Abstract
Setd8/PR-Set7/KMT5a-dependent mono-methylation of histone H4 at lysine 20 is essential for mitosis of cultured cells; yet, the functional roles of Setd8 in complex mammalian tissues are unknown. We use skin as a model system to explore how Setd8 may regulate cell division in vivo. Deletion of Setd8 in undifferentiated layers of the mouse epidermis impaired both proliferation and differentiation processes. Long-lived epidermal progenitor cells are lost in the absence of Setd8, leading to an irreversible loss of sebaceous glands and interfollicular epidermis. We show that Setd8 is a transcriptional target of c-Myc and an essential mediator of Myc-induced epidermal differentiation. Deletion of Setd8 in c-Myc-overexpressing skin blocks proliferation and differentiation and causes apoptosis. Increased apoptosis may be explained by our discovery that p63, an essential transcription factor for epidermal commitment is lost, while p53 is gained upon removal of Setd8. Both overexpression of p63 and deletion of p53 rescue Setd8-induced apoptosis. Thus, Setd8 is a crucial inhibitor of apoptosis in skin and its activity is essential for epidermal stem cell survival, proliferation and differentiation.
Keywords: c-Myc, Setd8/PR-Set7/Set8/KMT5a, skin, stem cells
Introduction
Epigenetic modifications, such as DNA methylation and post-translational modifications of histones, play an important role in chromatin structure and promoter activity, and have been implicated in a wide range of biological processes including development, reprogramming, aging and cancer (Fraga and Esteller, 2007; Richly et al, 2010; Shafa et al, 2010). Although the functional roles of histone modifications, for instance methylation, have been extensively studied during embryonic stem cell differentiation and reprogramming, little is known about the importance of histone methylation in multipotent stem cells of adult mammalian tissues.
Skin is the best-characterized mammalian tissue containing epithelial stem cells, and thus offers an ideal model to study the functional roles of histone methylation in vivo. The epidermis is a stratified epithelium that forms the outermost protective layer of the skin and comprises the interfollicular epidermis (IFE) and its appendages, including hair follicles (HFs) and sebaceous glands (SGs), all of which are maintained by resident skin stem cell pools (Blanpain and Fuchs, 2009; Watt and Jensen, 2009). During normal homeostasis, IFE and SGs are continuously regenerated and their overall proliferation rate is comparatively low. In contrast, HFs undergo cycles of growth, and their proliferation increases periodically in stages of anagen. Anagen replenishes terminally differentiated cells of the HF, and is followed by phases of regression (catagen) and rest (telogen) (Fuchs, 2009).
Recent studies in skin showed that epigenetic factors controlling histone H3 methylation are involved in regulating epidermal stem cell survival, proliferation and differentiation (Sen et al, 2008; Eckert et al, 2011; Ezhkova et al, 2011). Epidermal stem cells have also been shown to undergo global changes in histone modifications during differentiation, and one such modification is the mono-methylation of histone H4 at lysine 20 (H4K20me1) (Frye et al, 2007). Although global changes in H4K20 methylation are a common hallmark of cancer (Fraga et al, 2005), the functional roles of methylated H4K20 in normal adult mammalian tissue are unknown.
In mammalian cells, Setd8/PR-Set7/KMT5a is the sole enzyme required to catalyse the formation of H4K20me1 (Xiao et al, 2005). Embryonic deletion of Setd8 is lethal in flies and mice (Nishioka et al, 2002; Karachentsev et al, 2005; Oda et al, 2009). Thus, most functional studies on Setd8 have been carried out in vitro. Conditional deletion of Setd8 in embryonic stem cells results in cell-cycle arrest, DNA damage and genomic instability (Oda et al, 2009). Knockout approaches in transformed cells confirm an essential function of Setd8 in cell-cycle progression and replication (Jorgensen et al, 2007; Shi et al, 2007; Tardat et al, 2007, 2010; Houston et al, 2008; Huen et al, 2008). Both, levels of Setd8 and deposition of H4K20me1, are cell-cycle dependent and highest at G2/M phase of the cell cycle (Abbas et al, 2010; Centore et al, 2010; Oda et al, 2010). Unlike other epigenetic regulators, Setd8 is associated with mitotic chromosomes during cell division, which may represent a mechanism by which the H4K20-methyl mark is epigenetically transmitted (Rice et al, 2002). Although Setd8 is clearly essential for cellular survival in the early embryo and cultured cells, its function in less proliferative environments such as adult tissues is unknown.
Here, we demonstrate for the first time that Setd8 is required for normal tissue homeostasis, in vivo. Inducible, conditional deletion of Setd8 in the undifferentiated layers of epidermis results in cell-cycle arrest and apoptosis of long-lived progenitor cells leading to irreversible loss of IFE and SGs. Setd8 is a target gene of c-Myc and required for Myc-induced epidermal differentiation. Setd8-depleted epidermal cells fail to express p63 but gain p53, and thus exhibit an impaired terminal differentiation programme and undergo apoptosis instead.
Results
Setd8 is weakly expressed in skin but upregulated with proliferation
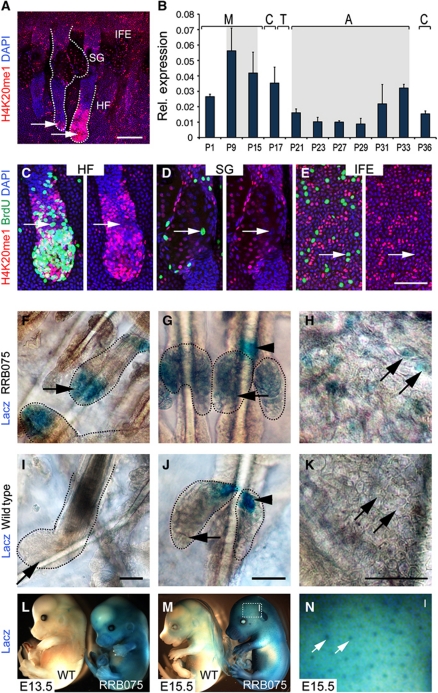
The histone methyltransferase Setd8 is specifically responsible for the mono-methylation of histone 4 at lysine 20 (H4K20me1). In skin, nuclei with high levels of H4K20me1 can be found in the basal undifferentiated layer of the IFE, the SG and in the growing anagen HF (Figure 1A; Frye et al, 2007). The accumulation of H4K20me1-positive nuclei in the bulb of HFs (Figure 1A, arrows) suggested that Setd8 activity might be highest in dividing skin progenitor cells. To confirm that Setd8 expression correlated with proliferation, we performed quantitative RT–PCR (QPCR) in skin after birth and during the first synchronized hair cycle. During morphogenesis (M), expression of Setd8 was highest at P9, when HFs are in anagen (Figure 1B). In adult skin, the first synchronized hair cycle begins with anagen (A) at P21. Setd8 RNA levels gradually increased from P21 until P33 and dropped at P36, when the destructive phase (catagen; C) of the hair cycle begins (Figure 1B).
Figure 1.
Endogenous expression of Setd8 in skin correlates with proliferation. (A) Detection of H4K20me1 (red)-positive nuclei in the interfollicular epidermis (IFE), sebaceous glands (SGs) and hair follicles (HFs) in a skin whole mount. Arrows indicate anagen hair follicles. (B) QPCR for Setd8 RNA during morphogenesis (M) and the first adult hair cycle of catagen (C), telogen (T) and anagen (A) at indicated postnatal days (P). Grey indicates anagen stages. (C–E) Co-labelling for H4K20me1-positive nuclei (red) with BrdU (green) in HF (C), SG (D) and IFE (E). Arrows indicate BrdU-positive nuclei lacking H4K20me1. Nuclei are counterstained with DAPI (blue) (A–E). (F–K) Detection of β-galactosidase in wild-type and LacZ-reporter mice (RRB075). Arrows indicate the HF (F, I), SG (G, J), IFE (H, K) and arrowhead marks unspecific staining in SG (G, J). (L–N) Whole mount LacZ staining of wild-type (wt) and RRB075 (RB) mouse embryos at indicated embryonic days (E). (N) Higher magnification of boxed area in (M). Arrow indicates hair follicles. Scale bars: 100 μm (A, F, G, I, J); 50 μm (C–E, H, K).
To investigate whether H4K20me1 generally marked dividing cells, we labelled mouse skin with BrdU and co-stained the nuclei for H4K20me1 (Figure 1C–E). BrdU labelling requires cells to be in S phase at the time of pulse; and we found that BrdU and H4K20me1 labelling was mutually exclusive in the HF, SGs and IFE (Figure 1C–E; arrows). Thus, in line with recent studies showing that Setd8 protein is degraded in S phase (Oda et al, 2010), H4K20me1 was also absent in S phase of the cell cycle. However, labelling for H4K20me1 and BrdU overlapped in the region of the bulb of anagen HFs where committed progenitor cells reside (Figure 1C).
