transferase Mediated dUTP Nick End Labeling
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: The authors have nothing to disclose.
Author contributions:
Jessica I. Cohen: study concept and design; acquisition of data; analysis and interpretation of data; drafting of the manuscript; statistical analysis;
Sanjoy Roychowdhury: acquisition of data; analysis and interpretation of data; statistical analysis
Megan R. McMullen: acquisition of data; analysis and interpretation of data; study supervision
Abram B. Stavitsky: study concept and design; critical revision of the manuscript for important intellectual content.
Laura E. Nagy: study concept and design; analysis and interpretation of data; drafting of the manuscript; critical revision of the manuscript for important intellectual content; statistical analysis; obtained funding; study supervision
Reference List
- 1.Vidali M, Hietala J, Occhino G, et al. Immune responses against oxidative stress-derived antigens are associated with increased circulating tumor necrosis factor-alpha in heavy drinkers. Free Radic Biol Med. 2008;45:306–11. doi: 10.1016/j.freeradbiomed.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 2.Albano E, Vidali M. Immune mechanisms in alcoholic liver disease. Genes Nutr. 2009 doi: 10.1007/s12263-009-0151-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gasque P. Complement: a unique innate immune sensor for danger signals. Mol Immunol. 2004;41:1089–98. doi: 10.1016/j.molimm.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Jarvelainen HA, Vakeva A, Lindros KO, et al. Activation of complement components and reduced regulator expression in alcohol-induced liver injury in the rat. Clin Immunol. 2002;105:57–63. doi: 10.1006/clim.2002.5267. [DOI] [PubMed] [Google Scholar]
- 5.Pritchard MT, McMullen MR, Stavitsky AB, et al. Differential contributions of C3, C5, and decay-accelerating factor to ethanol-induced fatty liver in mice. Gastroenterology. 2007;132:1117–26. doi: 10.1053/j.gastro.2007.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roychowdhury S, McMullen MR, Pritchard MT, et al. An early complement-dependent and TLR-4-independent phase in the pathogenesis of ethanol-induced liver injury in mice. Hepatology. 2009;49:1326–34. doi: 10.1002/hep.22776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bykov I, Junnikkala S, Pekna M, et al. Complement C3 contributes to ethanol-induced liver steatosis in mice. Ann Med. 2006;38:280–6. doi: 10.1080/07853890600664608. [DOI] [PubMed] [Google Scholar]
- 8.Paidassi H, Tacnet-Delorme P, Garlatti V, et al. C1q binds phosphatidylserine and likely acts as a multiligand-bridging molecule in apoptotic cell recognition. J Immunol. 2008;180:2329–38. doi: 10.4049/jimmunol.180.4.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korb LC, Ahearn JM. C1q binds directly and specifically to surface blebs of apoptotic human keratinocytes: complement deficiency and systemic lupus erythematosus revisited. J Immunol. 1997;158:4525–8. [PubMed] [Google Scholar]
- 10.Elward K, Griffiths M, Mizuno M, et al. CD46 plays a key role in tailoring innate immune recognition of apoptotic and necrotic cells. J Biol Chem. 2005;280:36342–54. doi: 10.1074/jbc.M506579200. [DOI] [PubMed] [Google Scholar]
- 11.Mevorach D, Mascarenhas JO, Gershov D, et al. Complement-dependent clearance of apoptotic cells by human macrophages. J Exp Med. 1998;188:2313–20. doi: 10.1084/jem.188.12.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trouw LA, Blom AM, Gasque P. Role of complement and complement regulators in the removal of apoptotic cells. Mol Immunol. 2008;45:1199–207. doi: 10.1016/j.molimm.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Higuchi H, Kurose I, Kato S, et al. Ethanol-induced apoptosis and oxidative stress in hepatocytes. Alcohol Clin Exp Res. 1996;20:340A–346A. [PubMed] [Google Scholar]