Detection of endogenous Setd8 protein in tissues is hampered by the lack of suitable antibodies. To localize Setd8 in vivo, we generated a reporter mouse carrying the β-galactosidase gene as a GeneTrap in intron 3 (RRB075) (Huen et al, 2008). Only in RRB075 mice, we detected high levels of β-galactosidase in the bulb of anagen HFs (Figure 1F and I). Whereas the base of the SGs stained unspecific for LacZ in wild-type and RRB075 mice (Figure 1G and J; arrowheads), the lower part of the SGs exhibited β-galactosidase activity only in RRB075 mice (Figure 1G and J; arrows). Expression of LacZ was weak in the IFE but we detected a patchy β-galactosidase activity in the reporter mice (Figure 1H and K; arrows). Staining for LacZ during late embryonic development at E13.5 and E15.5 demonstrated that Setd8 was highly expressed throughout the developing epidermis and HFs (Figure 1L–N).
In conclusion, we found a widespread but weak expression of Setd8 in SGs, IFE and the HF that correlated well with the occurrence of H4K20me1-positive nuclei, and increased with proliferative phases of the skin.
Skin cannot develop or be maintained in the absence of Setd8
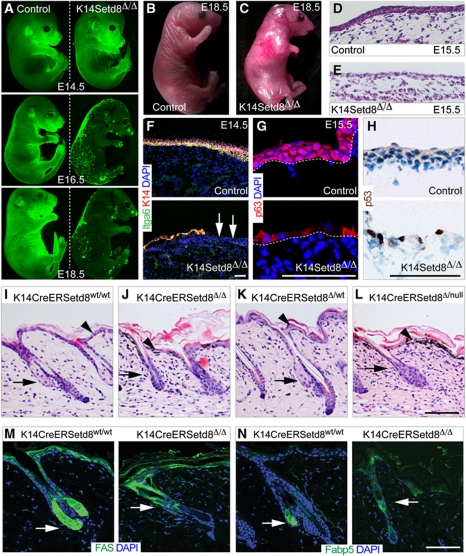
The expression pattern of Setd8 during morphogenesis indicated that Setd8 might be required for skin development. To test this hypothesis, we conditionally deleted Setd8 in the basal, undifferentiated layers of the developing epidermis (K14Setd8Δ/Δ) (Materials and methods; Supplementary Figure S1A). Mice with deleted Setd8 from E14.5, when the keratin 14 (K14) promoter is active died shortly after birth. To follow the fate of Setd8-depleted epidermal cells during development, we crossed the K14Setd8Δ/Δ mice with a green fluorescent protein (GFP)-reporter line for Cre-recombinase (Materials and methods; Figure 2A; Kawamoto et al, 2000). As soon as Setd8 was deleted at E14.5, we noted the disappearance of GFP-positive epidermal cells (Figure 2A). Further analyses of Setd8-depleted embryos at E18.5 showed that limb development was impaired and the skin was indeed absent (Figure 2B and C). Histological analysis of section obtained from embryos at E15.5 confirmed the lack of a developing epidermis (Figure 2D and E). When we labelled embryonic skin for markers of undifferentiated epidermis, we found that some areas in E14.5 embryos still had a single layer of epithelial cells, which was largely lost at E15.5 (Figure 2F and G; Supplementary Figure S1B and C). In rare cases, we found single epidermal cells at E15.5 in K14Setd8Δ/Δ mice, these cells lacked nuclear p63 but expressed keratin 8 (K8) (Figure 2G; Supplementary Figure S1B and C). Since, we detected p53-positive cells in Setd8-depleted epidermis at E15.5 (Figure 2H), we concluded that loss of Setd8 caused apoptosis rather than inhibiting cell differentiation.
Figure 2.
Embryonic and adult skin is disrupted in the absence of Setd8. (A) Visualization of GFP as a reporter for Cre-activity in control (left hand panels) and K14CreSetd8Δ/Δ (right hand panels) embryos at the indicated embryonic days (E). (B, C) Mouse embryos at E18.5 with normal expression of Setd8 (control) (B) and deleted Setd8 (K14CreSetd8Δ/Δ) in skin (C). (D, E) Haematoxylin & Eosin staining of skin sections from control (D) and K14CreSetd8Δ/Δ (E) littermates at E15.5. (F–H) Skin sections of control (upper panels) and K14CreSetd8Δ/Δ (lower panels) littermates at E14.5 (F) and E15.5 (G, H) labelled with antibodies to Itga6 (green) and keratin 14 (K14; red) (F) or p63 (red) (G) or p53 (brown) (H). Arrows in (F) indicate the loss of an epithelial cell layers starting from E14.5. (I–L) Histological sections of adult skin expressing inducible Cre-recombinase under control of the K14 promoter (K14CreER) crossed with wild-type mice (Setd8wt/wt) (I), or animals heterozygous (Setd8Δ/wt) (K), homozygous (Setd8Δ/Δ) (J) for the floxed Setd8 allele, or mice carrying one floxed and one null allele for Setd8 (Setd8Δ/null) (L). (M, N) Labelling for FAS and Fabp5 (green) in the presence (K14CreERSetd8wt/wt) (left hand panels) or absence of Setd8 (K14CreERSetd8Δ/Δ) (right hand panels). Nuclei are counterstained with DAPI (blue) in (F, G, M, N). Arrows in (I–N) indicate sebaceous glands and arrowheads (I–L) point to IFE. Scale bars: 100 μm (F–N).
Because the deletion of Setd8 during development was lethal, we generated an inducible conditional knockout mouse model for Setd8 in skin (K14CreERSetd8Δ/Δ) (Materials and methods). Setd8 RNA expression decreased (Supplementary Figure S1D) and mono-methylation of H4K20 was lost when Setd8 was deleted (Supplementary Figure S1E and F). To confirm that Cre-recombinase was uniformly activated in the epidermis we crossed the K14CreER mice to a GFP-reporter mouse (Supplementary Figure S2A and B; Kawamoto et al, 2000). Whereas we detected homogenous Cre-recombinase activity in the IFE and SGs, the HFs showed only a patchy staining for GFP (Supplementary Figure S2A). A time course of treatment with 4-hydroxytmaoxifen (4-OHT) on the GFP-reporter mice further revealed that efficient recombination was only achieved by 9 days in tail skin (Supplementary Figure S2B), we therefore treated the mice a minimum of 14 days with 4-OHT.
Control mice carrying the wild-type Setd8 alleles or being heterozygous for Setd8 deletion did not show any phenotype when treated with 4-OHT (Figure 2I and K). In contrast, when both alleles of Setd8 were deleted, either by breeding the mice to homozygosity for the floxed Setd8 alleles or by crossing the floxed mice with mice carrying one deleted allele (null) for Setd8 the epidermis was severely disrupted. The cellularity of the IFE was reduced (Figure 2I–L; arrowheads). Treatment of the tissue sections with DOPA showed that the black granules in the IFE of transgenic mice were melanin (Figure 2L, arrowhead; data not shown), indicating that melanocytes were recruited to damaged areas of the skin. Furthermore, SGs (arrows) were absent when Setd8 was deleted (Figure 2J and L). We confirmed the lack of functional SGs by labelling sections of back skin for fatty acid synthase (FAS) and the fatty acid binding protein (Fabp5) (Figure 2M and N; arrows), which are prominent markers for SGs (Collins and Watt, 2008; Lo Celso et al, 2008).
The morphology of HFs was not affected by deletion of Setd8 (Figure 2I–L). However, GFP-reporter mice demonstrated that Setd8 might not be efficiently deleted in the HFs (Supplementary Figure S2A and B) and we therefore focused our further studies on the IFE and SGs.
Proliferation and differentiation are impaired by loss of Setd8
Since epidermal morphology varies at different body sites we chose to measure the effect of Setd8 deletion on mouse tail and back skin. We first confirmed by whole mount staining for keratin 14 (K14) that deletion of Setd8 in tail skin resulted in the same phenotype as back skin; SGs were degenerated or lost and the integrity of the IFE was severely impaired (Supplementary Figure S3A and B; arrows).
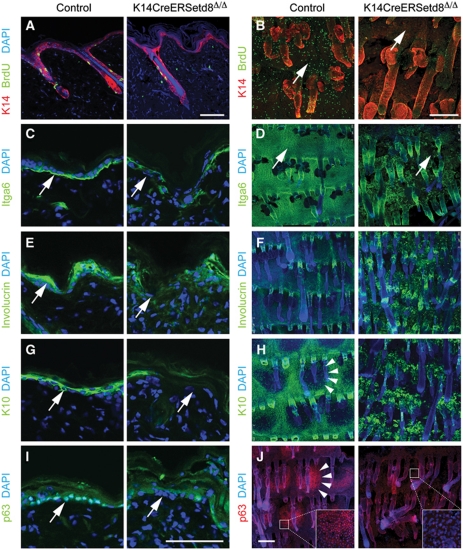
We next investigated markers for the IFE in back and tail skin (Figure 3). We began by testing the proliferative capacity of cells in the IFE. BrdU incorporation was four-fold reduced in the IFE in back and tail skin in the absence of Setd8 (Figure 3A and B, arrows; Supplementary Figure S3C). Itga6, a marker for basal, undifferentiated epidermal cells, was reduced or lost in some areas of the IFE when Setd8 was ablated (Figure 3C and D; arrows). Deposition of collagen IV remained unchanged when Setd8 was deleted (Supplementary Figure S3D and E). GFP-reporter mice (Materials and methods) confirmed that Setd8-deleted epidermis exhibited lower expression of Itga6 and incorporated BrdU to a lesser extent (Supplementary Figure S4A and B).
Figure 3.
Deletion of Setd8 impairs epidermal proliferation and differentiation. (A–J) Immunofluorescence labelling for K14 (red) and BrdU (green) (A, B) and Itga6 (green) (C, D) as markers for undifferentiated skin, Involucrin (green) (E, F) and K10 (green) (G, H) as markers for differentiated skin, and p63 (green) (I) red (J) in sections (A, C, E, G, I) and whole mounts (B, D, F, H, J) from control and K14CreERSetd8Δ/Δ mice. Arrows in (B–E, G, I) point to IFE and arrowheads in (H, J) indicate a parakeratotic scale. Inserts in (J) are a higher magnification of the IFE. Nuclei are counterstained with DAPI. Scale bars: 100 μm (A, C, E, G, I) and 250 μm (B, D, F, H, J).
To test whether the low number of proliferating cells in Setd8-depleted skin impaired differentiation processes, we labelled skin for the terminal differentiation markers keratin 10 (K10) and Involucrin (Ivl) (Figure 3E–H). Similarly to Itga6, we detected areas with disrupted expression of both differentiation markers in the back skin (Figure 3E and G). A mis-localization of differentiation markers was even more apparent in tail skin (Figure 3F and H). Skin of the mouse tail exhibits a regular, ordered pattern of parakeratotic scale epidermis that alternates with orthokeratotic interscale epidermis (Schweizer et al, 1987). Parakeratotic scales, which are negative for K10 but express high levels of p63 in wild-type mice (Figure 3H and J; left hand panels; arrowheads), were lost when S was depleted (Figure 3H and J; right hand panels). Since undifferentiated and proliferating cell populations were compromised in the epidermis, the reduction of expression of differentiation markers might be an indirect effect of Setd8 deletion.
The transcription factor p63 is one of the most important regulators of skin homeostasis and, similar to conditional deletion of Setd8 in E18.5 embryos (Figure 2A–H), embryos lacking p63 fail to undergo epidermal commitment (Koster, 2010). We asked whether impaired proliferation and differentiation processes in the IFE might be due to altered p63 levels. In the absence of Setd8, p63-positive nuclei were lost in the IFE of both back and tail epidermis (Figure 3I, arrows; Figure 3J, insert). QPCR confirmed that p63 RNA levels decreased in Setd8-ablated skin (Supplementary Figure S4C). We concluded that Setd8's essential function for epidermal stratification might at least in part be mediated by p63.
Stem/progenitor cells of IFE and SGs are lost in Setd8-depleted skin
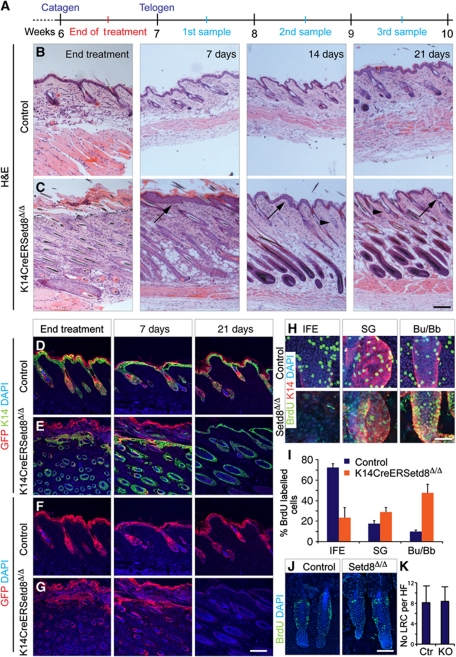
The depletion of SGs and IFE in the absence of Setd8 indicated that epidermal stem cells might require Setd8 for cell division and survival. We speculated that if ablation of Setd8 eliminated resident stem cell pools, the phenotype should be irreversible and SGs and IFE should not regenerate, even when we stopped treatment with 4-OHT. To test our hypothesis, we treated the back skin of K14CreERSetd8Δ/Δ and control mice with 4-OHT for 14 days and let the mice recover for 3 weeks. Skin samples were taken at four time points: the end of the treatment and after 1, 2 and 3 weeks of recovery (Figure 4A).
Figure 4.
Long-lived progenitors of IFE and sebaceous glands are irreversibly lost in K14CreERSetd8Δ/Δ mice and recovered skin derives from hair follicles (HFs). (A) Schematic overview of the 4-OHT-treatment regime. (B, C) Haematoxylin & Eosin (H&E) staining of skin section from control (B) and K14CreERSetd8Δ/Δ (C) mice at the end of treatment with 4-OHT and after 7, 14 and 21 days of recovery. Arrows point to IFE and arrowheads to sebaceous glands. (D–G) Co-labelling of control (D, F) and K14CreERSetd8Δ/Δ epidermis (E, G) for K14 (green) and GFP (red) (D, E) or GFP (red) only (F, G) after treatment with 4-OHT (end treatment) or after 7 and 21 days of recovery. (H) BrdU incorporation (BrdU; green) and labelling for K14 (red) in the IFE, sebaceous glands (SGs) and HF bulges/bulbs (Bu/Bb) in control (upper panels) and K14CreERSetd8Δ/Δ (Setd8Δ/Δ; lower panels) epidermis. (I) Quantification of BrdU-positive cells from (H). (J) Detection of quiescent label-retaining cells (LRCs) (BrdU; green) in bulges of control and K14CreERSetd8Δ/Δ (Setd8Δ/Δ) mice. (K) Quantification of (J). KO: K14CreERSetd8Δ/Δ. Nuclei are counterstained with DAPI (blue) in (D–H, J). Scale bars: 250 μm (A–G); 50 μm (H) and 75 μm (J).
Skin of control mice was not affected by treatment with 4-OHT during the course of the experiment and also skin of K14CreERSetd8Δ/Δ mice recovered both epidermal compartments, the IFE (arrows) and SGs (arrowhead) (Figure 4B and C). However, two aspects of recovered Setd8-depleted skin were unusual. First, skin of K14CreERSetd8Δ/Δ mice never exited anagen (Figure 4C). Second, dividing cells at 7 days of recovery mainly localized to the upper part of the HF (Supplementary Figure S5A and B, 7 days; arrow and line). One explanation for these observations was that deletion of Setd8 induced a wound healing reaction, in which HF stem cells contribute to formation of IFE, a process that can be associated with prolonged or induced anagen (Ito et al, 2005; Ansell et al, 2011). Thus, K14CreERSetd8Δ/Δ skin may be recovered by HF stem cells that escaped Setd8 deletion (Supplementary Figure S2A).
To determine the epidermal population responsible for the recovery of IFE and SGs in the absence of Setd8, we repeated the experiment using control- and K14CreERSetd8Δ/Δ-GFP-reporter mice (Figure 4D–G). After 21 days of recovery, control skin still expressed GFP, demonstrating that we successfully labelled long-lived progenitor cells of the IFE and SGs (Figure 4D and F; 21 days). In contrast, we did not detect any GFP-positive epidermal cell in skin of K14CreERSetd8Δ/Δ-GFP-reporter mice (Figure 4E and G; 21 days). These results indicated that the newly formed IFE and SGs were derived from HFs.
Our hypothesis that newly formed IFE might derive from upwards migrating HF cells was supported by the observation that proliferation in Setd8-depleted skin was only decreased in the IFE but increased in HFs (Bu/Bb) (Figure 4H and I). We next asked whether the recovery of IFE in the absence of Setd8 required the activation of quiescent HF bulge stem cells. One characteristic of bulge stem cells is that if they incorporate BrdU in neonatal epidermis they retain the label in adulthood; such cells are known as label-retaining cells (LRCs; Bickenbach and Chism, 1998; Braun et al, 2003). However, the number of LRC in the bulge of K14CreERSetd8Δ/Δ and control skin remained unchanged (Figure 4J and K), indicating that the recovered IFE in the absence of Setd8 does not require the activation of quiescent bulge stem cells.