- 14.Cohen JI, Roychowdhury S, Dibello PM, et al. Exogenous thioredoxin prevents ethanol-induced oxidative damage and apoptosis in mouse liver. Hepatology. 2009;49:1709–1717. doi: 10.1002/hep.22837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pastorino JG, Shulga N, Hoek JB. TNF-alpha-induced cell death in ethanol-exposed cells depends on p38 MAPK signaling but is independent of Bid and caspase-8. Am J Physiol Gastrointest Liver Physiol. 2003;285:G503–16. doi: 10.1152/ajpgi.00442.2002. [DOI] [PubMed] [Google Scholar]
- 16.McVicker BL, Tuma DJ, Kubik JL, et al. Ethanol-induced apoptosis in polarized hepatic cells possibly through regulation of the Fas pathway. Alcohol Clin Exp Res. 2006;30:1906–15. doi: 10.1111/j.1530-0277.2006.00235.x. [DOI] [PubMed] [Google Scholar]
- 17.Botto M, Dell'Agnola C, Bygrave AE, et al. Homozygous C1q deficiency causes glomerulonephritis associated with multiple apoptotic bodies. Nat Genet. 1998;19:56–9. doi: 10.1038/ng0598-56. [DOI] [PubMed] [Google Scholar]
- 18.Taylor PR, Carugati A, Fadok VA, et al. A hierarchical role for classical pathway complement proteins in the clearance of apoptotic cells in vivo. J Exp Med. 2000;192:359–66. doi: 10.1084/jem.192.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pritchard MT, Roychowdhury S, McMullen MR, et al. Early growth response-1 contributes to galactosamine/lipopolysaccharide-induced acute liver injury in mice. Am J Physiol Gastrointest Liver Physiol. 2007;293:G1124–33. doi: 10.1152/ajpgi.00325.2007. [DOI] [PubMed] [Google Scholar]
- 20.Wieckowska A, Zein NN, Yerian LM, et al. In vivo assessment of liver cell apoptosis as a novel biomarker of disease severity in nonalcoholic fatty liver disease. Hepatology. 2006;44:27–33. doi: 10.1002/hep.21223. [DOI] [PubMed] [Google Scholar]
- 21.Mastellos D, Prechl J, Laszlo G, et al. Novel monoclonal antibodies against mouse C3 interfering with complement activation: description of fine specificity and applications to various immunoassays. Mol Immunol. 2004;40:1213–21. doi: 10.1016/j.molimm.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 22.Lu JH, Teh BK, Wang L, et al. The classical and regulatory functions of C1q in immunity and autoimmunity. Cell Mol Immunol. 2008;5:9–21. doi: 10.1038/cmi.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kishore U, Ghai R, Greenhough TJ, et al. Structural and functional anatomy of the globular domain of complement protein C1q. Immunol Lett. 2004;95:113–28. doi: 10.1016/j.imlet.2004.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Botto M, Walport MJ. C1q, autoimmunity and apoptosis. Immunobiology. 2002;205:395–406. doi: 10.1078/0171-2985-00141. [DOI] [PubMed] [Google Scholar]
- 25.Roychowdhury S, McMullen MR, Pritchard MT, et al. Formation of gamma-ketoaldehyde-protein adducts during ethanol-induced liver injury in mice. Free Radic Biol Med. 2009;47:1526–38. doi: 10.1016/j.freeradbiomed.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deaciuc IV, Fortunato F, D'Souza NB, Hill DB, McClain CJ. Chronic alcohol exposure of rats exacerbates apoptosis in hepatocytes and sinusoidal endothelial cells. Hepatol Res. 2001;19:306–324. doi: 10.1016/s1386-6346(00)00112-1. [DOI] [PubMed] [Google Scholar]
- 27.Miller AM, Wang H, Park O, Horiguchi N, Lafdil F, Mukhopadhyay P, Moh A, Fu XY, Kunos G, Pacher P, Gao B. Anti-Inflammatory and Anti-Apoptotic Roles of Endothelial Cell STAT3 in Alcoholic Liver Injury. Alcohol Clin Exp Res. 2010 doi: 10.1111/j.1530-0277.2009.01141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bajtay Z, Jozsi M, Banki Z, et al. Mannan-binding lectin and C1q bind to distinct structures and exert differential effects on macrophages. Eur J Immunol. 2000;30:1706–13. doi: 10.1002/1521-4141(200006)30:6<1706::AID-IMMU1706>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 29.Yamada M, Oritani K, Kaisho T, et al. Complement C1q regulates LPS-induced cytokine production in bone marrow-derived dendritic cells. Eur J Immunol. 2004;34:221–30. doi: 10.1002/eji.200324026. [DOI] [PubMed] [Google Scholar]