Together, our data demonstrated that long-lived progenitor cells of the IFE and the SGs required Setd8 for survival.
Setd8 mediates functions of c-Myc in skin
So far our data showed that the loss of long-lived progenitor cells in Setd8-depleted skin was the likely result of a failure to undergo cell divisions. c-Myc is one important transcription factor in skin that induces epidermal stem cells to exit the stem cell compartment and stimulates their proliferation as progenitors (Arnold and Watt, 2001; Frye et al, 2003). Furthermore, Myc-induced exit from the epidermal stem cell niche is accompanied by transient accumulation of nuclei with high levels of H4K20me1 (Frye et al, 2007). Thus, we asked whether Setd8 might mediate c-Myc's function on epidermal stem cells.
We confirmed that expression of Setd8 RNA was increased in skin when c-Myc was overexpressed (K14MycER) (Figure 5A and B). In K14MycER transgenic mice, c-Myc can be activated by topical application of 4-OHT, and we found a time-dependent upregulation of Setd8 RNA over 6 days of treatment in K14MycER skin relative to wild-type epidermis (Figure 5B). We detected several putative Myc-binding sites in the Setd8 promoter using TESS (Transcription Element Search System) (Figure 5C, insert; Schug, 2008). Chromatin immunoprecipitations (ChIPs) using a c-Myc antibody confirmed a 15-fold enrichment of the Setd8 promoter in K14MycER epidermis compared with wild-type skin (Figure 5C). Nucleolin (Ncl1) is direct target gene of c-Myc and served as positive control (Figure 5C; Greasley et al, 2000). ChIP on chip experiments further confirmed a transcriptional regulation of Setd8 by c-Myc (Supplementary Figure S6A).
Figure 5.
Setd8 is transcriptionally regulated by c-Myc and required to mediate skin-specific functions of c-Myc. (A) Relative expression of Setd8 RNA in wild-type (WT) and K14MycER transgenic (TG) mice measured by QPCR. (B) Log2 fold-change (FC) of Setd8 RNA expression in K14MycER epidermis relative to wild-type skin in time course of treatment with 4-OHT for the days indicated. Data are obtained from gene expression arrays. (C) ChIP for Setd8 and Nucleolin (Ncl1) promoters using a c-Myc antibody in skin of WT and K14MycER TG animals using two different sets of primers (Setd8.1; Setd8.2). Ctr indicates the negative control. The insert is a graphical overview of the Setd8 promoter and the putative c-Myc binding sites. Red lines indicate putative c-Myc binding sites amplified by QPCR. (D, E) Measuring proliferation, using Ki67 as a marker (D) or BrdU incorporation (E) in control, K14CrERSetd8Δ/Δ, K14MycER animals and K14MycER mice with deleted Setd8 alleles (K14MycER-Setd8Δ/Δ). K14 (red) was used as counterstain in (E). Lines in (D, E: K14MycER-Setd8Δ/Δ) indicate loss of cycling cells. Areas outside the lines may have not deleted Setd8 due to short treatment with 4-OHT (6 days) (Supplementary Figure S2C and D). (F–K) Labelling for K10 (F), p63 (G), K8 (H), 14-3-3σ (I) (red) and p53 (J) (brown), or TUNEL (green) (K). Arrows in (F) mark cells of the basal layer and arrows in (H) indicate K8-positive cells in the IFE. Nuclei are counterstained with DAPI (blue) in (E–I, K). Scale bars: 100 μm (D, E) and 50 μm (F–K). (L) QPCR for p63 RNA levels in the indicated transgenic lines. (M) Quantification of (J) as percentage of p53-positive cells in the IFE. (N) Colour code for transgenic lines used in (L–N) and survival of epidermal cells in culture for the indicated hours. Y axis shows relative survival of the 4-OHT-treated lines to their untreated controls.
To test whether Setd8 was an essential functional mediator of c-Myc in skin, we deleted Setd8 in K14MycER transgenic mice (Figure 5D–N). Ki67-labelling and BrdU incorporation assays showed that c-Myc's promoting effect on cellular proliferation was reduced or absent when Setd8 was deleted (Figure 5D and E, lines; Supplementary Figure S6B). The decrease of proliferation in K14MycER skin that lacked Setd8 led to an inhibition of Myc-induced differentiation into IFE (Figure 5F). Premature expression of the terminal differentiation marker K10 in the basal layers of K14MycER epidermis was largely reversed in the absence of Setd8 (Figure 5F; arrows).
Loss of Setd8 dominates over Myc's function to induce epidermal differentiation
To validate whether overexpression of c-Myc might revert Setd8-depleted skin to normal, we tested whether expression of p63 might be restored. However, we found that K14MycER epidermis showed low levels of p63 when Setd8 was deleted (Figure 5G and L). To maintain stratified epithelia, p63 is required to repress K8, a marker for simple epithelia (Truong et al, 2006); indeed, we find an accumulation of K8-positive cells in skin that overexpressed c-Myc but lacked Setd8 (Figure 5H; arrows). We confirmed that K8 was absent in wild-type epidermis and, as described previously, only found in Merkel cells (Supplementary Figure S6C–G; Moll et al, 1984). Furthermore, K14MycER-Setd8Δ/Δ skin exhibited increased expression of 14-3-3σ and p21 (Figure 5I; data not shown), both of which are direct targets of p63-mediated gene repression (Westfall et al, 2003).
Because of the high degree of structural and functional similarity between p63 and p53, we asked whether p53 might also be involved in reducing proliferation in Setd8-depleted epidermis (Dotsch et al, 2010). In epidermal cells, c-Myc-induced activation of p53 causes a progressive reduction in growth rate, without inducing apoptosis, but a marked stimulation of terminal differentiation (Gandarillas and Watt, 1997; Dazard et al, 2000; Arnold and Watt, 2001). Although c-Myc-overexpressing epidermal cells upregulated p53, the number of p53-positive cells remained the same when Setd8 was deleted (Figure 5J and M).
Thus, deletion of Setd8 affected expression of p63 and p53 very differently. Expression of p63 was low in Setd8-depleted epidermis, independent of the presence or absence of c-Myc (Figures 3I and 5G). In contrast, the number of p53-positive nuclei was not affected by deletion of Setd8 in K14MycER skin (Figure 5J and M). We speculated that c-Myc-induced terminal differentiation, possibly mediated by p53, was dependent on expression of Setd8. If our hypothesis was correct, c-Myc-induced activation of p53 led to cell survival rather than apoptosis because the presence of Setd8 allowed p63 expression and increased genomic stability during cell divisions. Indeed, we only find a marked increase in DNA damage and apoptosis in epidermis that overexpressed c-Myc but lacked Setd8 (Figure 5K; Supplementary Figure S6H and I). Cell-cycle analyses confirmed that the effect of Setd8 deletion in c-Myc-overexpressing cells was rather due to apoptosis than cell-cycle arrest (Supplementary Figure S6J). Finally, overexpression of c-Myc did not increase survival of Setd8-depleted epidermal cells in culture (Figure 5N).
We concluded that Myc-induced epidermal proliferation and differentiation required expression of Setd8.
Setd8-mediated apoptosis is p53 dependent
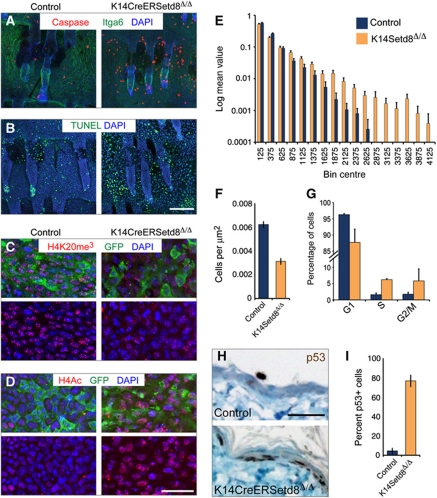
Our data indicated so far that epidermal progenitors undergo apoptosis in the absence of Setd8 independent of whether c-Myc was overexpressed or not (Figures 5K, 6A and B; Supplementary Figures S6H, I and S7A). Depletion of Setd8 in embryonic stem cells causes improper chromosome condensation (Oda et al, 2009), and also Setd8-depleted epidermal cells lacked histone H4 tri-methylation but gained acetylation (Figure 6C and D). Up-regulation of global acetylation might lead to de-condensation of chromatin, and we indeed find increased nuclear size in the absence of Setd8 (Figure 6E; Supplementary Figure S7B). Due to apoptosis, we also measured less cells per area in the IFE (Figure 6F); and in contrast to c-Myc-overexpressing cells, epidermal cells with normal levels of c-Myc underwent cell-cycle arrest when Setd8 was deleted (Figure 6G; Supplementary Figure S6J). As expected, the majority of Setd8-depleted epidermal cell stained positive for p53 (Figure 6H and I).
Figure 6.
Depletion of Setd8 in skin results in apoptosis, altered histone modifications and increased nuclear size. (A, B) Whole mounts of tail skin labelled for cleaved Caspase-3 (red) and Itga6 (green) (A) or TUNEL (green) (B). (C, D) Tri-methylated H4K20 (red) (C) is lost and global H4 acetylation (red) (D) is gained in GFP-positive cells of K14CreERSetd8Δ/Δ mice (right hand panels) compared with controls (left hand panels). (E) Size distribution of control (blue) and Setd8-depleted (yellow) nuclei in the IFE. For raw data examples see Supplementary Figure S7B. (F) Occurrence of nuclei per μm2. (G) Cell-cycle profile of control (blue) and K14CreERSetd8Δ/Δ mice (yellow). (H) Sections of back skin of control and K14CreERSetd8Δ/Δ mice labelled for p53 (brown). (I) Quantification of (H) shown as percentage of p53-positive cells. Nuclei are counterstained with DAPI (blue) in (A–D). Scale bars: 250 μm (A, B) and 50 μm (C, D, H).
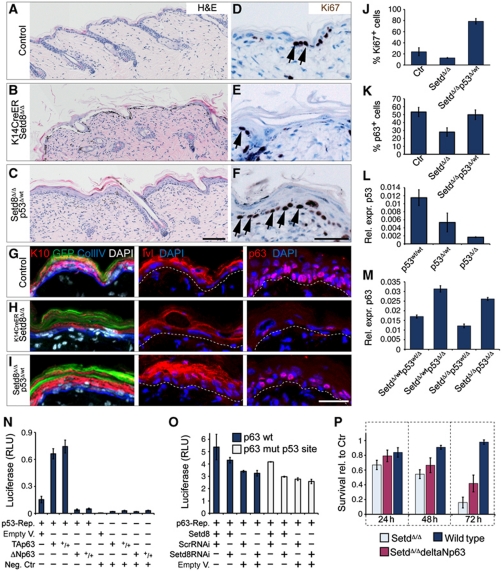
To test whether Setd8-induced apoptosis in the epidermis was dependent on p53, we deleted p53 in K14CreERSetd8Δ/Δ mice (Setd8Δ/Δp53wt/Δ). Ablation of one allele of p53 was sufficient to rescue loss of Setd8 (Figure 7A–C). Restoration of Setd8-depleted IFE in the absence of p53 was due to increased proliferation (Figure 7D–F and J). Setd8-induced erosion of the IFE was reversed when p53 levels were reduced as shown by staining for the terminal differentiation markers K10 and Ivl; and the number of p63-positive nuclei increased (Figure 7G–I and K). We confirmed that expression levels of p53 were reduced in p53Δ/wt and p53Δ/Δ epidermal cells and that ablation of p53 was sufficient to induce RNA expression of p63 (Figure 7L and M).
Figure 7.
Setd8-induced erosion of the IFE can be rescued by decreasing levels of p53 and survival of Setd8-depleted cells is enhanced by overexpression of ΔNp63. (A–I) Staining for Haematoxylin & Eosin (A–C), Ki67 (D–F), K10 (red), GFP (green) and collagen IV (Coll IV) (blue) (G–I; left hand panels), and Ivl (G–I; middle panels), and p63 (red) (G–I; right hand panels) in epidermis from control (A, D, G), K14CreERSetd8Δ/Δ (B, E, H) and K14CreERSetd8Δ/Δ mice with a deleted p53 allele (Setd8Δ/Δp53Δ/wt) (C, F, I). Arrows in (D–F) indicate Ki67-positive nuclei. Dotted lines in (G–I) mark the basement membrane. Nuclei are counterstained with DAPI (white, blue) (G–I). Scale bars: 100 μm (A–C) and 50 μm (G–I). (J, K) Quantification of Ki67 (D–F) and p63 (G–I; right hand panels) positive nuclei in control (ctr), K14CreERSetd8Δ/Δ (Set8Δ/Δ) and Setd8Δ/Δp53Δ/wt mice. (L, M) Relative expression (rel. expr.) of p53 (L) and p63 (M) in epidermal cells isolated from the indicated lines. Cells in (M) were treated with 4-OHT. (N, O) Luciferase assays using a p53 reporter (N) or the ΔNp63 promoter (O). (+) Indicates presence and (−) absence of the indicated plasmids. (+/+) Indicates double concentration of the plasmid. Empty V.: empty vector control; Neg. Ctr: negative control; Scr: scrambled RNAi construct; p63wt: wild-type p63 promoter construct; p63 mut p53 site: p63 promoter with mutated p53 binding site. (P) Survival of cultured epidermal cells isolated from wild-type and K14CreERSetd8Δ/Δ mice for the indicated hours. Cells were either transfected with the empty vector as control (wild-type, Setd8Δ/Δ) or with ΔNp63 construct (Setd8Δ/ΔdeltaNp63). Y axis shows relative survival of the 4-OHT-treated lines to their untreated controls (Ctr).
Of the two p63 isoforms (TAp63 and ΔNp63) expressed in skin, ΔNp63 is the most abundant. In contrast to TAp63 ΔNp63 lacks the transactivation domain and fails to induce apoptosis by inhibiting p53 transcriptional activity (Yang et al, 1998). Accordingly, in p53-reporters assays, ΔNp63 reduced and TAp63 increased p53 transactivation (Figure 7N). Our data indicated that Setd8 expression in skin was required for cell survival by stabilizing p63 but reducing p53 levels. To test whether Setd8 might induce ΔNp63 expression in the skin to repress p53, we performed luciferase assays using a 10.3-kb promoter regulating ΔNp63 (Figure 7O; Harmes et al, 2003). Overexpression of Setd8 led to a slight but reproducible increase of ΔNp63 promoter activity (Figure 7O), an effect that was independent of a p53-binding element within the ΔNp63 promoter (Figure 7O; mut p53 site). Accordingly, overexpression of ΔNp63 in Setd8-depleted epidermal cells increased their survival (Figure 7P).
In summary, Setd8 is an essential histone methyltransferase that is required for survival of stem and progenitor cells in skin. Setd8 mediates survival by simultaneously inducing ΔNp63 and repressing p53 levels in epidermal cells.
Discussion
This report is the first to assess the functional consequences of deregulated Setd8 in a mammalian adult tissue. We show through generation of skin-specific knockout mouse models that Setd8 is not only required during epidermal morphogenesis but also for adult skin homeostasis. In developing embryos, deletion of Setd8 entirely blocked the formation of epidermis. In adult skin, ablation of Setd8 led to an irreversible loss of IFE and SGs due to failure to maintain short- and long-lived progenitor cells and disrupted skin homeostasis via cell death.
Role of Setd8 in regulating transcription and DNA compaction
The robust phenotype of cell-cycle arrest and apoptosis upon removal of Setd8 complicates analysing its direct transcriptional functions. Accordingly, the general role of Setd8 in regulating gene transcription remains controversial. Mono-methylated H4K20 is located in active genes, suggesting a positive role in transcription (Vakoc et al, 2006; Barski et al, 2007). Setd8 activates expression of PPARγ, and thereby promotes adipogenesis (Wakabayashi et al, 2009). Setd8 has also been implicated in Wnt- and oestrogen receptor α-targeted gene activation (Li et al, 2011a, 2011b). In contrast, H4K20me1 is recognized by the malignant brain tumour domain of L3MBTL1, and results in local compaction of the chromatin, thus promoting transcriptional repression (Kalakonda et al, 2008). Here, we show that deletion of Setd8 in skin led to a deregulation of key regulators of skin homeostasis, including the loss of p63 versus upregulation of p53.
Loss of Setd8 causes cellular apoptosis and cell-cycle arrest in S and G2/M stages of the cell cycle because of its essential role in regulating chromosome stability during mitosis (Jorgensen et al, 2007; Tardat et al, 2007, 2010; Houston et al, 2008; Huen et al, 2008; Oda et al, 2009). However, dissecting the direct functional roles of Setd8 in DNA compaction in the epidermis is hindered by the fact that loss of Setd8-mediated deposition of H4K20me1 led to further changes of other histone modifications: H4 acetylation was induced and deposition of H4K20me3 was blocked. Mono-, di- and tri-methyl groups at H4K20 each confer a distinct functional state at the cellular level. For instance, di-methylated H4K20 serves as a binding site for p53BP1 and is involved in DNA repair pathways (Sanders et al, 2004; Botuyan et al, 2006); and tri-methylated H4K20 is enriched in inactive pericentric heterochromatin (Schotta et al, 2004).