- 30.Fraser DA, Arora M, Bohlson SS, et al. Generation of inhibitory NFkappaB complexes and phosphorylated cAMP response element-binding protein correlates with the anti-inflammatory activity of complement protein C1q in human monocytes. J Biol Chem. 2007;282:7360–7. doi: 10.1074/jbc.M605741200. [DOI] [PubMed] [Google Scholar]
- 31.Hoek JB, Pastorino JG. Ethanol, oxidative stress, and cytokine-induced liver cell injury. Alcohol. 2002;27:63–8. doi: 10.1016/s0741-8329(02)00215-x. [DOI] [PubMed] [Google Scholar]
- 32.Ziol M, Tepper M, Lohez M, et al. Clinical and biological relevance of hepatocyte apoptosis in alcoholic hepatitis. J Hepatol. 2001;34:254–60. doi: 10.1016/s0168-8278(00)00047-7. [DOI] [PubMed] [Google Scholar]
- 33.Hines IN, Wheeler MD. Recent advances in alcoholic liver disease III. Role of the innate immune response in alcoholic hepatitis. Am J Physiol Gastrointest Liver Physiol. 2004;287:G310–4. doi: 10.1152/ajpgi.00094.2004. [DOI] [PubMed] [Google Scholar]
- 34.Galluzzi L, Kroemer G. Necroptosis: a specialized pathway of programmed necrosis. Cell. 2008;135:1161–3. doi: 10.1016/j.cell.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 35.Ullman E, Fan Y, Stawowczyk M, Chen HM, Yue Z, Zong WX. Autophagy promotes necrosis in apoptosis-deficient cells in response to ER stress. Cell Death Differ. 2008;15:422–5. doi: 10.1038/sj.cdd.4402234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White E. Autophagic cell death unraveled: Pharmacological inhibition of apoptosis and autophagy enables necrosis. Autophagy. 2008;4:399–401. doi: 10.4161/auto.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Witek RP, Stone WC, Karaca FG, Syn WK, Pereira TA, Agboola KM, Omenetti A, Jung Y, Teaberry V, Choi SS, Guy CD, Pollard J, Charlton P, Diehl AM. Pan-caspase inhibitor VX-166 reduces fibrosis in an animal model of nonalcoholic steatohepatitis. Hepatology. 2009;50:1421–30. doi: 10.1002/hep.23167. [DOI] [PubMed] [Google Scholar]
- 38.Malhi H, Gores GJ. Cellular and molecular mechanisms of liver injury. Gastroenterology. 2008;134:1641–54. doi: 10.1053/j.gastro.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feldstein AE, Gores GJ. Apoptosis in alcoholic and nonalcoholic steatohepatitis. Front Biosci. 2005;10:3093–9. doi: 10.2741/1765. [DOI] [PubMed] [Google Scholar]
- 40.Rensen SS, Slaats Y, Driessen A, Peutz-Kootstra CJ, Nijhuis J, Steffensen R, Greve JW, Buurman WA. Activation of the complement system in human nonalcoholic fatty liver disease. Hepatology. 2009;50:1809–17. doi: 10.1002/hep.23228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1: Characteristics of ethanol- and pair-fed wild-type and C1qa-/- mice. Pair-fed n=4, ethanol-fed n=6. No statistical differences were found between genotypes, p>0.05.
Supplemental Figure 1: TUNEL-positive cells in mouse livers after short-term ethanol feeding. Wild-type mice were allowed free access to ethanol-containing diets (4d/11% ethanol) or pair-fed control diets. TUNEL-positive nuclei were visualized by immunohistochemistry in formalin-fixed liver sections with peroxidase labelling. Slides were counterstained with hematoxylin. Images are representative of at least 3 mice per group. Areas in the red boxes are enlarged. Red arrows point to cells with macrophage-like morphology that are TUNEL-positive and white arrows point to cells with macrophage-like morphology that are TUNEL-negative.