Thus, although it is difficult to speculate how exactly transcription and/or DNA compaction is directly affected by loss of Setd8, our study clearly demonstrated that Setd8 is essential for the proper deposition of histone modifications at H4 and required for survival, proliferation and differentiation of epidermal stem cells.
c-Myc and Setd8 cooperate to maintain skin homeostasis
Activation of the transcription factor c-Myc in skin triggers epidermal stem cells to exit their niche, induces proliferation of progenitors and subsequently stimulates lineage-specific differentiation into IFE and SGs (Watt et al, 2008). How c-Myc mechanistically stimulates epidermal cells to differentiate, as opposed to undergo apoptosis for example, is unknown. Here, we provide evidence that Setd8 is a transcriptional target of c-Myc and functionally required to mediate c-Myc-induced proliferation and differentiation in skin. Without Setd8, even c-Myc-overexpressing cells fail to proliferate or differentiate and undergo apoptosis instead, a mechanism that might directly be linked to expression p63 and p53.
As in other tissues, overexpression of c-Myc in epidermal cells induces activation of the tumour suppressive transcription factor p53, leading to a progressive reduction in growth rate; however, instead of inducing apoptosis c-Myc stimulates terminal differentiation (Gandarillas and Watt, 1997; Dazard et al, 2000; Arnold and Watt, 2001). Since overexpression of c-Myc causes apoptosis in many other tissues, its function to induce epidermal differentiation must be tissue-specifically regulated. A strong candidate to inhibit Myc-induced apoptosis via activation of p53 is p63.
p63 and p53 are structural and functional protein homologues, and due to high sequence identity in their transactivation domains, p63 can transactivate p53-responsive genes (Dotsch et al, 2010). However, the p63 gene encodes several protein isoforms generated by alternative splicing. The most abundant isoform in the epidermis (ΔNp63α) lacks the transactivation domain, and accordingly fails to induce apoptosis and inhibits p53 transcriptional activity (Yang et al, 1998). We show that c-Myc-overexpressing skin is highly enriched in both p53- and p63-positive nuclei, and ΔNp63 might exert a dominant-negative function over p53 to induce differentiation.
Deletion of Setd8 in Myc-overexpressing skin downregulates p63 but expression of p53 is maintained. Thus, the inhibitory function of p63 is lost and the p53-dependent apoptotic pathway induced. Indeed, we only detect apoptosis in c-Myc-overexpressing skin when Setd8 is removed and p63 is absent. The direct functional link between Setd8 and p63 remains elusive but recent studies showed that Setd8 methylates p53 at lysine 382 and thereby suppresses p53-mediated transcription in vitro (Shi et al, 2007). It is intriguing to speculate that p63 may also be a direct target for Setd8-dependent methylation.
Setd8-mediated epidermal survival, proliferation and differentiation via p63
The transcription factor p63 is a key regulator required for epidermal stratification and differentiation (Truong et al, 2006), and p63 is absent in Setd8-depleted skin. Notably, Setd8 and p63 appear to have similar cellular functional roles in skin. Epidermal stem cells lacking p63 have decreased proliferative capacity and undergo apoptosis (Liefer et al, 2000; Senoo et al, 2007). Furthermore, UV-induced apoptosis requires the downregulation of both Setd8 and p63 (Liefer et al, 2000; Oda et al, 2009; Abbas et al, 2010; Centore et al, 2010). Finally, loss of p63 during embryonic development causes a similar epidermal phenotype as removal of Setd8. Ablation of p63 leads to the formation of truncated limbs and a block of ectodermal differentiation (Mills et al, 1999; Yang et al, 1999). In conclusion, lack of p63 might explain why Setd8-depleted cells upregulate p53 and undergo cycle arrest and apoptosis.
Thus, Setd8 may act as a key regulator in balancing the negative feedback between p63 and p53 in skin. Our data show that Setd8-induced erosion in the IFE can be rescued by decreasing the levels of p53, indicating that expression of Setd8 is required to suppress p53 in epidermal stem and progenitor cells. Setd8 might suppress p53 indirectly by stabilizing ΔNp63 levels. Expression of Setd8 had a positive effect on the ΔNp63 promoter and overexpression of ΔNp63 increased survival of Setd8-depleted epidermal cells.
In sum, our data show that the function of Setd8 to promote epidermal survival and differentiation is at least in part regulated by p63. Furthermore, Setd8 is required to repress apoptosis in skin and essential to mediate c-Myc-induced epidermal stem cell proliferation and differentiation processes.
Materials and methods
Ethics statement
All mouse husbandry and experiments were carried out according to the local ethics committee under the terms of a UK Home Office license.
Transgenic mouse lines, genotyping and 4-OHT treatment
To conditionally delete Setd8 in skin, mice containing floxed alleles of the SETD8 gene (kindly provided by Danny Reinberg) were crossed with KRT14-cre mice (The Jackson Laboratory). In KRT14-cre mice, Cre-recombinase is expressed under the control of the keratin 14 (K14)—promoter leading to deletion of Setd8 in all basal, undifferentiated cells of the epidermis (K14Setd8Δ/Δ). To generate the inducible, conditional knockout lines K14CreERSetd8Δ/Δ and K14CreSetd8Δ/null, mice containing floxed alleles or one floxed and one null allele of the SETD8 gene (kindly provided by Danny Reinberg) were crossed with KRT14-cre/ERT mice (K14CreER) (The Jackson Laboratory). In these transgenic mice, Cre-recombinase is fused to a mutated oestrogen receptor domain and can be activated by application of 4-OHT. Genotyping for Cre-recombinase was performed as recommended (http://jaxmice.jax.org/protocolsdb) and genotyping for floxed and null SETD8 alleles was performed as previously described (Oda et al, 2009). To delete p53, K14CreSetd8Δ/Δ mice were crossed to p53+/Δ mice (Jacks et al, 1994).
To activate K14CreER, ∼5-week-old mice were treated topically with 1.4 mg 4-OHT (Sigma) dissolved in acetone or acetone alone as a control every other day. If not otherwise stated mouse back skin was treated for 14 days and tail skin for 21 days.
To generate GFP-reporter lines for Cre-recombinase, the respective lines were crossed with CAG-CAT-EGFP mice, expressing enhanced GFP (EGFP) upon Cre-mediated recombination (kindly provided by Jun-ichi Miyazaki) (Kawamoto et al, 2000). These mice were genotyped using primers specific for EGFP SK1 (TGA ACC GCA TCG AGC TGA AGG G) and SK2 (TCC AGC AGG ACC ATG TGA TCG C).
To generate a LacZ-reporter mouse for endogenous Setd8 expression, the embryonic stem cell clone RRB075, carrying the β-galactosidase gene as a GeneTrap in intron 3 (Huen et al, 2008), was obtained from BayGenomics and injected into C57BL/6 blastocysts to generate chimeric mice. The chimeras were then crossed with F1 females, and heterozygous mice with successful germ line transmission of the targeted allele were used for LacZ stainings.
The mouse line K14MycER was kindly provided by Fiona Watt and genotyped and treated with 4-OHT as described previously (Arnold and Watt, 2001). To delete Setd8 in Myc-overexpressing skin, K14MycER mice were crossed with K14CreERSetd8Δ/Δ animals (K14MycER-Setd8Δ/Δ).
BrdU labelling
To measure proliferation, mice were injected with 50 mg of BrdU per kg body weight and killed after 2 h. DNA LRCs were generated by repeated BrdU injections of neonatal mice at P10 and animals were chased for a minimum of 7 weeks (Braun et al, 2003).
Cell-cycle analysis and flow sorting GFP-positive cells
Epidermal cells were isolated as described previously (Jensen et al, 2010). The cells were sorted with a DakoCytomation MoFlo sorter using 533 filter and the GFP+ cells were then fixed with ethanol and labelled with propidium iodide (PI). The DNA content measured using Dako CyAn ADP analyser. The data analysis was done using FlowJo program.
Measuring nuclear size and abundance
Z volumes of DAPI labelled IFE were imaged at random using a Leica SP5 confocal microscope. These were analysed using Volocity software (Perkin-Elmer). Nuclei were identified by segmentation and their volume measured. A frequency distribution of nuclear size was generated using 250 μm3 bins, and each distribution normalized to the total number of identified nuclei per sample (n=11). A mean frequency distribution was generated for wild-type and transgenic mice and plotted on a logarithmic scale to clearly display the disparity of larger nuclear volume.
Tissue staining and antibodies
Tissue samples were either fixed overnight in 4% PFA and then embedded in paraffin or frozen unfixed, in OCT compound (Miles). Tail whole mounts were prepared following a previously published protocol (Braun et al, 2003). In all, 6–10 μm paraffin and cryosections of back skin were used for immunostainings. After citrate epitope retrieval of paraffin sections, tissues were permeabilized for 5 min with 0.2% Triton X-100 at room temperature, blocked for 1 h with 5% FCS and incubated overnight with the appropriate antibody dilution. Stainings of cryosections were performed as for paraffin but after fixation for 10 min in 4% paraformaldehyde at room temperature. Tail epidermal whole mounts were prepared and immunolabelled as described previously.
Primary antibodies against the following proteins were used: Collagen IV (1:600; AB769, Millipore), FAS (1:100; G-11, Santa Cruz), Itga6 (1:200; cd49f, AbD Serotec), Involucrin (1:800; SY-5, kindly provided by Fiona Watt), Ki67 (1:100; SF6, Vector Laboratories Ltd.), FABP5 (1:100; BAF1476, R&D Systems), p63 (1:100; H-137, Santa Cruz), cleaved Caspase-3 Asp175 (1:100; 9661, Cell Signaling), Phospho-H2A. XSer139 (γ-H2AX) (1:400; 20E3 Cell Signaling), H4K20me1 (1:500; 39175/39180 Active Motif), H4K20me3 (1:500; ab9051/ab9053 Abcam), K14 (1:1000; AF64, Covance), K10 (1:500; PRB-159P, Covance), K8 (1:800; ab59400, Abcam), K8 TROMA-1 (1:600; DSHB), 14-3-3σ (1:100; ab14123 Abcam), p53 (1:200; CM5, Vector Laboratories LTD), H4Ac (1:200; 06-598 Upstate), BrdU (1:100; BU1/75 (ICR1), Abcam) and GFP (1:500; ab13970, Abcam).
Secondary antibodies Alexa-Fluor 488-, 594-, 647-conjugated goat, chicken or donkey anti-rabbit, anti-mouse, anti-chicken or anti-rat IgG (Invitrogen) were added at a dilution of 1:500 for 1 h at room temperature together with DAPI to label nuclei. Slides were mounted with Mowiol (madein house from Mowiol® 4-88 Polysciences, Inc.).
For LacZ staining, freshly obtained skin samples from 3-week-old RRB075 mice were fixed for ∼20 min at room temperature in buffer containing 0.1 M phosphate buffer, 5 mM EGTA, 2 mM MgCl2 and 0.2% glutaraldehyde. Samples were then washed three times for 15 min each in wash buffer (2 mM MgCl2 and 0.1% Nonidet P40 in 0.1 M phosphate buffer) and stained for up to 12 h in a solution consisting of 1 mg/ml X-gal (Melford), 5 mM K3Fe(CN)6 and 5 mM K4Fe(CN)6 in wash buffer.
To detect apoptotic cells in skin section, we used DeadEnd™ Fluorometric TUNEL System (Promega) according to the manufactures instructions.
White field images were acquired using an Olympus IX80 microscope and a DP50 camera. Confocal images were acquired on a Leica TCS SP5 confocal microscope. Z-stacks were acquired at 100 Hz with an optimal stack distance and 1024 × 1024 dpi resolutions. Z-stack projections were generated using the LAS AF software package (Leica Microsystems). All the images were processed with Photoshop CS4 (Adobe) software.
Mouse keratinocyte culture, transfection and luciferase assays
Epidermal cells were isolated from mouse back skin and cultured as described previously (Frye et al, 2003). Cell survival was measured using CellTiter-Glo® Luminescent Cell Viability Assay (Promega) according to the manufacturer's instructions post activation with 4-OHT. Epidermal and HEK293H cells were transfected using Attractene (Qiagen) according to the manufacturer's instructions. The cytomegalovirus-driven expression vectors contained full-length cDNA of Setd8 or TAp63 and ΔNp63 (kindly provided by Dennis Roop). Luciferase activity was measured 36–48 h after the transfection of HEK293H using the Dual-Luciferase Reporter Assay System (Promega) on Glomax (Promega). Each transfection was carried out in triplicate and the experiment was repeated twice. p53-reporter assays (Qiagen) were performed according to the manufacturer's instructions using HEK293H cells. The ΔNp63 promoter construct was a kind gift of James DiRenzo (Harmes et al, 2003).
RNA extraction, QPCR and ChIP
RNA was extracted from the back skin of experimental animals or cultured epidermal cells using Trizol Reagent (Invitrogen) according to the manufacturers’ instructions. Following RNA extraction, cDNA was made using SuperScript® III Reverse Transcriptase (Invitrogen). RT–PCR was run using the standard protocol for TaqMan® Fast Universal PCR Master Mix (2 × ) or Fast SYBR® Green Master Mix using the 7900HT Fast Real-Time PCR System or StepOne™ Real-Time PCR System (Applied Biosystems). Primers used for SYBR Green QPCR were as follows: GFP forward (AGC AAG GGC GAG GAG CTG TT) and GFP reverse (GTA GGT CAG GGT GGT CAC GA), Setd8 forward (GTG TGA TCG CTA CCA AGC AGT TCT) and Setd8 reverse (ATA GTA CAT GTA GCA GCC AGT GGA GG), and GAPDH forward (GTC TCC TGC GAC TTC AAC AGC) and GAPDH reverse (TCA TTG TCA TAC CAG GAA ATG AGC). RNA levels were determined using the ΔCT method and relative expression levels were normalized to GAPDH.
ChIPs followed by QPCR using SYBR Green were performed as described previously (Hussain et al, 2009). Primers to amplify genomic DNA surrounding the Myc-binding sites were as follows: Setd8.1 forward (GGT CAA ACC TAA GGC AAA CCG GAA G), Setd8.1 reverse (TCA GCA ATC TGC GTC CTG), Setd8.2 forward (CCA GGG AAC GGT TGT TAC TG), Setd8.2 reverse (TAG TGT TAA ACT TTG CAT GAT GGT GTA TGG C), nucleolin forward (TGG GGG AGT GTC TGT AGT ACC) and nucleolin reverse (AAA CCC AGT TAA AGG GAT CCC). As a negative control, a DNA region of around 7000 bp downstream the Myc-binding sites was amplified: negative control forward (GCT GGC CTC AAA CTC AGA AA) and negative control reverse (GGC GCA CAC CTT TAA TCC). Enrichment of Myc binding in promoter regions was quantified by normalization to the whole cell lysate of each sample.
Supplementary Material
Acknowledgments
We thank David Beck and Duncan Odom for advice and very helpful comments on this manuscript. We further thank Danny Reinberg for providing the floxed/null Setd8 mice, Jun-ichi Miyazaki for the GFP-reporter lines and BayGenomics for the ES cell line RRB075. Thank you to Fiona Watt for the K14MycER and David Tuveson for the p53+/Δ mice; and Dennis Roop and James DiRenzo for reagents. We gratefully acknowledge the support of the University of Cambridge and Stephen Evans-Freke. This work was funded by Cancer Research UK and the Medical Research Council (MRC). SB was supported by the Ramón Areces Spanish Foundation and EMBO.
Author contributions: ID performed and designed experiments. HO performed experiments and advised on manuscript. SB performed experiments. EN performed experiments. PH analysed data and MF designed experiments and wrote manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A (2010) CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell 40: 9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansell DM, Kloepper JE, Thomason HA, Paus R, Hardman MJ (2011) Exploring the ‘hair growth-wound healing connection’: anagen phase promotes wound re-epithelialization. J Invest Dermatol 131: 518–528 [DOI] [PubMed] [Google Scholar]
- Arnold I, Watt FM (2001) c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr Biol 11: 558–568 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bickenbach JR, Chism E (1998) Selection and extended growth of murine epidermal stem cells in culture. Exp Cell Res 244: 184–195 [DOI] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E (2009) Epidermal homeostasis: a balancing act of stem cells in the skin. Nat Rev Mol Cell Biol 10: 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G (2006) Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127: 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun KM, Niemann C, Jensen UB, Sundberg JP, Silva-Vargas V, Watt FM (2003) Manipulation of stem cell proliferation and lineage commitment: visualisation of label-retaining cells in whole mounts of mouse epidermis. Development (Cambridge, England) 130: 5241–5255 [DOI] [PubMed] [Google Scholar]
- Centore RC, Havens CG, Manning AL, Li JM, Flynn RL, Tse A, Jin J, Dyson NJ, Walter JC, Zou L (2010) CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol Cell 40: 22–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, Watt FM (2008) Dynamic regulation of retinoic acid-binding proteins in developing, adult and neoplastic skin reveals roles for beta-catenin and Notch signalling. Dev Biol 324: 55–67 [DOI] [PubMed] [Google Scholar]
- Dazard JE, Piette J, Basset-Seguin N, Blanchard JM, Gandarillas A (2000) Switch from p53 to MDM2 as differentiating human keratinocytes lose their proliferative potential and increase in cellular size. Oncogene 19: 3693–3705 [DOI] [PubMed] [Google Scholar]
- Dotsch V, Bernassola F, Coutandin D, Candi E, Melino G (2010) p63 and p73, the ancestors of p53. Cold Spring Harb Perspect Biol 2: a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert RL, Adhikary G, Rorke EA, Chew YC, Balasubramanian S (2011) Polycomb group proteins are key regulators of keratinocyte function. J Invest Dermatol 131: 295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E (2011) EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev 25: 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, Iyer NG, Perez-Rosado A, Calvo E, Lopez JA, Cano A, Calasanz MJ, Colomer D, Piris MA, Ahn N, Imhof A et al. (2005) Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 37: 391–400 [DOI] [PubMed] [Google Scholar]
- Fraga MF, Esteller M (2007) Epigenetics and aging: the targets and the marks. Trends Genet 23: 413–418 [DOI] [PubMed] [Google Scholar]
- Frye M, Fisher AG, Watt FM (2007) Epidermal stem cells are defined by global histone modifications that are altered by Myc-induced differentiation. PLoS ONE 2: e763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M, Gardner C, Li ER, Arnold I, Watt FM (2003) Evidence that Myc activation depletes the epidermal stem cell compartment by modulating adhesive interactions with the local microenvironment. Development (Cambridge, England) 130: 2793–2808 [DOI] [PubMed] [Google Scholar]
- Fuchs E (2009) The tortoise and the hair: slow-cycling cells in the stem cell race. Cell 137: 811–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandarillas A, Watt FM (1997) c-Myc promotes differentiation of human epidermal stem cells. Genes Dev 11: 2869–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greasley PJ, Bonnard C, Amati B (2000) Myc induces the nucleolin and BN51 genes: possible implications in ribosome biogenesis. Nucleic Acids Res 28: 446–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmes DC, Bresnick E, Lubin EA, Watson JK, Heim KE, Curtin JC, Suskind AM, Lamb J, DiRenzo J (2003) Positive and negative regulation of deltaN-p63 promoter activity by p53 and deltaN-p63-alpha contributes to differential regulation of p53 target genes. Oncogene 22: 7607–7616 [DOI] [PubMed] [Google Scholar]
- Houston SI, McManus KJ, Adams MM, Sims JK, Carpenter PB, Hendzel MJ, Rice JC (2008) Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J Biol Chem 283: 19478–19488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Sy SM, van Deursen JM, Chen J (2008) Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication. J Biol Chem 283: 11073–11077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Benavente SB, Nascimento E, Dragoni I, Kurowski A, Gillich A, Humphreys P, Frye M (2009) The nucleolar RNA methyltransferase Misu (NSun2) is required for mitotic spindle stability. J Cell Biol 186: 27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Liu Y, Yang Z, Nguyen J, Liang F, Morris RJ, Cotsarelis G (2005) Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med 11: 1351–1354 [DOI] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA (1994) Tumor spectrum analysis in p53-mutant mice. Curr Biol 4: 1–7 [DOI] [PubMed] [Google Scholar]
- Jensen KB, Driskell RR, Watt FM (2010) Assaying proliferation and differentiation capacity of stem cells using disaggregated adult mouse epidermis. Nat Protoc 5: 898–911 [DOI] [PubMed] [Google Scholar]
- Jorgensen S, Elvers I, Trelle MB, Menzel T, Eskildsen M, Jensen ON, Helleday T, Helin K, Sorensen CS (2007) The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol 179: 1337–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalakonda N, Fischle W, Boccuni P, Gurvich N, Hoya-Arias R, Zhao X, Miyata Y, Macgrogan D, Zhang J, Sims JK, Rice JC, Nimer SD (2008) Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1. Oncogene 27: 4293–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karachentsev D, Sarma K, Reinberg D, Steward R (2005) PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev 19: 431–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto S, Niwa H, Tashiro F, Sano S, Kondoh G, Takeda J, Tabayashi K, Miyazaki J (2000) A novel reporter mouse strain that expresses enhanced green fluorescent protein upon Cre-mediated recombination. FEBS Lett 470: 263–268 [DOI] [PubMed] [Google Scholar]
- Koster MI (2010) p63 in skin development and ectodermal dysplasias. J Invest Dermatol 130: 2352–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sun L, Zhang Y, Wang D, Wang F, Liang J, Gui B, Shang Y (2011a) The histone modifications governing TFF1 transcription mediated by estrogen receptor. J Biol Chem 286: 13925–13936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Nie F, Wang S, Li L (2011b) From the Cover: Feature Article: Histone H4 Lys 20 monomethylation by histone methylase SET8 mediates Wnt target gene activation. Proc Natl Acad Sci USA 108: 3116–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liefer KM, Koster MI, Wang XJ, Yang A, McKeon F, Roop DR (2000) Down-regulation of p63 is required for epidermal UV-B-induced apoptosis. Cancer Res 60: 4016–4020 [PubMed] [Google Scholar]
- Lo Celso C, Berta MA, Braun KM, Frye M, Lyle S, Zouboulis CC, Watt FM (2008) Characterization of bipotential epidermal progenitors derived from human sebaceous gland: contrasting roles of c-Myc and beta-catenin. Stem Cells (Dayton, Ohio) 26: 1241–1252 [DOI] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A (1999) p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398: 708–713 [DOI] [PubMed] [Google Scholar]
- Moll R, Moll I, Franke WW (1984) Identification of Merkel cells in human skin by specific cytokeratin antibodies: changes of cell density and distribution in fetal and adult plantar epidermis. Differentiation 28: 136–154 [DOI] [PubMed] [Google Scholar]
- Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, Lis JT, Allis CD, Reinberg D (2002) PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell 9: 1201–1213 [DOI] [PubMed] [Google Scholar]
- Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D (2010) Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol Cell 40: 364–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, Torres-Padilla ME, Heard E, Reinberg D (2009) Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol 29: 2278–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JC, Nishioka K, Sarma K, Steward R, Reinberg D, Allis CD (2002) Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes Dev 16: 2225–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly H, Lange M, Simboeck E, Di Croce L (2010) Setting and resetting of epigenetic marks in malignant transformation and development. Bioessays 32: 669–679 [DOI] [PubMed] [Google Scholar]
- Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T (2004) Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119: 603–614 [DOI] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T (2004) A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 18: 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug J (2008) Using TESS to predict transcription factor binding sites in DNA sequence. Curr Protoc Bioinformatics Chapter 2: Unit 2.6. [DOI] [PubMed] [Google Scholar]
- Schweizer J, Furstenberger G, Winter H (1987) Selective suppression of two postnatally acquired 70 kD and 65 kD keratin proteins during continuous treatment of adult mouse tail epidermis with vitamin A. J Invest Dermatol 89: 125–131 [DOI] [PubMed] [Google Scholar]
- Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA (2008) Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev 22: 1865–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo M, Pinto F, Crum CP, McKeon F (2007) p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell 129: 523–536 [DOI] [PubMed] [Google Scholar]
- Shafa M, Krawetz R, Rancourt DE (2010) Returning to the stem state: epigenetics of recapitulating pre-differentiation chromatin structure. Bioessays 32: 791–799 [DOI] [PubMed] [Google Scholar]
- Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, Dutta S, Appella E, Gozani O (2007) Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell 27: 636–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat M, Brustel J, Kirsh O, Lefevbre C, Callanan M, Sardet C, Julien E (2010) The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat Cell Biol 12: 1086–1093 [DOI] [PubMed] [Google Scholar]
- Tardat M, Murr R, Herceg Z, Sardet C, Julien E (2007) PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. J Cell Biol 179: 1413–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA (2006) p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev 20: 3185–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc CR, Sachdeva MM, Wang H, Blobel GA (2006) Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol 26: 9185–9195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi K, Okamura M, Tsutsumi S, Nishikawa NS, Tanaka T, Sakakibara I, Kitakami J, Ihara S, Hashimoto Y, Hamakubo T, Kodama T, Aburatani H, Sakai J (2009) The peroxisome proliferator-activated receptor gamma/retinoid X receptor alpha heterodimer targets the histone modification enzyme PR-Set7/Setd8 gene and regulates adipogenesis through a positive feedback loop. Mol Cell Biol 29: 3544–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt FM, Frye M, Benitah SA (2008) MYC in mammalian epidermis: how can an oncogene stimulate differentiation? Nat Rev 8: 234–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt FM, Jensen KB (2009) Epidermal stem cell diversity and quiescence. EMBO Mol Med 1: 260–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall MD, Mays DJ, Sniezek JC, Pietenpol JA (2003) The Delta Np63 alpha phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Mol Cell Biol 23: 2264–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Jing C, Kelly G, Walker PA, Muskett FW, Frenkiel TA, Martin SR, Sarma K, Reinberg D, Gamblin SJ, Wilson JR (2005) Specificity and mechanism of the histone methyltransferase Pr-Set7. Genes Dev 19: 1444–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F (1998) p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 2: 305–316 [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F (1999) p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398: 714–718 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.